Deep Sequencing and Screening of Differentially Expressed MicroRNAs Related to Milk Fat Metabolism in Bovine Primary Mammary Epithelial Cells
Abstract
:1. Introduction
2. Results
2.1. Cultures of Bovine pMECs and Comparisons of Triglyceride Content
| pMEC-HH | pMEC-LL | |
|---|---|---|
| Passage 2 | 179.3441 ± 12.68155 A | 84.0345 ± 2.76037 A |
| Passage 3 | 162.9136 ± 3.81292 A | 69.0142 ± 11.83466 A |
| Passage 5 | 112.2417 ± 9.79188 B | 109.2433 ± 10.54467 B |
2.2. Sequence Data Analysis
2.3. Identification of Known and Novel miRNAs in Bovine pMECs
| miRNA | miRNA* | miRNA-5p | miRNA-3p | pre-miRNA | |
|---|---|---|---|---|---|
| Known miRNAs in miRase 18.0 | 579 | 42 | 28 | 27 | 662 |
| pMEC-HH | 263 | 17 | 19 | 17 | 320 |
| pMEC-LL | 292 | 21 | 19 | 20 | 355 |
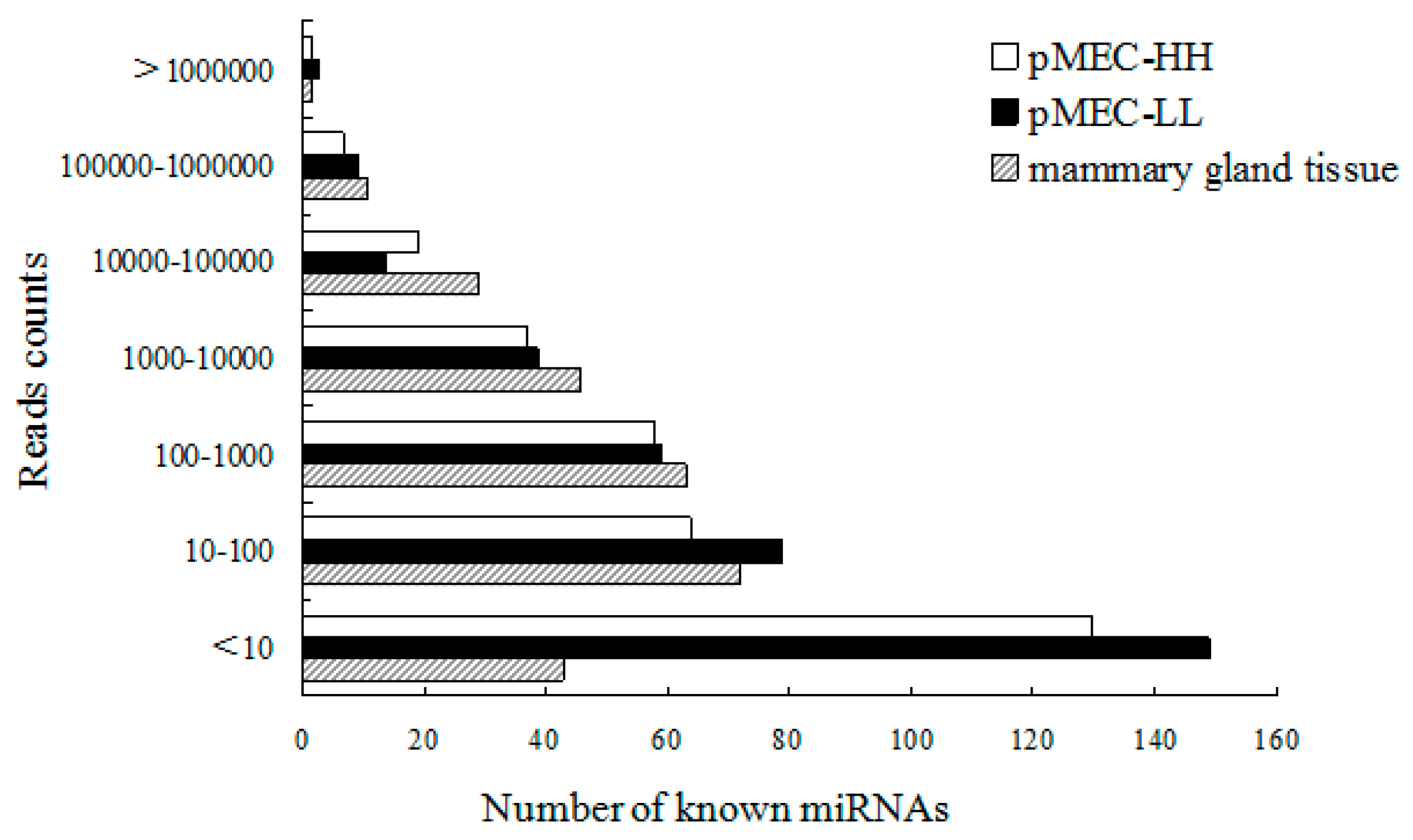
2.4. Validation of miRNA Expression Patterns at Both Cellular and Tissular Levels


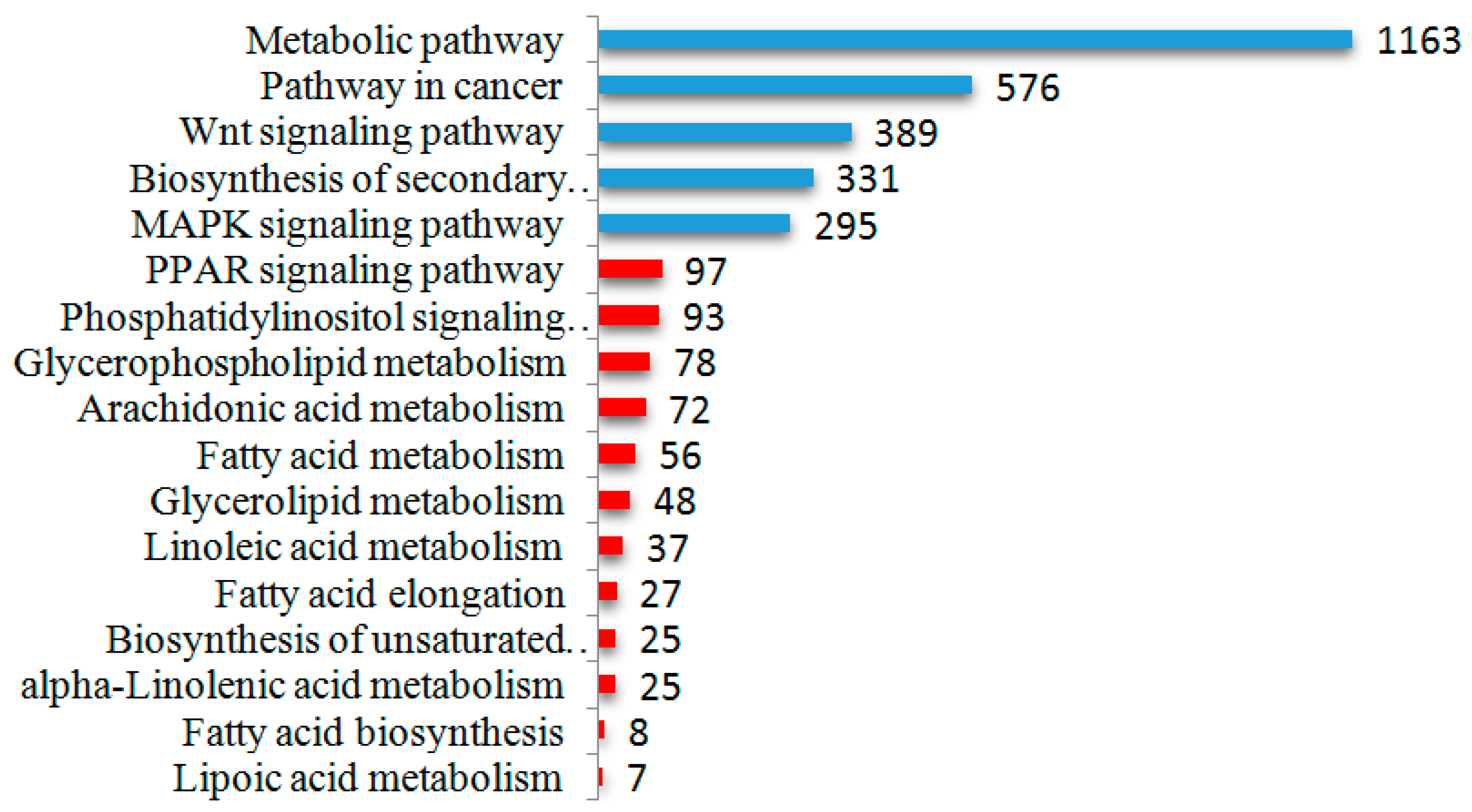
2.5. Differential Expression of miRNAs and Functional Analysis of Their Predicted Target Genes
| miR-Name | pMEC-HH-std | pMEC-LL-std | Fold-Change | p Value | FDR | Sig-Label |
|---|---|---|---|---|---|---|
| bta-miR-124a | 0.0100 | 14.8710 | 10.5383 | 5.50 × 10−43 | 0.0813 | ** |
| bta-miR-224 | 1.9114 | 15.8505 | 3.05183 | 1.22 × 10−27 | 0.2259 | ** |
| bta-miR-452 | 11.4682 | 70.0808 | 2.6114 | 6.56 × 10−100 | 0.3946 | ** |
| bta-miR-877 | 39.8015 | 175.7807 | 2.1429 | 8.77 × 10−197 | 0.2389 | ** |
| bta-miR-2344 | 40.4761 | 169.7254 | 2.0681 | 8.17 × 10−182 | 0.8923 | ** |
| bta-miR-29b | 588.2531 | 97.5966 | −2.5915 | 0 | 0.9213 | ** |
| bta-miR-152 | 1231.1490 | 189.5831 | −2.6991 | 0 | 0.4573 | ** |
| bta-miR-30b-5p | 20.9127 | 1.8700 | −3.4833 | 1.68 × 10−43 | 0.1364 | ** |
| bta-miR-193a-3p | 265.4560 | 20.2139 | −3.7151 | 0 | 0.8523 | ** |
| bta-miR-33a | 32.3809 | 1.4248 | −4.5063 | 1.09 × 10−80 | 0.4039 | ** |
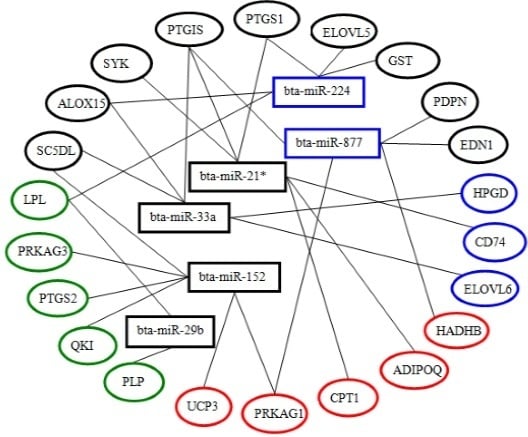
2.6. Screening and Identification of miRNAs Related to Milk Fat Metabolism in Dairy Cattle
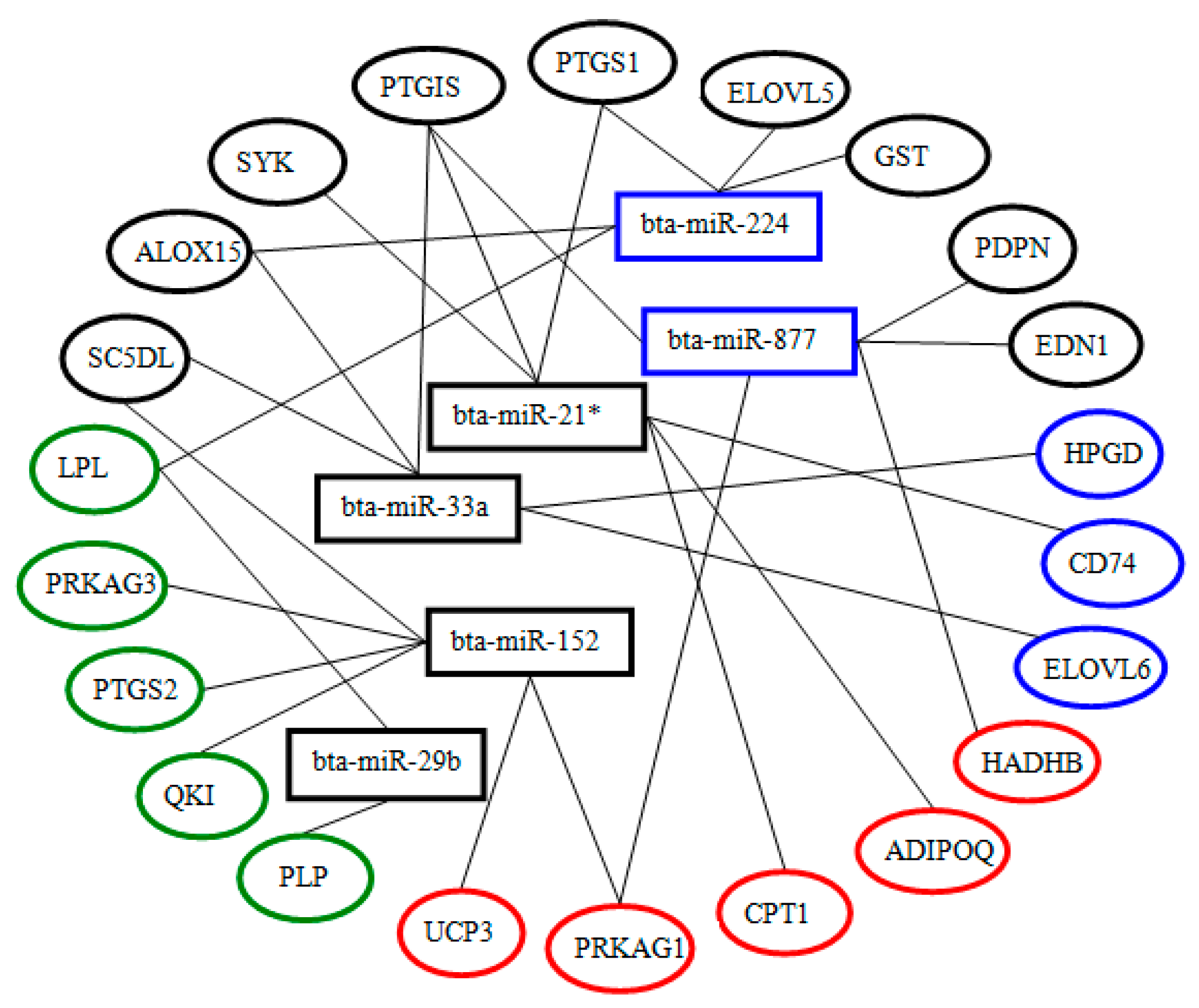

3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Animals and Sample Collection
4.3. Cell Culture, Triglyceride Content Detecting and RNA Extraction
