Reference Genes in the Pathosystem Phakopsora pachyrhizi/ Soybean Suitable for Normalization in Transcript Profiling
Abstract
:1. Introduction
2. Results and Discussion
2.1. Choice of Samples
2.2. Choice of Candidate Reference Genes
| Gene Designation | Description | GenBank Accession/Name | Reference |
|---|---|---|---|
| Act | actin | GACM01001510/TSA:GACM01:Pp_contig05019 | [18] |
| ASUB | ATP synthase β subunit | PpGI_Contig363rc a | - |
| aTub | α tubulin | GACM01003080/TSA:GACM01:Pp_contig00787 | [20] |
| CytB | cytochrome b | GQ332420 | [18,19] |
| Elf1a | elongation factor 1 α | GACM01002155/TSA:GACM01:Pp_contig07365 | - |
| Elf3 | elongation factor 3 | GACM01003209/TSA:GACM01:Pp_contig00583 | - |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | GACM01002868/TSA:GACM01:Pp_contig00008 | [20] |
| PDH | pyruvate dehydrogenase | GACM01002893/TSA:GACM01:Pp_contig00239 | - |
| PDK | pyruvate dehydrogenase kinase | GACM01003388/TSA:GACM01:Pp_contig02726 | - |
| RPS9 | 40S ribosomal protein S9 | GACM01002779/TSA:GACM01:Pp_contig00421 | [19] |
| RPS11 | 40S ribosomal protein S11 | GACM01003017/TSA:GACM01:Pp_contig00682 | [19] |
| RPS14 | 40S ribosomal protein S14 | GACM01002939/TSA:GACM01:Pp_contig00153 | [19] |
| SDH | succinate dehydrogenase | GACM01002923/TSA:GACM01:Pp_contig00241 | [21] |
| Ubc | ubiquitin | GACM01002338/TSA:GACM01:Pp_contig00942 | [21] |
| UbcE2 | ubiquitin conjugated enzyme | GACM01000888/TSA:GACM01:Pp_contig06751 | [21] |
| Gene Designation a | Description | Gene ID | Reference |
|---|---|---|---|
| cons7 | metalloprotease | AW310136 b | [23] |
| cons15 | CDPK-related protein kinase | AW396185 b | [23] |
| CYP2 | cyclophilin | TC224926 c | [22,24] |
| ELF1B | elongation factor 1 β | TC203623 c | [22,24] |
