SOCS3 Methylation Predicts a Poor Prognosis in HBV Infection-Related Hepatocellular Carcinoma
Abstract
:1. Introduction
2. Results and Discussion
2.1. Results
2.1.1. Suppressor of Cytokine Signaling 3 (SOCS3) CpG Island (CGI) Is Hypermethylated in the Region Close to the Translational Start Site
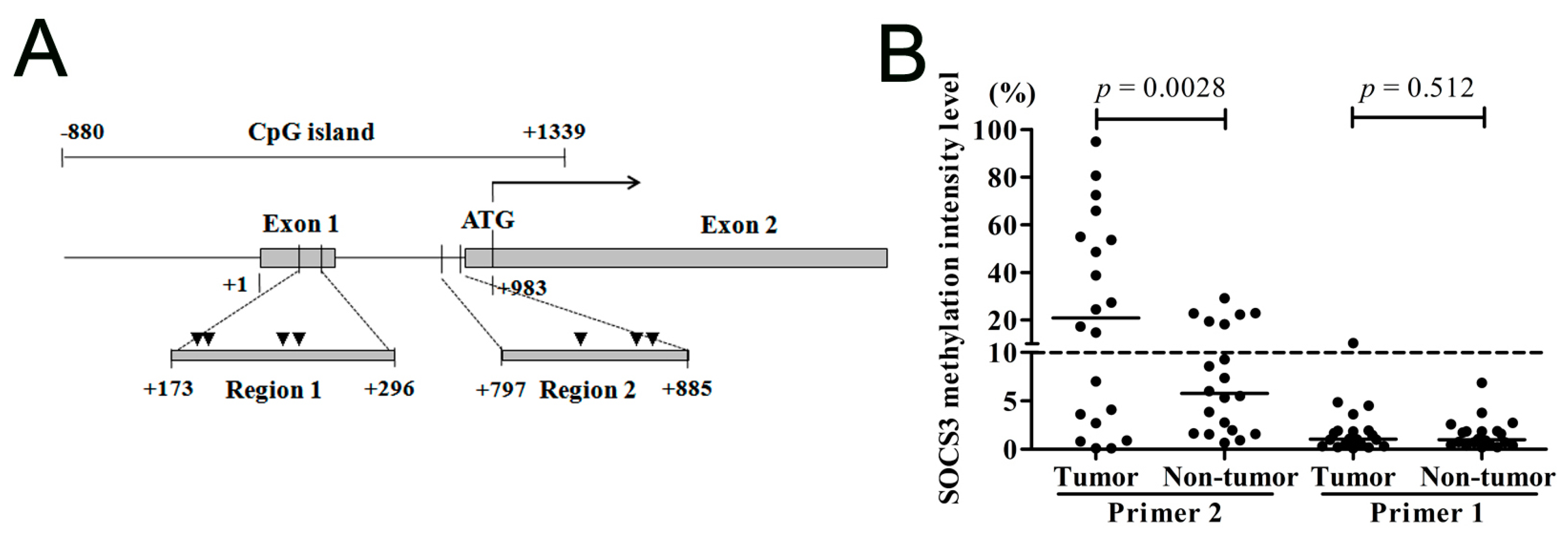
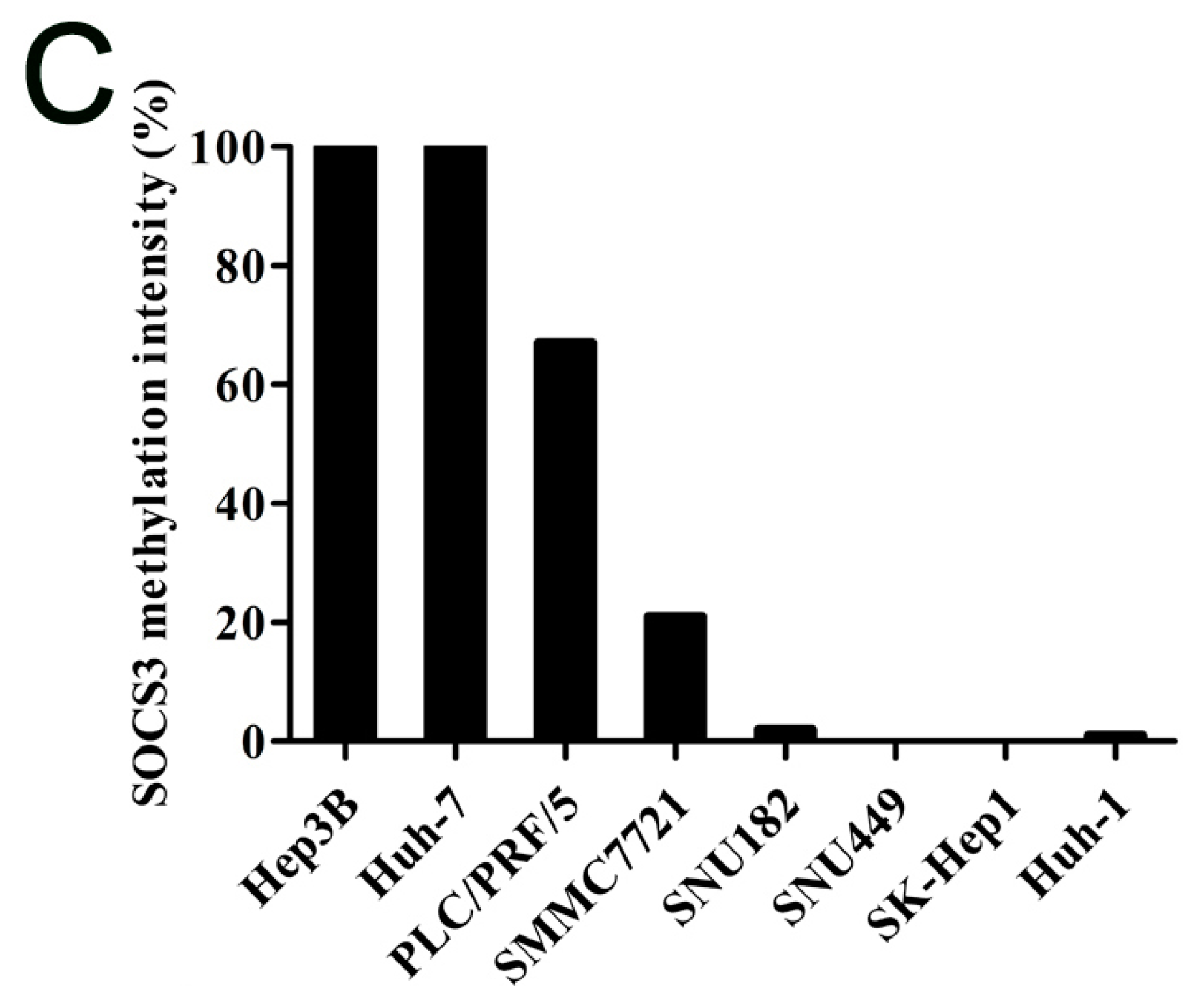
2.1.2. SOCS3 Methylation Is Associated with Altered mRNA Expression in Hepatocellular Carcinoma (HCC) Cell Lines

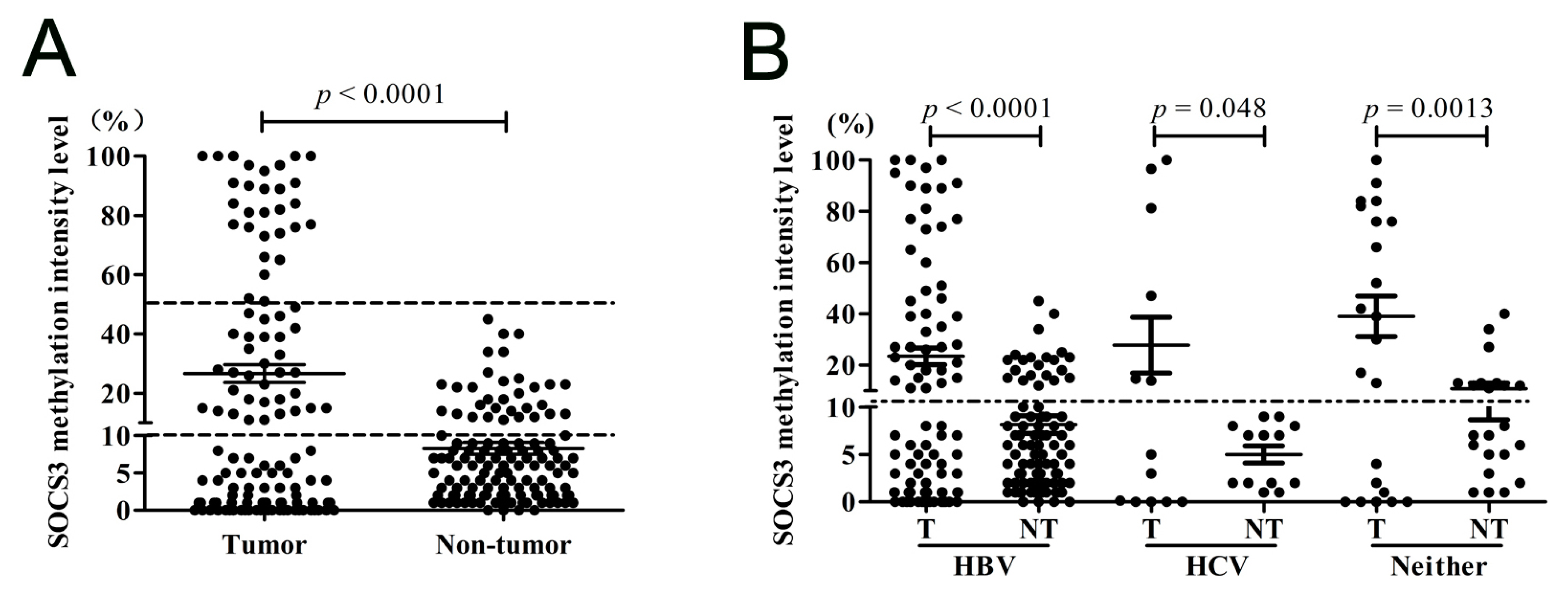
2.1.3. SOCS3 CGI Is Hypermethylated in Primary HCC Tumor Tissues
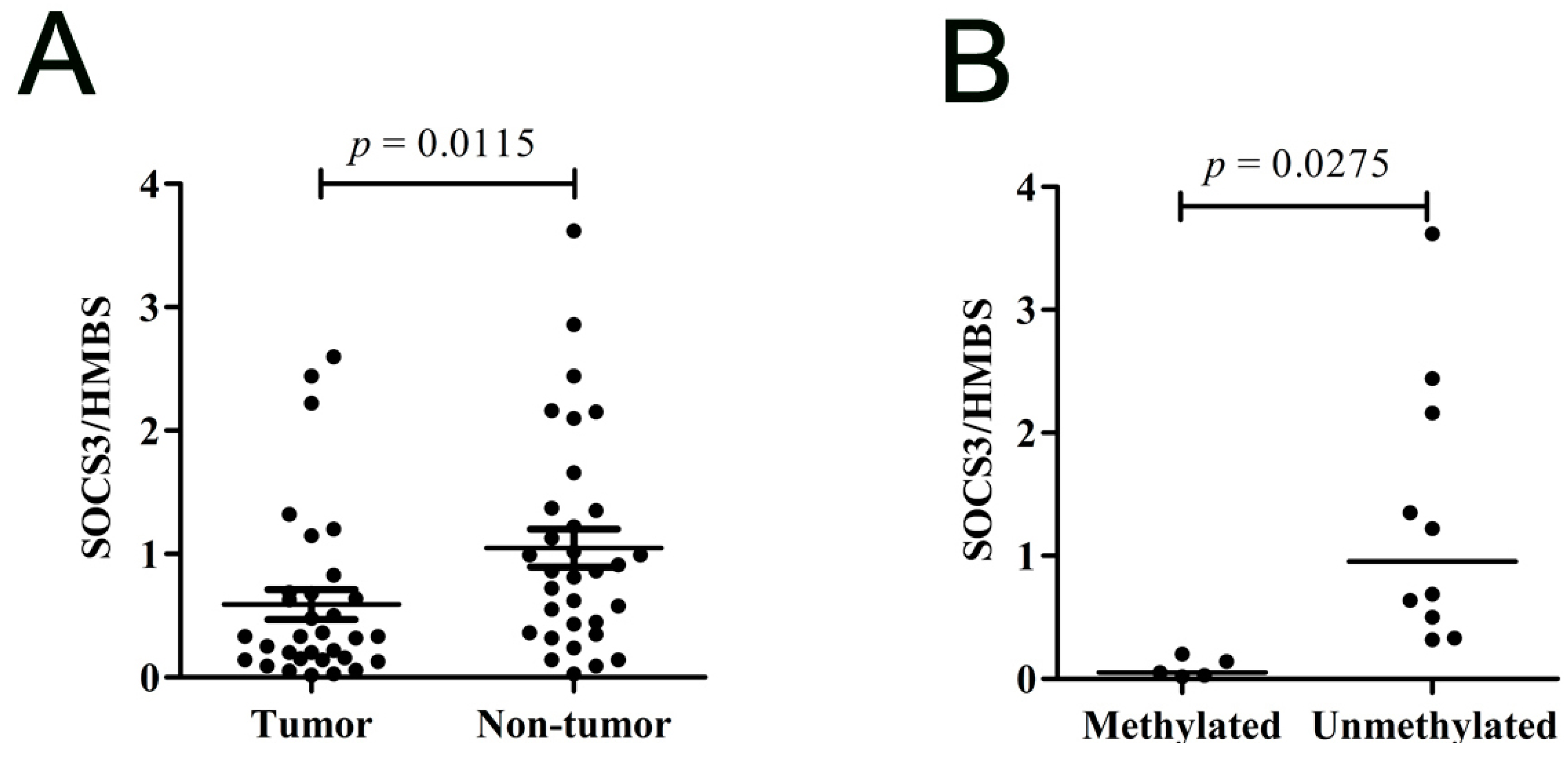
2.1.4. SOCS3 Is down-Regulated in Primary HCC Tumor Tissues
2.1.5. Methylation of SOCS3 Is Associated with the Poor Prognosis of HCC with Hepatitis B Virus (HBV) Infection
| Feature | Hypermethylation (n = 61) | Unmethylation (n = 66) | p-Value |
|---|---|---|---|
| Gender | |||
| Male | 46 (52.87%) | 41 (47.13%) | 0.107 |
| Female | 15 (37.5%) | 25 (62.5%) | |
| Age | |||
| ≥55 | 42 (54.5%) | 35 (45.5%) | 0.068 |
| <55 | 19 (38%) | 31 (62%) | |
| Cirrhosis | |||
| Yes | 47 (47.47%) | 52 (52.53%) | 0.813 |
| No | 14 (50%) | 14 (50%) | |
| TNM stage | |||
| I–II | 22 (48.89%) | 23 (51.11%) | 0.886 |
| III–IV | 39 (47.56%) | 43 (52.44%) | |
| Portal vein tumor thrombosis | |||
| Present | 9 (40.91%) | 13 (59.09%) | 0.267 |
| Absent | 52 (50.98%) | 50 (49.02%) | |
| Tumor size | |||
| ≥5 cm | 46 (47.92%) | 50 (52.08%) | 0.964 |
| <5 cm | 15 (48.39%) | 16 (51.61%) | |
| Tumor encapsulation | |||
| Complete | 52 (50%) | 52 (50%) | 0.800 |
| Incomplete | 6 (37.5%) | 10 (62.5%) | |