4.4. Solexa Sequencing and Bioinformatic Analysis of Small RNAs
4.5. miRNA Validation via Stem-Loop Real-Time qPCR Both at the Cellular and Tissular Level
4.6. Target Gene and Function Prediction of Differentially Expressed MicroRNAs
4.7. Screening of miRNAs Related to Milk Fat Metabolism in Dairy Cattle
4.8. qPCR of Predicted Genes Involved in Lipid Metabolism
4.9. Western Blot Analysis of Predicted Proteins Involved in Lipid Metabolism
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Harvatine, K.J.; Bauman, D.E. SREBP1 and thyroid hormone responsive spot 14 (S14) are involved in the regulation of bovine mammary lipid synthesis during diet-induced milk fat depression and treatment with CLA. J. Nutr. 2006, 136, 2468–2474. [Google Scholar] [PubMed]
- Kadegowda, A.K.; Bionaz, M.; Piperova, L.S.; Erdman, R.A.; Loor, J.J. Peroxisome proliferator-activated receptor-gamma activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents. J. Dairy Sci. 2009, 92, 4276–4289. [Google Scholar] [CrossRef] [PubMed]
- Pawar, A.; Botolin, D.; Mangelsdorf, D.J.; Jump, D.B. The role of liver X receptor-α in the fatty acid regulation of hepatic gene expression. J. Biol. Chem. 2003, 278, 40736–40743. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Blott, S.; Kim, J.J.; Moisio, S.; Schmidt-Kuntzel, A.; Cornet, A.; Berzi, P.; Cambisano, N.; Ford, C.; Grisart, B.; Johnson, D.; et al. Molecular dissection of a quantitative trait locus: A phenylalanine-to-tyrosine substitution in the transmembrane domain of the bovine growth hormone receptor is associated with a major effect on milk yield and composition. Genetics 2003, 163, 253–266. [Google Scholar] [PubMed]
- Sun, D.; Jia, J.; Ma, Y.; Zhang, Y.; Wang, Y.; Yu, Y.; Zhang, Y. Effects of DGAT1 and GHR on milk yield and milk composition in the Chinese dairy population. Anim. Genet. 2009, 40, 997–1000. [Google Scholar] [CrossRef] [PubMed]
- Davalos, A.; Goedeke, L.; Smibert, P.; Ramirez, C.M.; Warrier, N.P.; Andreo, U.; Cirera-Salinas, D.; Rayner, K.; Suresh, U.; Pastor-Pareja, J.C.; et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 9232–9237. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Nicolas, E.; Marks, D.; Sander, C.; Lerro, A.; Buendia, M.A.; Xu, C.; Mason, W.S.; Moloshok, T.; Bort, R.; et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004, 1, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Drosatos, K.; Hiyama, Y.; Goldberg, I.J.; Zannis, V.I. MicroRNA-370 controls the expression of microRNA-122 and Cpt1α and affects lipid metabolism. J. Lipid Res. 2010, 51, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Gerin, I.; Bommer, G.T.; McCoin, C.S.; Sousa, K.M.; Krishnan, V.; MacDougald, O.A. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E198–E206. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Gao, Z.; Alarcon, R.M.; Ye, J.; Yun, Z. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009, 276, 2348–2358. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, M.; Guan, J.; Li, P.; Wang, H.; Guo, Y.; Shuai, S.; Li, X. MicroRNAs miR-27a and miR-143 regulate porcine adipocyte lipid metabolism. Int. J. Mol. Sci. 2011, 12, 7950–7959. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, N.; Nakagawa, Y.; Tokushige, N.; Aoki, N.; Matsuzaka, T.; Ishii, K.; Yahagi, N.; Kobayashi, K.; Yatoh, S.; Takahashi, A.; et al. The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem. Biophys. Res. Commun. 2009, 385, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Luo, J.; Zhang, L.; Wang, W.; Gou, D. MiR-103 controls milk fat accumulation in goat (Capra hircus) mammary gland during lactation. PLoS ONE 2013, 8, e79258. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, H.; Jin, X.; Lo, L.; Liu, J. Expression profiles of microRNAs from lactating and non-lactating bovine mammary glands and identification of miRNA related to lactation. BMC Genom. 2012, 13, 731. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Wang, G.; Xie, Z.; Wang, J.; Zhang, C.; Dong, F.; Chen, C. Identification of novel and differentially expressed microRNAs of dairy goat mammary gland tissues using Solexa sequencing and bioinformatics. PLoS ONE 2012, 7, e49463. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, L.L.; Matukumalli, L.K.; Sonstegard, T.S.; van Tassell, C.P.; Gasbarre, L.C.; Capuco, A.V.; Smith, T.P. Discovery and profiling of bovine microRNAs from immune-related and embryonic tissues. Physiol. Genom. 