| SKIP16 | F-box protein | CD397253 b | [23,24] |
| TIP41 | TAP42 interacting protein-signaling | EV263725 b | [24] |
| Ukn1 | hypothetical protein/possibly ABC transporter | BU578186 b | [23,24] |
| Ukn2 | hypothetical protein | BE330043 b | [24] |
2.3. Reference Genes for P. pachyrhizi: All Tested Genes Vary in Expression for in Vitro Structures but Are Stable in in planta Samples
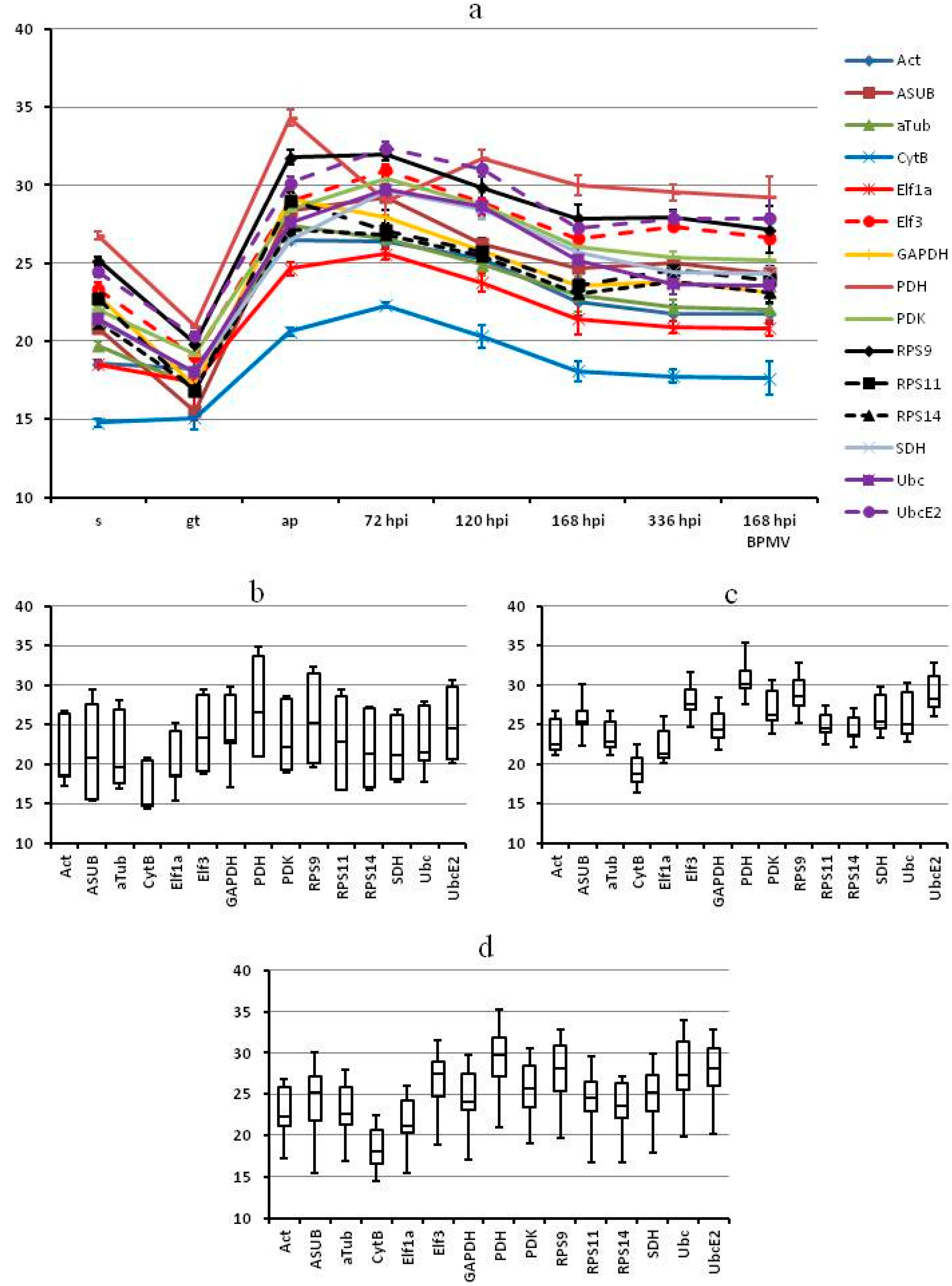
| Name | Gene | Sequence | Reference | Amplicon (bp) | Efficiency (%) |
|---|---|---|---|---|---|
| ActinDis 1 f (KES 1461 fw) | Act | acagtttcaccacaaccgcc | [18] | 144 | 93 |
| ActinDis 1 r (KES 1462 rv) | tgaccgtcgggaagttcg | ||||
| qPpGI363fw | ASUB | tgtcccaaccctttgctgtt | this study | 146 | 86 |
| qPpGI363rv | ggcaccgaccatgtaaaacg | ||||
| AtubDis 1 f (KES 1463 fw) | aTub | ctgcgaacaactatgctcgtc | [18] | 116 | 90 |
| AtubDis 1 r (KES 1464 fw) | cacgaagaagccttggagtcc | ||||
| CytB 1 f (KES 1688 fw) | CytB | tcaagacgcatccaattctaggtc | [28] | 138 | 85 |
| CytB 1 r (KES 1689 rv) | gtgttacacccgtgataatctgaatgat | ||||
| Elf1a 1 f | Elf1a | gtgagcgtggtatcaccatc | this study | 143 | 92 |
| Elf1a 1 r | cagaatggcgcaatcagc | ||||
| q00583fw | Elf3 | aatgcgtggtctctctggtg | this study | 89 | 95 |
| q00583rv | gctcgtccaagatcaccaca | ||||
| GAPDH 1 f (KES 1465 fw) | GAPDH | ggtatggctttccgagttcca | [18] | 157 | 90 |
| GAPDH 1 r (KES 1466 rv) | tcagttgataccaaatcatcctcag | ||||
| q00239fw | PDH | gcggaaggaaaggataagggg | this study | 136 | 95 |
| q00239rv | tccgatccttagtctggcct | ||||
| q02726fw | PDK | acctcccgttcagctagtct | this study | 138 | 94 |
| q02726rv | aattcatcagagtcggcccc | ||||
| RibPro 3 f | RPS9 | gtgaatgggagaccaatctcag | this study | 128 | 94 |
| RibPro 3 r | ttgcctcctccatgagtcag | ||||
