A Perturbed MicroRNA Expression Pattern Characterizes Embryonic Neural Stem Cells Derived from a Severe Mouse Model of Spinal Muscular Atrophy (SMA)
Abstract
:1. Introduction
2. Results
2.1. SMNΔ7 Stem Cells Derived from E13.5 Spinal Cords form Neurospheres and Express Specific Regional Markers

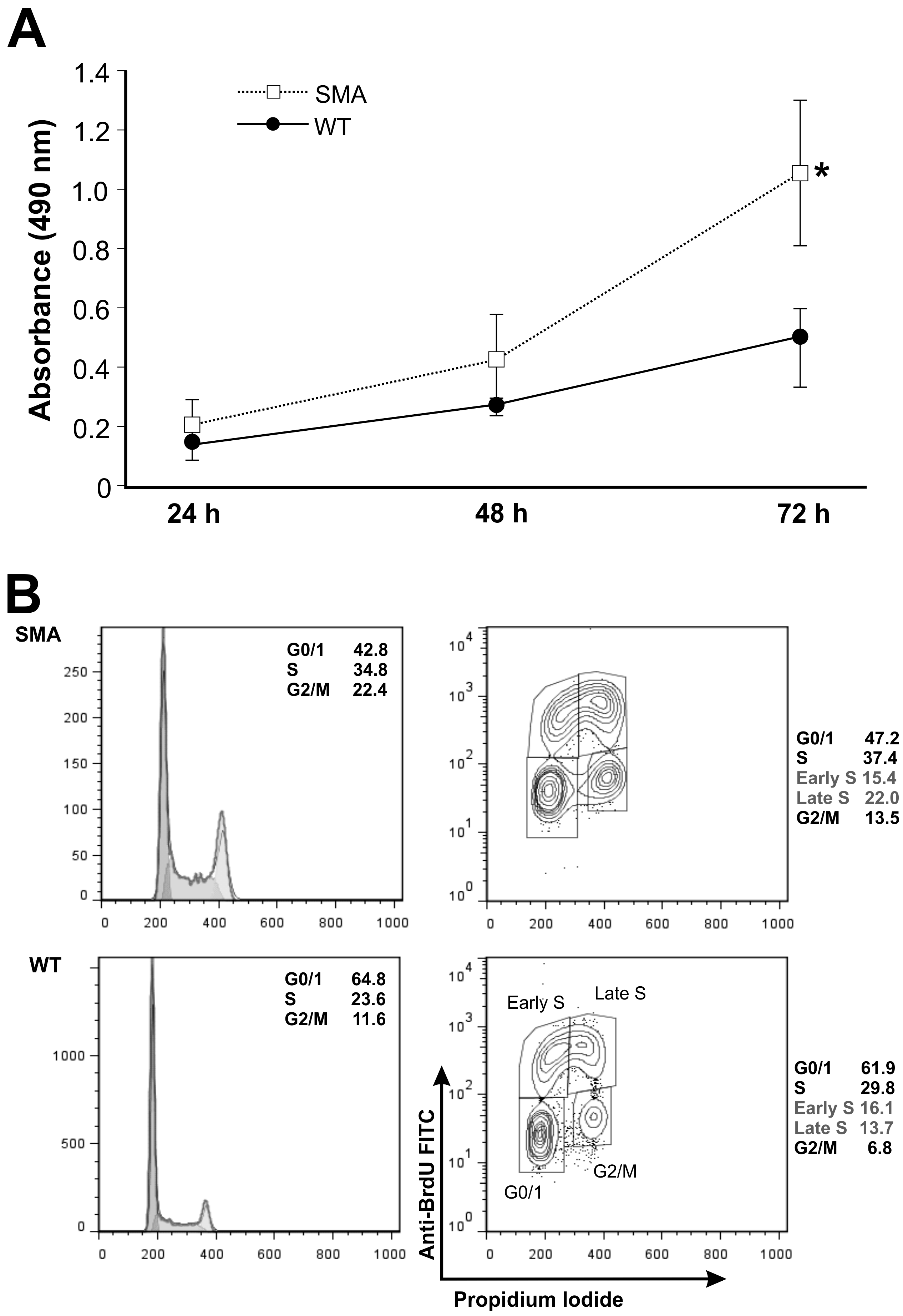
2.2. SMNΔ7 Neural Stem Cells Show a Perturbed Cell Cycle Phase Distribution and Increased Proliferation Rate Compared to Wild Type Cells
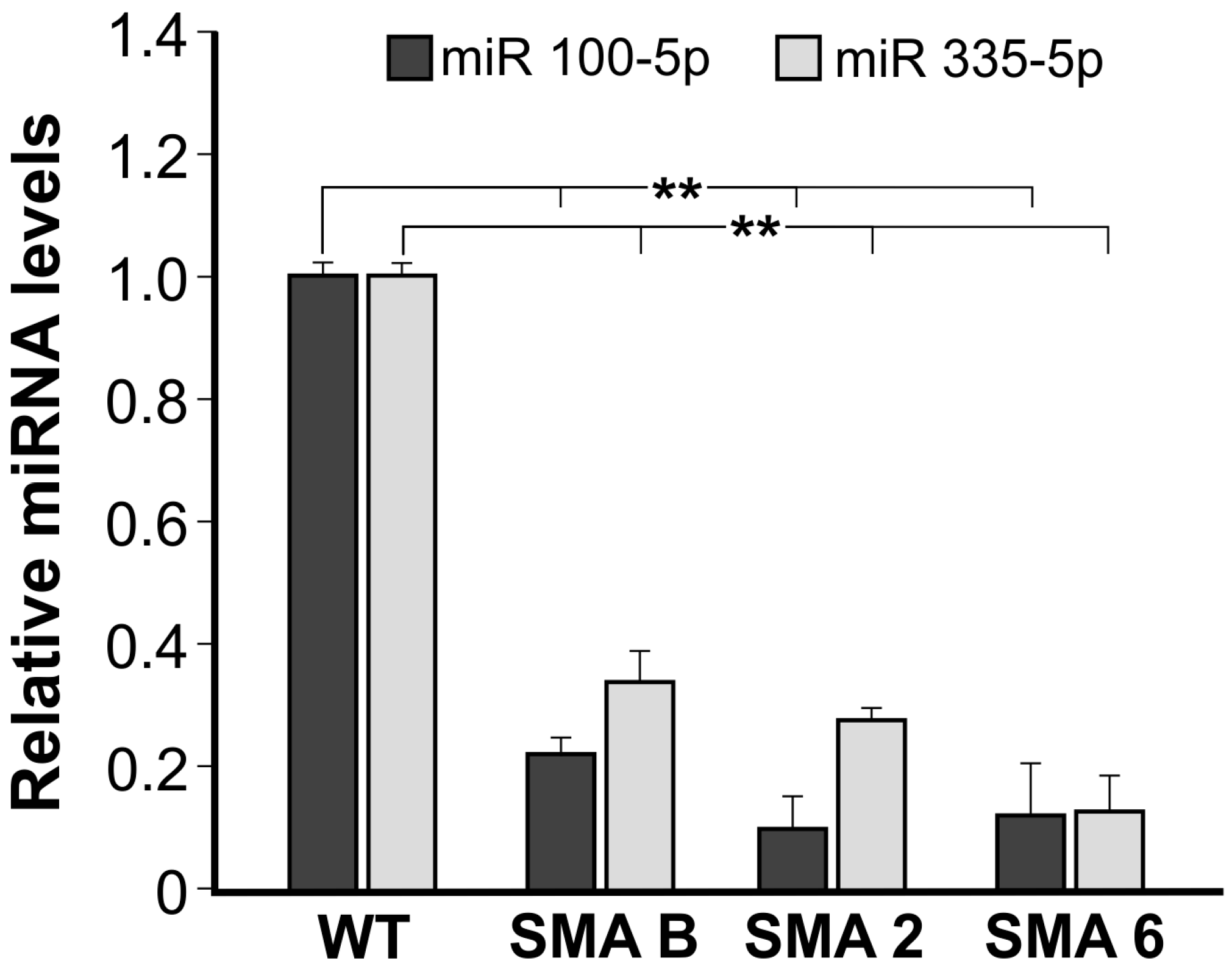
2.3. The Expression of Specific microRNAs Is Affected in SMNΔ7 Neural Stem Cells
| Underexpressed in SMA vs. WT | ||
| miRNA | p-Value | Log2 (SMA/WT) |
| mmu-miR-20b-5p | 4.73 × 10−3 | −1.81 |
| mmu-miR-6944-5p | 2.79 × 10−2 | −1.78 |
| mmu-miR-100-5p | 3.22 × 10−2 | −1.74 |
| mmu-miR-335-5p | 1.70 × 10−2 | −1.69 |
| mmu-let-7k | 4.76 × 10−2 | −1.31 |
| mmu-miR-6931-5p | 3.88 × 10−2 | −1.15 |
| mmu-miR-1224-5p | 3.77 × 10−2 | −1.13 |
| mmu-miR-3960 | 1.00 × 10−2 | −0.81 |
| mmu-miR-26b-5p | 1.65 × 10−2 | −0.76 |
| mmu-miR-2861 | 3.47 × 10−2 | −0.72 |
| mmu-miR-7047-5p | 2.01 × 10−2 | −0.52 |
| mmu-miR-106a-5p | 4.90 × 10−2 | −0.52 |
| mmu-miR-23b-3p | 3.35 × 10−2 | −0.51 |
| Overexpressed in SMA vs. WT | ||
| miRNA | p-Value | Log2 (SMA/WT) |
| mmu-miR-466q | 3.98 × 10−2 | 0.80 |
| mmu-miR-1187 | 1.32 × 10−2 | 0.64 |
| mmu-miR-466g | 2.58 × 10−2 | 0.64 |
3. Discussion
4. Experimental Section
4.1. Animal Models
4.2. Isolation and Culture of Neural Stem Cells (NSCs)
4.3. RT-PCR Analysis
| Gene | Forward | Reverse |
|---|---|---|
| Gapdh | CATGGCCTTCCGTGTTCCTA | GCGGCACGTCAGATCCA |
| Hoxb4 | TCACGTGAGCACGGTAAACCC | GCGTCAGGTAGCGATTGTAGTGAA |
| Hoxb9 | ATTTGCGAAGGAAGCGAGGACA | TAGCTCCAGCGTCTGGTATTTGGT |
| Olig2 | GTTCTCCTCCGCAGCGAG | CCTTCTTTTTTCAACCTTCCGA |
| Pax6 | CAGCTCCAGCATGCAGAACA | CCGCCCGTTGACAAAGAC |
| Sox2 | AGAAGAGGAGAGAGAAAGAAAGGGAGAGA | GAGAGAGGCAAACTGGAATCAGGATCAAA |
| Nestin | AGGCTGAGAACTCTCGCTTGC | GGTGCTGGTCCTCTGGTATCC |
| U1 | CCTGGCAGGGGAGATACCATGAT | TGCAGTCGAGTTTCCCGCATTT |
| U2 | CGGCCTTTTGGCTAAGATCAAGTG | TCCTCGGATAGAGGACGTATCAGA |
| U4 | GAGGTTTATCCGAGGCGCGATTAT | CACGGCGTATTGGGAAAAGTT |
| U11 | CGTGCGGAATCGACATCAAGAGA | CAACGATCACCAGCTGCCCAATTA |
| U12 | AACTTATGAGTAAGGAAAATAACGATTCG | CCGCYCAAAAATTCTTCTCACA |
| U4atac | TTTCTTGGGGTTGCGCTACTGT | AAAGCAGAGCTCTAACCGATGCAG |
| 5.8S | GCGCTAGCTGCGAGAATTAAT | CAAGTGCGTTCGAAGTGTCGA |
| SMN FL | TGCTCCGTGGACCTCATTTCT | TGGCTTTCCTGGTCCTAATCC |
4.4. Expression Analyses by RT-qPCR