2.2. Discussion
3. Experimental Section
3.1. Tissue Samples
| Clinicopathological Parameter Variables | Cases n (%) n = 127 |
|---|---|
| Age | |
| Median (Range) | 57 (11–80) |
| Gender | |
| Male | 87 (68.50) |
| Female | 40 (31.50) |
| Liver cirrhosis | |
| Yes | 99 (77.95) |
| No | 28 (22.05) |
| TNM stage | |
| I–II | 45 (35.43) |
| III–IV | 82 (64.57) |
| Portal vein tumor thrombosis | |
| Present | 22 (17.32) |
| Absent | 102 (80.32) |
| N/A | 3 (2.36) |
| Tumor size | |
| ≥5 cm | 96 (75.59) |
| <5 cm | 31 (24.41) |
| Tumor encapsulation | |
| Complete | 104 (81.89) |
| Incomplete | 16 (12.60) |
| N/A | 7 (5.51) |
3.2. Cell Culture and 5-Aza-2ʹ-Deoxycytidine Treatment
3.3. DNA Extraction and Methylation Assay
3.4. RNA Extraction and Real-Time RT-qPCR
3.5. Statistical Analyses
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Portolani, N.; Coniglio, A.; Ghidoni, S.; Giovanelli, M.; Benetti, A.; Tiberio, G.A.; Giulini, S.M. Early and late recurrence after liver resection for hepatocellular carcinoma: Prognostic and therapeutic implications. Ann. Surg. 2006, 243, 229–235. [Google Scholar] [PubMed]
- Wang, H.Q.; Yang, J.; Yan, L.N.; Zhang, X.W.; Yang, J.Y. Liver resection in hepatitis B related-hepatocellular carcinoma: Clinical outcomes and safety in elderly patients. World J. Gastroenterol. 2014, 20, 6620–6625. [Google Scholar] [CrossRef]
- Yeo, W.; Mo, F.K.; Chan, S.L.; Leung, N.W.; Hui, P.; Lam, W.Y.; Mok, T.S.; Lam, K.C.; Ho, W.M.; Koh, J.; et al. Hepatitis B viral load predicts survival of HCC patients undergoing systemic chemotherapy. Hepatology 2007, 45, 1382–1389. [Google Scholar] [CrossRef] [PubMed]