| q00682fw | RPS11 | ggactgggcttcaagactcc | this study | 97 | 86 |
| q00682rv | gaatcctgcccctgatcgag | ||||
| q00153fw | RPS14 | agttgctcgagtgactggtg | this study | 86 | 89 |
| q00153rv | catcttgggcagccaacatg | ||||
| q00241fw | SDH | caatcgcctgaggaccgtaa | this study | 80 | 88 |
| q00241rv | ctggggcaacttgtagagca | ||||
| Ubc 1 f | Ubc | cggaccagtacccttacaaatc | this study | 128 | 93 |
| Ubc 1 r | atcaaacatcggcgaccag | ||||
| UbcE2 3 f | UbcE2 | gtcgaactgtgacgagtttg | this study | 117 | 89 |
| UbcE2 3 r | acggccttagtcttcgatg |

2.4. Reference Genes for G. max: Ukn2 and cons7 Can Be Used for Normalizing RT-qPCR in Tissues Infected with Soybean Rust and Also for Superinfection of Soybean Rust on BPMV
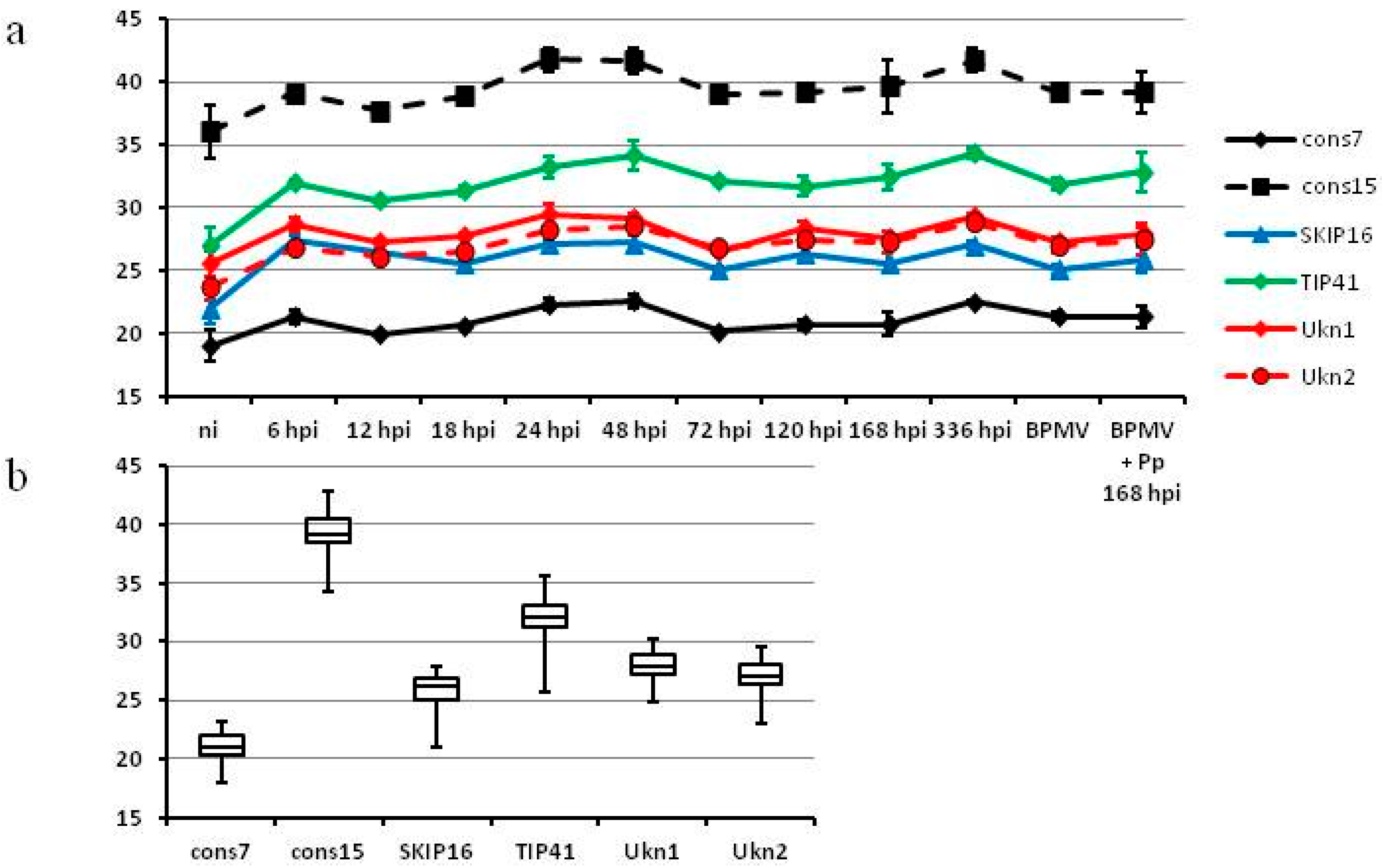
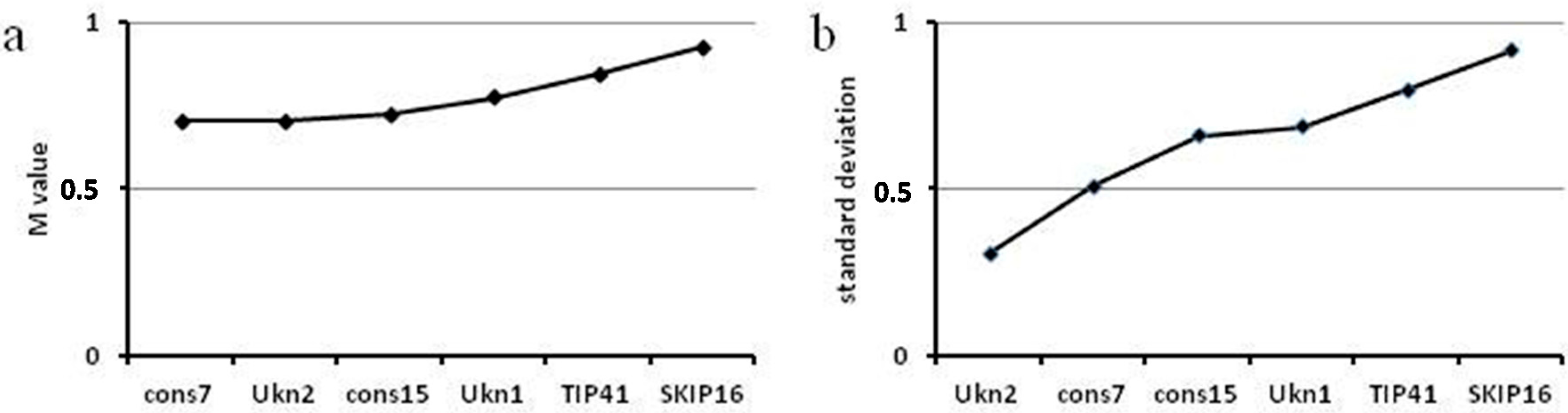
2.5. Number of Reference Genes Needed for Efficient Normalization
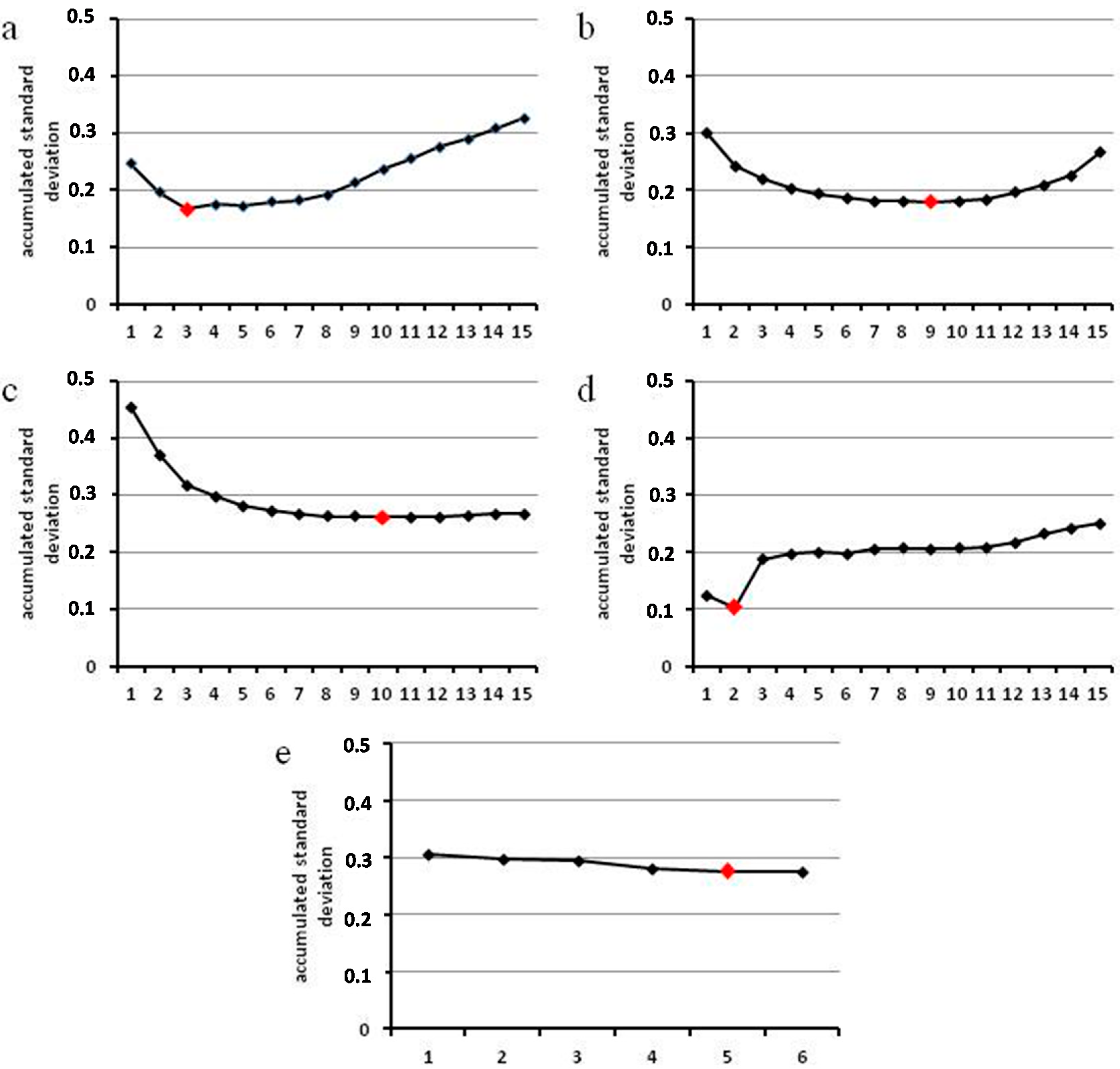
3. Experimental Section
3.1. Plant Material, Fungal Isolate, Inoculation with Virus and Rust Fungus
3.1.1. Plant Material
3.1.2. Fungal Isolate, Plant Inoculation and in Vitro Structures
3.1.3. BPMV Inoculation
3.2. Sample Homogenization and RNA Isolation
3.3. RT-qPCR
3.3.1. Reverse Transcription
3.3.2. Machinery, Chemistry, and Reaction Conditions of Real Time PCR
3.3.3. Data Analysis
3.4. Primers
| Name | Gene | Sequence | Reference | Amplicon [bp] | Efficiency [%] |
|---|---|---|---|---|---|
| Gm cons7 f | cons7 | atgaatgacggttcccatgta | [23] | 114 | 81 |
| Gm cons7 r | ggcattaaggcagctcactct | ||||
| Gm cons15 f | cons15 | taaagagcaccatgcctatcc | [23] | 97 | 106 |
| Gm cons15 r | tggttatgtgagcagatgcaa | ||||
| Gm CYP2 f | CYP2 | cgggaccagtgtgcttcttca | [22] | 154 | na |
| Gm CYP2 r | cccctccactacaaaggctcg | ||||
| Gm ELF1B f | ELF1B | gttgaaaagccaggggaca | [22] | 118 | na |