2007, 29, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. MicroRNAs in breastmilk and the lactating Breast: Potential immunoprotectors and developmental regulators for the infant and the mother. Int. J. Environ. Res. Public Health 2015, 12, 13981–14020. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; TatLai, C.; Hartmann, P.E.; Geddes, D.T.; Kakulas, F. Human milk miRNAs primarily originate from the mammary gland resulting in unique miRNA profiles of fractionated milk. Sci. Rep. 2016, in press. [Google Scholar]
- Modepalli, V.; Kumar, A.; Hinds, L.A.; Sharp, J.A.; Nicholas, K.R.; Lefevre, C. Differential temporal expression of milk miRNA during the lactation cycle of the marsupial tammar wallaby (Macropus eugenii). BMC Genom. 2014, 15, 1012. [Google Scholar] [CrossRef] [PubMed]
- Juan, W.F.W.; Shenyuan, W. Primary culture of Bovine mammary epithelial cells. J. Agric. Sci. Technol. 2009, 10, 119–123. [Google Scholar]
- Rohlfs, E.M.; Louie, D.S.; Zeisel, S.H. Lipid synthesis and secretion by primary cultures of rat mammary epithelial cells. J. Cell. Physiol. 1993, 157, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Chunyan, L.B.S.; Zhihui, Z. Establishment of two types of mammary epithelial cell lines from Chinese Holstein dairy cow. J. Anim. Vet. Adv. 2012, 11, 1166–1172. [Google Scholar]
- Jensen, R.G. Composition of bovine milk lipids. J. Am. Oil Chem. Soc. 1973, 50, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.E.; Hemati, N.; Longo, K.A.; Bennett, C.N.; Lucas, P.C.; Erickson, R.L.; MacDougald, O.A. Inhibition of adipogenesis by Wnt signaling. Science 2000, 289, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Bauman, D.; Davis, C. Biosynthesis of milk fat. Lact. Compr. Treatise 2013, 2, 31–75. [Google Scholar]
- Sorg, D.; Potzel, A.; Beck, M.; Meyer, H.H.; Viturro, E.; Kliem, H. Effects of cell culture techniques on gene expression and cholesterol efflux in primary bovine mammary epithelial cells derived from milk and tissue. In Vitro Cell. Dev. Biol. Anim. 2012, 48, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Alsaweed, M.; Hepworth, A.R.; Lefevre, C.; Hartmann, P.E.; Geddes, D.T.; Hassiotou, F. Human milk MicroRNA and total RNA differ depending on milk fractionation. J. Cell. Biochem. 2015, 116, 2397–2407. [Google Scholar] [CrossRef] [PubMed]