- Mikeska, T.; Bock, C.; Do, H.; Dobrovic, A. DNA methylation biomarkers in cancer: Progress towards clinical implementation. Expert Rev. Mol. Diagn. 2012, 12, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Pogribny, I.P.; Rusyn, I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 2014, 342, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Sproul, D.; Meehan, R.R. Genomic insights into cancer-associated aberrant CpG island hypermethylation. Brief. Funct. Genom. 2013, 12, 174–190. [Google Scholar] [CrossRef] [PubMed]
- Paziewska, A.; Dabrowska, M.; Goryca, K.; Antoniewicz, A.; Dobruch, J.; Mikula, M.; Jarosz, D.; Zapala, L.; Borowka, A.; Ostrowski, J. DNA methylation status is more reliable than gene expression at detecting cancer in prostate biopsy. Br. J. Cancer 2014, 111, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Calvisi, D.F.; Ladu, S.; Gorden, A.; Farina, M.; Lee, J.S.; Conner, E.A.; Schroeder, I.; Factor, V.M.; Thorgeirsson, S.S. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J. Clin. Investig. 2007, 117, 2713–2722. [Google Scholar] [CrossRef] [PubMed]
- Dhar, K.; Rakesh, K.; Pankajakshan, D.; Agrawal, D.K. SOCS3 promotor hypermethylation and STAT3-NF-κB interaction downregulate SOCS3 expression in human coronary artery smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 2013, 304, H776–H785. [Google Scholar] [CrossRef] [PubMed]