| Gm ELF1B r | tcttaccccttgagcgtgg | ||||
| Gm SKIP16 f | SKIP16 | gagcccaagacattgcgagag | [24] | 60 | 87 |
| Gm SKIP16 r | cggaagcggagaactgaacc | ||||
| Gm TIP41 f | Tip41 | aggatgaactcgctgataatgg | [24] | 88 | 95 |
| Gm TIP41 r | cagaaacgcaacagaagaaacc | ||||
| Gm UKN1 f | Ukn1 | tggtgctgccgctatttactg | [24] | 74 | 90 |
| Gm UKN1 r | ggtggaaggaactgctaacaatc | ||||
| Gm UKN2 f | Ukn2 | gcctctggatacctgctcaag | [24] | 79 | 96 |
| Gm UKN2 r | acctcctcctcaaactcctctg |
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Goellner, K.; Loehrer, M.; Langenbach, C.; Conrath, U.; Koch, E.; Schaffrath, U. Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol. Plant Pathol. 2010, 11, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Voegele, R.T.; Hahn, M.; Mendgen, K. The Uredinales: Cytology, biochemistry, and molecular biology. In The Mycota V—Plant Relationships, 2nd ed.; Deising, H., Ed.; Springer: Berlin, Germany; Heidelberg, Germany, 2009; pp. 69–98. [Google Scholar]
- Posada-Buitrago, M.L.; Frederick, R.D. Expressed sequence tag analysis of the soybean rust pathogen Phakopsora pachyrhizi. Fungal Genet. Biol. 2005, 42, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.; Mendgen, K. Characterization of in planta-induced rust genes isolated from a haustorium-specific cDNA library. Mol. Plant Microbe Interact. 1997, 10, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Duplessis, S.; Cuomo, C.A.; Lin, Y.C.; Aerts, A.; Tisserant, E.; Veneault-Fourrey, C.; Joly, D.L.; Hacquard, S.; Amselem, J.; Cantarel, B.L.; et al. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA 2011, 108, 9166–9171. [Google Scholar] [CrossRef] [PubMed]