4.5. MTS Cell Viability Assay
4.6. Flow Cytometry
4.7. miRNA Microarray Assay
4.8. Validation of miRNA Expression
4.9. snRNP (Small Nuclear Ribonucleoproteins) Level Analysis
4.10. Immunofluorescence
4.11. Western Blot Analysis
4.12. Statistical Analyses
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
References
- Crawford, T.O.; Pardo, C.A. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996, 3, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, S.; Bürglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M.; et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef]
- Lorson, C.L.; Hahnen, E.; Androphy, E.J.; Wirth, B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. USA 1999, 96, 6307–6311. [Google Scholar] [CrossRef] [PubMed]
- Monani, U.R.; Lorson, C.L.; Parsons, D.W.; Prior, T.W.; Androphy, E.J.; Burghes, A.H.; McPherson, J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999, 8, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Burnett, B.G.; Munoz, E.; Tandon, A.; Kwon, D.Y.; Sumner, C.J.; Fishbeck, K.H. Regulation of SMN protein stability. Mol. Cell. Biol. 2009, 29, 1107–1115. [Google Scholar] [CrossRef] [PubMed]
- Frugier, T.; Nicole, S.; Cifuentes-Diaz, C.; Melki, J. The molecular bases of spinal muscular atrophy. Curr. Opin. Genet. Dev. 2002, 12, 294–298. [Google Scholar] [CrossRef]
- Lefebvre, S.; Burlet, P.; Liu, Q.; Bertrandy, S.; Clermont, O.; Munnich, A.; Dreyfuss, G.; Melki, J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997, 16, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Pellizzoni, L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007, 8, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Schrank, B.; Götz, R.; Gunnersen, J.M.; Ure, J.M.; Toyka, K.V.; Smith, A.G.; Sendtner, M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA 1997, 94, 9920–9925. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Dreyfuss, G. Characterization of functional domains of the SMN protein in vivo. J. Biol. Chem. 2001, 276, 45387–45393. [Google Scholar] [CrossRef] [PubMed]
- Monani, U.R. Spinal Muscular atrophy: A deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron 2005, 48, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Hsieh-Li, H.M.; Chang, J.G.; Jong, Y.J.; Wu, M.H.; Wang, N.M.; Tsai, C.H.; Li, H.A. Mouse model for spinal muscular atrophy. Nat. Genet. 2000, 24, 66–70. [Google Scholar] [PubMed]