- Iwahori, K.; Serada, S.; Fujimoto, M.; Nomura, S.; Osaki, T.; Lee, C.M.; Mizuguchi, H.; Takahashi, T.; Ripley, B.; Okumura, M.; et al. Overexpression of SOCS3 exhibits preclinical antitumor activity against malignant pleural mesothelioma. Int. J. Cancer 2011, 129, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Chinen, T.; Yoshida, T.; Kinjyo, I.; Takaesu, G.; Shiraishi, H.; Iida, M.; Kobayashi, T.; Yoshimura, A. Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-β1 production. Oncogene 2006, 25, 2520–2530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Badgwell, D.B.; Bevers, J.J., 3rd.; Schlessinger, K.; Murray, P.J.; Levy, D.E.; Watowich, S.S. IL-6 signaling via the STAT3/SOCS3 pathway: Functional analysis of the conserved STAT3 N-domain. Mol. Cell. Biochem. 2006, 288, 179–189. [Google Scholar] [CrossRef] [PubMed]
- He, B.; You, L.; Uematsu, K.; Zang, K.; Xu, Z.; Lee, A.Y.; Costello, J.F.; McCormick, F.; Jablons, D.M. SOCS-3 is frequently silenced by hypermethylation and suppresses cell growth in human lung cancer. Proc. Natl. Acad. Sci. USA 2003, 100, 14133–14138. [Google Scholar] [CrossRef] [PubMed]
- Inagaki-Ohara, K.; Kondo, T.; Ito, M.; Yoshimura, A. SOCS, inflammation, and cancer. JAKSTAT 2013, 2, e24053. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; de Haar, C.; Chen, M.; Deuring, J.; Gerrits, M.M.; Smits, R.; Xia, B.; Kuipers, E.J.; van der Woude, C.J. Disease-related expression of the IL6/STAT3/SOCS3 signalling pathway in ulcerative colitis and ulcerative colitis-related carcinogenesis. Gut 2010, 59, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.B.; Ao, L.; Zhou, Z.Y.; Cui, Z.H.; Zhou, Y.H.; Yuan, X.Y.; Xiang, Y.L.; Cao, J.; Liu, J.Y. CpG island hypermethylation of multiple tumor suppressor genes associated with loss of their protein expression during rat lung carcinogenesis induced by 3-methylcholanthrene and diethylnitrosamine. Biochem. Biophys. Res. Commun. 