- Link, T.I.; Voegele, R.T. Secreted proteins of Uromyces fabae: Similarities and stage specificity. Mol. Plant Pathol. 2008, 9, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Voegele, R.T.; Schmid, A. RT real-time PCR-based quantification of Uromyces fabae in planta. FEMS Microbiol. Lett. 2011, 322, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Voegele, R.T.; Mendgen, K.W. Nutrient uptake in rust fungi: How sweet is parasitic life? Euphytica 2011, 179, 41–55. [Google Scholar] [CrossRef]
- Link, T.I.; Lang, P.; Scheffler, B.E.; Duke, M.V.; Graham, M.A.; Cooper, B.; Tucker, M.L.; van de Mortel, M.; Voegele, R.T.; Mendgen, K.; et al. The haustorial transcriptomes of Uromyces appendiculatus and Phakopsora pachyrhizi and their candidate effector families. Mol. Plant Pathol. 2014, 15, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rafiqi, M.; Gan, P.H.; Hardham, A.R.; Jones, D.A.; Ellis, J.G. Effectors of biotrophic fungi and oomycetes: Pathogenicity factors and triggers of host resistance. New Phytol. 2009, 183, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Petre, B.; Joly, D.L.; Duplessis, S. Effector proteins of rust fungi. Front. Plant Sci. 2014, 5, 416. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, G.J.; Dodds, P.N.; Ellis, J.G. Transformation of the flax rust fungus, Melampsora lini: Selection via silencing of an avirulence gene. Plant J. 2010, 61, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 2010, 22, 3130–3141. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.S.; Nam, H.; Hopkins, R.G.; Morrison, R.F. Impact of reference gene selection for target gene normalization on experimental outcome using real-time qRT-PCR in adipocytes. PLoS ONE 2010, 5, e15208. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J.; Oh, E.; Lee, S.; Roh, M.R.; Kim, S.E.; Lee, Y.; Choi, Y.L.; In, Y.H.; Park, T.; Koh, S.S.; et al. Identification of novel reference genes using multiplatform expression data and their validation for quantitative gene expression analysis. PLoS ONE 2009, 4, e6162. [Google Scholar] [CrossRef] [PubMed]