- Munch, E.M.; Harris, R.A.; Mohammad, M.; Benham, A.L.; Pejerrey, S.M.; Showalter, L.; Hu, M.; Shope, C.D.; Maningat, P.D.; Gunaratne, P.H.; et al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS ONE 2013, 8, e50564. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gao, C.; Li, H.; Huang, L.; Sun, Q.; Dong, Y.; Tian, C.; Gao, S.; Dong, H.; Guan, D.; et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010, 20, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, C.M.; Davalos, A.; Goedeke, L.; Salerno, A.G.; Warrier, N.; Cirera-Salinas, D.; Suarez, Y.; Fernandez-Hernando, C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2707–2714. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yoon, H.; Ramirez, C.M.; Lee, S.M.; Hoe, H.S.; Fernandez-Hernando, C.; Kim, J. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp. Neurol. 2012, 235, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Larson, B.L.; Smith, V.R. Lactation. A Comprehensive Treatise. Volume III. Nutrition and Biochemistry of Milk Maintenance; Academic Press: New York, NY, USA, 1974. [Google Scholar]
- Wang, C.; Li, Q. Identification of differentially expressed microRNAs during the development of Chinese murine mammary gland. J. Genet. Genom. 2007, 34, 966–973. [Google Scholar] [CrossRef]
- Chen, T.; Li, Z.; Tu, J.; Zhu, W.; Ge, J.; Zheng, X.; Yang, L.; Pan, X.; Yan, H.; Zhu, J. MicroRNA-29a regulates pro-inflammatory cytokine secretion and scavenger receptor expression by targeting LPL in oxLDL-stimulated dendritic cells. FEBS Lett. 2011, 585, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Guillou, H.; Zadravec, D.; Martin, P.G.; Jacobsson, A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 2010, 49, 186–199. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, M.; van der Sluis, R.J.; Kuiper, J.; van Berkel, T.J. Nonalcoholic fatty liver disease is associated with an altered hepatocyte microRNA profile in LDL receptor knockout mice. J. Nutr. Biochem. 2012, 23, 622–628. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Smith, L.D.; Chang, R.; Li, X.; Vockley, J. The role of sterol-C4-methyl oxidase in epidermal biology. Biochim. Biophys. Acta 2014, 1841, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Krishan, S.; Richardson, D.R.; Sahni, S. AMP kinase (PRKAA1). J. Clin. Pathol. 