2010, 402, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Pierconti, F.; Martini, M.; Pinto, F.; Cenci, T.; Capodimonti, S.; Calarco, A.; Bassi, P.F.; Larocca, L.M. Epigenetic silencing of SOCS3 identifies a subset of prostate cancer with an aggressive behavior. Prostate 2011, 71, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Tischoff, I.; Hengge, U.R.; Vieth, M.; Ell, C.; Stolte, M.; Weber, A.; Schmidt, W.E.; Tannapfel, A. Methylation of SOCS-3 and SOCS-1 in the carcinogenesis of Barrett’s adenocarcinoma. Gut 2007, 56, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Hengge, U.R.; Bardenheuer, W.; Tischoff, I.; Sommerer, F.; Markwarth, A.; Dietz, A.; Wittekind, C.; Tannapfel, A. SOCS-3 is frequently methylated in head and neck squamous cell carcinoma and its precursor lesions and causes growth inhibition. Oncogene 2005, 24, 6699–6708. [Google Scholar] [CrossRef] [PubMed]
- Niwa, Y.; Kanda, H.; Shikauchi, Y.; Saiura, A.; Matsubara, K.; Kitagawa, T.; Yamamoto, J.; Kubo, T.; Yoshikawa, H. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene 2005, 24, 6406–6417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, J.; Cheng, J.; Ding, S.; Li, M.; Sun, S.; Zhang, L.; Liu, S.; Chen, X.; Zhuang, H.; et al. An integrated analysis of SOCS1 down-regulation in HBV infection-related hepatocellular carcinoma. J. Viral Hepat. 2013. [Google Scholar] [CrossRef] [PubMed]