- Van de Mortel, M.; Recknor, J.C.; Graham, M.A.; Nettleton, D.; Dittman, J.D.; Nelson, R.T.; Godoy, C.V.; Abdelnoor, R.V.; Almeida, A.M.; Baum, T.J.; et al. Distinct biphasic mRNA changes in response to asian soybean rust infection. Mol. Plant Microbe Interact. 2007, 20, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, H.K.; Medeiros, C.A.; Craig, I.R.; Stammler, G. Sensitivity of Phakopsora pachyrhizi towards quinone-outside-inhibitors and demethylation-inhibitors, and corresponding resistance mechanisms. Pest Manag. Sci. 2014, 70, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.; Talhinhas, P.; Loureiro, A.; Duplessis, S.; Fernandez, D.; Silva Mdo, C.; Paulo, O.S.; Azinheira, H.G. Validation of RT-qPCR reference genes for in planta expression studies in Hemileia vastatrix, the causal agent of coffee leaf rust. Fungal Biol. 2011, 115, 891–901. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Veneault-Fourrey, C.; Delaruelle, C.; Frey, P.; Martin, F.; Duplessis, S. Validation of Melampsora larici-populina reference genes for in planta RT-quantitative PCR expression profiling during time-course infection of poplar leaves. Physiol. Mol. Plant Pathol. 2011, 75, 106–112. [Google Scholar] [CrossRef]
- Song, X.; Rampitsch, C.; Soltani, B.; Mauthe, W.; Linning, R.; Banks, T.; McCallum, B.; Bakkeren, G. Proteome analysis of wheat leaf rust fungus, Puccinia triticina, infection structures enriched for haustoria. Proteomics 2011, 11, 944–963. [Google Scholar] [CrossRef] [PubMed]