2014, 67, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Polekhina, G.; Gupta, A.; Michell, B.J.; van Denderen, B.; Murthy, S.; Feil, S.C.; Jennings, I.G.; Campbell, D.J.; Witters, L.A.; Parker, M.W.; et al. AMPK β subunit targets metabolic stress sensing to glycogen. Curr. Biol. 2003, 13, 867–871. [Google Scholar] [CrossRef]
- Carling, D. The AMP-activated protein kinase cascade—A unifying system for energy control. Trends Biochem. Sci. 2004, 29, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Abdelazeem, A.H.; Abdelatef, S.A.; El-Saadi, M.T.; Omar, H.A.; Khan, S.I.; McCurdy, C.R.; El-Moghazy, S.M. Novel pyrazolopyrimidine derivatives targeting COXs and iNOS enzymes; design, synthesis and biological evaluation as potential anti-inflammatory agents. Eur. J. Pharm. Sci. 2014, 62, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Ding, N.S.; Ren, D.R.; Guo, Y.M.; Ren, J.; Yan, Y.; Ma, J.W.; Chen, K.F.; Huang, L.S. Genetic variation of porcine prostaglandin-endoperoxide synthase 2 (PTGS2) gene and its association with reproductive traits in an Erhualian x Duroc F2 population. Yi Chuan Xue Bao 2006, 33, 213–219. [Google Scholar] [CrossRef]
- Choi, C.S.; Fillmore, J.J.; Kim, J.K.; Liu, Z.X.; Kim, S.; Collier, E.F.; Kulkarni, A.; Distefano, A.; Hwang, Y.J.; Kahn, M.; et al. Overexpression of uncoupling protein 3 in skeletal muscle protects against fat-induced insulin resistance. J. Clin. Investig. 2007, 117, 1995–2003. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef] [PubMed]
- Mattis, A.N.; Song, G.; Hitchner, K.; Kim, R.Y.; Lee, A.Y.; Sharma, A.D.; Malato, Y.; McManus, M.T.; Esau, C.C.; Koller, E.; et al. A screen in mice uncovers repression of lipoprotein lipase by microRNA-29a as a mechanism for lipid distribution away from the liver. Hepatology 2015, 61, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Ye, Y.F.; Chen, S.H.; Yu, C.H.; Liu, J.; Li, Y.M. MicroRNA expression pattern in different stages of nonalcoholic fatty liver disease. Dig. Liver Dis. 2009, 41, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Lieb, D.C.; Brotman, J.J.; Hatcher, M.A.; Aye, M.S.; Cole, B.K.; Haynes, B.A.; Wohlgemuth, S.D.; Fontana, M.A.; Beydoun, H.; Nadler, J.L.; et al. Adipose tissue 12/15 lipoxygenase pathway in human obesity and diabetes. J. Clin. Endocrinol. Metab. 