- Shire, A.; Lomberk, G.; Lai, J.P.; Zou, H.; Tsuchiya, N.; Aderca, I.; Moser, C.D.; Gulaid, K.H.; Oseini, A.; Hu, C.; et al. Restoration of epigenetically silenced SULF1 expression by 5-aza-2-deoxycytidine sensitizes hepatocellular carcinoma cells to chemotherapy-induced apoptosis. Med. Epigenet. 2015, 3, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.Y.; Li, J.; Wu, Z.S.; Zhang, C.L.; Meng, X.L.; Lobie, P.E. Prognostic significance of phosphorylated signal transducer and activator of transcription 3 and suppressor of cytokine signaling 3 expression in hepatocellular carcinoma. Exp. Ther. Med. 2011, 2, 647–653. [Google Scholar] [PubMed]
- Deng, J.; Jiao, X.; Liu, H.; Wu, L.; Zhang, R.; Wang, B.; Pan, Y.; Hao, X.; Liang, H. Lymph node metastasis is mediated by suppressor of cytokine signaling-3 in gastric cancer. Tumour Biol. 2013, 34, 3627–3636. [Google Scholar] [CrossRef] [PubMed]
- Stofas, A.; Levidou, G.; Piperi, C.; Adamopoulos, C.; Dalagiorgou, G.; Bamias, A.; Karadimou, A.; Lainakis, G.A.; Papadoukakis, S.; Stravodimos, K.; et al. The role of CXC-chemokine receptor CXCR2 and suppressor of cytokine signaling-3 (SOCS-3) in renal cell carcinoma. BMC Cancer 2014, 14, 149. [Google Scholar] [CrossRef] [PubMed]
- Baltayiannis, G.; Baltayiannis, N.; Tsianos, E.V. Suppressors of cytokine signaling as tumor repressors. Silencing of SOCS3 facilitates tumor formation and growth in lung and liver. J. BUON 2008, 13, 263–265. [Google Scholar] [PubMed]