- Jian, B.; Liu, B.; Bi, Y.; Hou, W.; Wu, C.; Han, T. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Thibivilliers, S.; Bilgin, D.D.; Radwan, O.; Benitez, M.; Clough, S.J.; Stacey, G. Identification of four soybean reference genes for gene expression normalization. Plant Genome 2008, 1, 44–54. [Google Scholar] [CrossRef]
- Hu, R.; Fan, C.; Li, H.; Zhang, Q.; Fu, Y.F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 2009, 10, 93. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acid Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1–research0034.11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, H.K. In Vivo und molekularbiologische Untersuchungen zur Sensitivität von Phakopsora pachyrhizi gegenüber Demethylierungs-Inhibitoren und Qo-Inhibitoren; Universität Hohenheim: Stuttgart, Germany, 2013; p. 41. (In Germany) [Google Scholar]
- Le, D.T.; Aldrich, D.L.; Valliyodan, B.; Watanabe, Y.; Ha, C.V.; Nishiyama, R.; Guttikonda, S.K.; Quach, T.N.; Gutierrez-Gonzalez, J.J.; Tran, L.S.; et al. Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS ONE 2012, 7, e46487. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Niu, H.; Liu, C.; Zhang, J.; Hou, C.; Wang, D. Expression stabilities of candidate reference genes for RT-qPCR under different stress conditions in soybean. PLoS ONE 2013, 8, e75271. [Google Scholar] [CrossRef] [PubMed]
- Miranda, V.D.J.; Coelho, R.R.; Viana, A.A.; de Oliveira Neto, O.B.; Carneiro, R.M.; Rocha, T.L.; de Sa, M.F.; Fragoso, R.R. Validation of reference genes aiming accurate normalization of qPCR data in soybean upon nematode parasitism and insect attack. BMC Res. Notes 2013, 6, 196. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.; Mittapelly, P.; Cassone, B.J.; Mamidala, P.; Redinbaugh, M.G.; Michel, A. Recommended reference genes for quantitative PCR analysis in soybean have variable stabilities during diverse biotic stresses. PLoS ONE 2015, 10, e0134890. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Bradshaw, J.D.; Whitham, S.A.; Hill, J.H. The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiol. 2010, 153, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yang, C.; Whitham, S.A.; Hill, J.H. Development and use of an efficient DNA-based viral gene silencing vector for soybean. Mol. Plant Microbe Interact. 2009, 22, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirschburger, D.; Müller, M.; Voegele, R.T.; Link, T. Reference Genes in the Pathosystem Phakopsora pachyrhizi/ Soybean Suitable for Normalization in Transcript Profiling. Int. J. Mol. Sci. 2015, 16, 23057-23075. https://doi.org/10.3390/ijms160923057
Hirschburger D, Müller M, Voegele RT, Link T. Reference Genes in the Pathosystem Phakopsora pachyrhizi/ Soybean Suitable for Normalization in Transcript Profiling. International Journal of Molecular Sciences. 2015; 16(9):23057-23075. https://doi.org/10.3390/ijms160923057
Chicago/Turabian StyleHirschburger, Daniela, Manuel Müller, Ralf T. Voegele, and Tobias Link. 2015. "Reference Genes in the Pathosystem Phakopsora pachyrhizi/ Soybean Suitable for Normalization in Transcript Profiling" International Journal of Molecular Sciences 16, no. 9: 23057-23075. https://doi.org/10.3390/ijms160923057
APA StyleHirschburger, D., Müller, M., Voegele, R. T., & Link, T. (2015). Reference Genes in the Pathosystem Phakopsora pachyrhizi/ Soybean Suitable for Normalization in Transcript Profiling. International Journal of Molecular Sciences, 16(9), 23057-23075. https://doi.org/10.3390/ijms160923057