- Monani, U.R.; Sendtner, M.; Coovert, D.D.; Parsons, D.W.; Andreassi, C.; Le, T.T.; Jablonka, S.; Schrank, B.; Rossoll, W.; Prior, T.W.; et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn−/− mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000, 9, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Pham, L.T.; Butchbach, M.E.; Zhang, H.L.; Monani, U.R.; Coovert, D.D.; Gavrilina, T.O.; Xing, L.; Bassell, G.J.; Burghes, A.H. SMNΔ7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005, 14, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Shafey, D.; Mac Kenzie, A.E.; Kothary, R. Neurodevelopmental abnormalities in neurosphere-derived neural stem cells from SMN-depleted mice. J. Neurosci. Res. 2008, 86, 2839–2847. [Google Scholar] [CrossRef] [PubMed]
- McKay, R. Stem cells in the central nervous system. Science 1997, 276, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Eacker, S.M.; Dawson, T.M.; Dawson, V.L. Understanding microRNAs in neurodegeneration. Nat. Rev. Neurosci. 2009, 10, 837–841. [Google Scholar] [CrossRef] [PubMed]
- Lau, P.; de Strooper, B. Dysregulated microRNAs in neurodegenerative disorders. Cell. Dev. Biol. 2010, 21, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Stappert, L.; Roese-Koerner, B.; Brustle, O. The role of microRNAs in human stem cells, neuronal differentiation and subtype specification. Cell Tissue Res. 2015, 359, 47–64. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.A.; Huang, Y.P.; Mazzoni, E.O.; Tan, G.C.; Zavadil, J.; Wichterle, H. miR-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron 2011, 69, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, C.; Zhang, C.; Li, P.; Wang, F.; Zhang, Z. Comparative profiling of microRNA expression between neural stem cells and motor neurons in embryonic spinal cord in rat. Int. J. Dev. Neurosci. 2010, 28, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Andersson, T.; Rahman, S.; Sansom, S.N.; Alsio, J.M.; Kaneda, M.; Smith, J.; O’Carrol, D.; Tarakhovsky, A.; Livesey, F.J. Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS ONE 2010, 5, e13453. [Google Scholar] [CrossRef] [PubMed]
- Farrel, B.C.; Power, E.M.; Mc Dermott, K.W. Developmentally regulated expression of Sox9 and microRNAs 124, 128 and 23 in neuroepithelial stem cells in the developing spinal cord. Int. J. Dev. Neurosci. 2011, 29, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Onorati, M.; Binetti, M.; Conti, L.; Camnasio, S.; Calabrese, G.; Albieri, I.; de Febo, F.; Toselli, M.; Biella, G.; Martynoga, B.; et al. Preservation of positional identity in fetus-derived neural stem (NS) cells from different mouse central nervous system compartments. Cell. Mol. Life Sci. 2011, 68, 1769–1783. [Google Scholar] [CrossRef] [PubMed]