- Wu, W.Y.; Kim, H.; Zhang, C.L.; Meng, X.L.; Wu, Z.S. Loss of suppressors of cytokine signaling 3 promotes aggressiveness in hepatocellular carcinoma. J. Investig. Surg. 2014, 27, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Yao, M.; Chen, X.; Lu, W.; Lv, Q.; Wang, K.; Zhang, L.; Lu, F. Reduced red blood cell count predicts poor survival after surgery in patients with primary liver cancer. Medicine 2015, 94, e577. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, J.; Xu, C.; Liu, S.; Jiang, S.; Xu, Q.; Chen, X.; Zhuang, H.; Lu, F. Quantitative methylation analysis reveals gender and age differences in p16INK4a hypermethylation in hepatitis B virus-related hepatocellular carcinoma. Liver Int. 2012, 32, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhu, P.; Zhang, L.; Ding, S.; Zheng, S.; Wang, Y.; Lu, F. Selection of reference genes for RT-qPCR analysis in tumor tissues from male hepatocellular carcinoma patients with hepatitis B infection and cirrhosis. Cancer Biomark. 2013, 13, 345–349. [Google Scholar] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; You, Q.; Zhang, X.; Chen, X. SOCS3 Methylation Predicts a Poor Prognosis in HBV Infection-Related Hepatocellular Carcinoma. Int. J. Mol. Sci. 2015, 16, 22662-22675. https://doi.org/10.3390/ijms160922662
Zhang X, You Q, Zhang X, Chen X. SOCS3 Methylation Predicts a Poor Prognosis in HBV Infection-Related Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2015; 16(9):22662-22675. https://doi.org/10.3390/ijms160922662
Chicago/Turabian StyleZhang, Xin, Qingshan You, Xiaolei Zhang, and Xiangmei Chen. 2015. "SOCS3 Methylation Predicts a Poor Prognosis in HBV Infection-Related Hepatocellular Carcinoma" International Journal of Molecular Sciences 16, no. 9: 22662-22675. https://doi.org/10.3390/ijms160922662