2014, 99, E1713–E1720. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Nie, D. Tumor-suppressing 15-lipoxygenase-2: Time for prime time? Cell Cycle 2014, 13, 1836–1837. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Liu, B.; Luo, W.; Zhang, Y.; Li, H.; Li, S.; Zhou, Y. Vasoconstrictor role of cyclooxygenase-1-mediated prostacyclin synthesis in non-insulin-dependent diabetic mice induced by high-fat diet and streptozotocin. Am. J. Physiol. Heart Circ. Physiol. 2014, 307, H319–H327. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Nutrient Requirements of Dairy Cattle; National Academies: Washington, DC, USA, 1978.
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, B.; Zhang, L.; Lian, C.; Lu, C.; Zhang, Y.; Pan, Q.; Yang, R.; Zhao, Z. Deep Sequencing and Screening of Differentially Expressed MicroRNAs Related to Milk Fat Metabolism in Bovine Primary Mammary Epithelial Cells. Int. J. Mol. Sci. 2016, 17, 200. https://doi.org/10.3390/ijms17020200
Shen B, Zhang L, Lian C, Lu C, Zhang Y, Pan Q, Yang R, Zhao Z. Deep Sequencing and Screening of Differentially Expressed MicroRNAs Related to Milk Fat Metabolism in Bovine Primary Mammary Epithelial Cells. International Journal of Molecular Sciences. 2016; 17(2):200. https://doi.org/10.3390/ijms17020200
Chicago/Turabian StyleShen, Binglei, Liying Zhang, Chuanjiang Lian, Chunyan Lu, Yonghong Zhang, Qiqi Pan, Runjun Yang, and Zhihui Zhao. 2016. "Deep Sequencing and Screening of Differentially Expressed MicroRNAs Related to Milk Fat Metabolism in Bovine Primary Mammary Epithelial Cells" International Journal of Molecular Sciences 17, no. 2: 200. https://doi.org/10.3390/ijms17020200
APA StyleShen, B., Zhang, L., Lian, C., Lu, C., Zhang, Y., Pan, Q., Yang, R., & Zhao, Z. (2016). Deep Sequencing and Screening of Differentially Expressed MicroRNAs Related to Milk Fat Metabolism in Bovine Primary Mammary Epithelial Cells. International Journal of Molecular Sciences, 17(2), 200. https://doi.org/10.3390/ijms17020200