- Conti, L.; Pollard, S.M.; Gorba, T.; Reitano, E.; Toselli, M.; Biella, G.; Sun, Y.; Sanzone, S.; Ying, Q.L.; Cattaneo, E.; et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005, 3, e283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Lotti, F.; Dittmar, K.; Younis, I.; Wan, L.; Kasim, M.; Dreyfuss, G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 2008, 133, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Gabanella, F.; Butchbach, M.E.; Saieva, L.; Carissimi, C.; Burghes, A.H.; Pellizzoni, L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE 2007, 2, e921. [Google Scholar] [CrossRef] [PubMed]
- Murdocca, M.; Malgieri, A.; Luchetti, A.; Saieva, L.; Dobrowolny, G.; de Leonibus, E.; Filareto, A.; Quitadamo, M.C.; Novelli, G.; Musarò, A.; et al. IPLEX administration improves motor neuron survival and ameliorates motor functions in a severe mouse model of spinal muscular atrophy. Mol. Med. 2012, 18, 1076–1085. [Google Scholar] [CrossRef] [PubMed]
- Haramati, S.; Chapnik, E.; Sztainberg, Y.; Eilam, R.; Zwang, R.; Gershoni, N.; McGlinn, E.; Heiser, P.W.; Wills, A.M.; Wirguin, I.; et al. miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. USA 2010, 107, 13111–13116. [Google Scholar] [CrossRef] [PubMed]
- Giachino, C.; Basak, O.; Taylor, V. Isolation and manipulation of mammalian neural stem cells in vitro. Methods Mol. Biol. 2009, 482, 143–158. [Google Scholar] [PubMed]
- Sun, T.; Wang, X.J.; Xie, S.S.; Zhang, D.L.; Wang, X.P.; Li, B.Q.; Ma, W.; Xin, H. A comparison of proliferative capacity and passaging potential between neural stem and progenitor cells in adherent and neurosphere cultures. Int. J. Dev. Neurosci. 2011, 29, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Luo, X.Q.; Han, B.W.; Duan, F.T.; Wei, P.P.; Chen, Y.Q. MicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br. J. Cancer 2013, 109, 2189–2198. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.N.; Schneider, J.S.; Qin, M.; Tyler, W.A.; Pintar, J.E.; Fraidenraich, D.; Wood, T.L.; Levison, S.W. IGF-II promotes stemness of neural restricted precursors. Stem Cells 2012, 30, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Dohi, O.; Yasui, K.; Gen, Y.; Takada, H.; Endo, M.; Tsuji, K.; Konishi, C.; Yamada, N.; Mitsuyoshi, H.; Yagi, N.; et al. Epigenetic silencing of miR-335 and its host gene MEST in hepatocellular carcinoma. Int. J. Oncol. 2013, 42, 411–418. [Google Scholar] [PubMed]
- Maeda, M.; Harris, A.W.; Kingham, B.F.; Lumpkin, C.J.; Opdenaker, L.M.; McChan, S.M.; Wang, W.; Butchbach, M.E. Transcriptome profiling of spinal muscular atrophy motor neuron derived from mouse embryonic stem cells. PLoS ONE 2014, 9, e106818. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Scarola, M.; Comisso, E.; Schneider, C.; Benetti, R. An Oct4-pRb axis, controlled by miR-335, integrates stem cell self-renewal and cell cycle control. Stem Cells 2013, 31, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Cikos, S.; Bukovská, A.; Koppel, J. Relative quantification of mRNA: Comparison of methods currently used for real-time PCR data analysis. BMC Mol. Biol. 2007, 8. [Google Scholar] [CrossRef] [PubMed]
- Bolstad, B.M.; Irizarry, R.A.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luchetti, A.; Ciafrè, S.A.; Murdocca, M.; Malgieri, A.; Masotti, A.; Sanchez, M.; Farace, M.G.; Novelli, G.; Sangiuolo, F. A Perturbed MicroRNA Expression Pattern Characterizes Embryonic Neural Stem Cells Derived from a Severe Mouse Model of Spinal Muscular Atrophy (SMA). Int. J. Mol. Sci. 2015, 16, 18312-18327. https://doi.org/10.3390/ijms160818312
Luchetti A, Ciafrè SA, Murdocca M, Malgieri A, Masotti A, Sanchez M, Farace MG, Novelli G, Sangiuolo F. A Perturbed MicroRNA Expression Pattern Characterizes Embryonic Neural Stem Cells Derived from a Severe Mouse Model of Spinal Muscular Atrophy (SMA). International Journal of Molecular Sciences. 2015; 16(8):18312-18327. https://doi.org/10.3390/ijms160818312
Chicago/Turabian StyleLuchetti, Andrea, Silvia Anna Ciafrè, Michela Murdocca, Arianna Malgieri, Andrea Masotti, Massimo Sanchez, Maria Giulia Farace, Giuseppe Novelli, and Federica Sangiuolo. 2015. "A Perturbed MicroRNA Expression Pattern Characterizes Embryonic Neural Stem Cells Derived from a Severe Mouse Model of Spinal Muscular Atrophy (SMA)" International Journal of Molecular Sciences 16, no. 8: 18312-18327. https://doi.org/10.3390/ijms160818312
APA StyleLuchetti, A., Ciafrè, S. A., Murdocca, M., Malgieri, A., Masotti, A., Sanchez, M., Farace, M. G., Novelli, G., & Sangiuolo, F. (2015). A Perturbed MicroRNA Expression Pattern Characterizes Embryonic Neural Stem Cells Derived from a Severe Mouse Model of Spinal Muscular Atrophy (SMA). International Journal of Molecular Sciences, 16(8), 18312-18327. https://doi.org/10.3390/ijms160818312









