Isolation and Heterologous Expression of a Polygalacturonase Produced by Fusarium oxysporum f. sp. cubense Race 1 and 4
Abstract
:1. Introduction
2. Results and Discussion
2.1. Purification of an Exo-Polygalacturonase (PGC3) from Fusarium oxysporum f. sp. cubense Race 4 (FOC4)
| Step | Total Protein (mg) | Total Activity (U a) | Yield (% activity) | Specific Activity (U a/mg) |
|---|---|---|---|---|
| Crude | 36.6 | 131.2476 | 100 | 3.586 |
| Ultrafiltration | 8.21 | 70.2604 | 53.53 | 8.5579 |
| Sephacryl S-100 | 1.85 | 26.35 | 20.08 | 14.2432 |
| Sepharose FF CM | 0.4 | 7.25 | 5.52 | 18.125 |
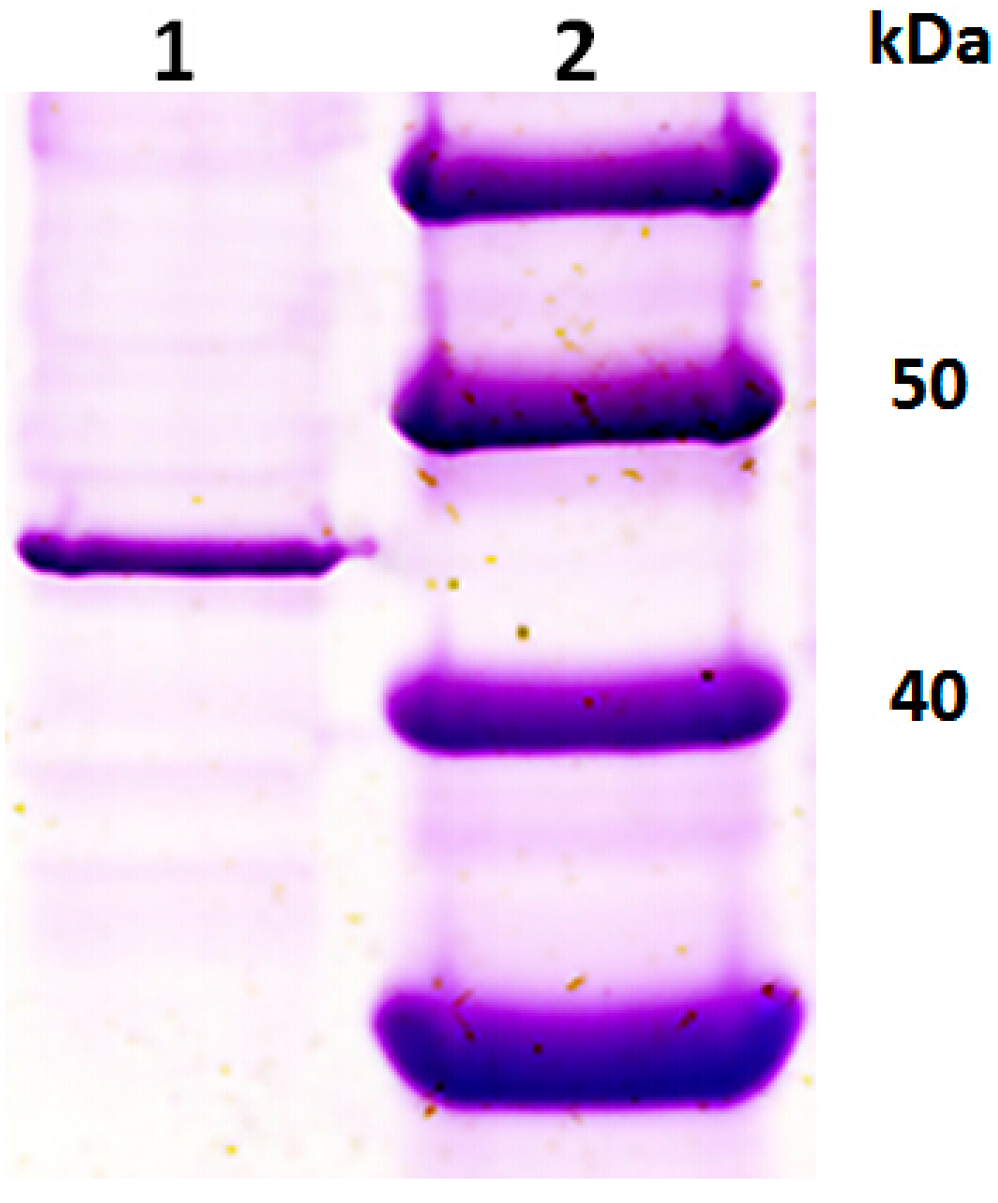
2.2. Isolation of Genes Encoding PGC3 from FOC1 and FOC4

2.3. Expression and Purification of Recombinant PGC3
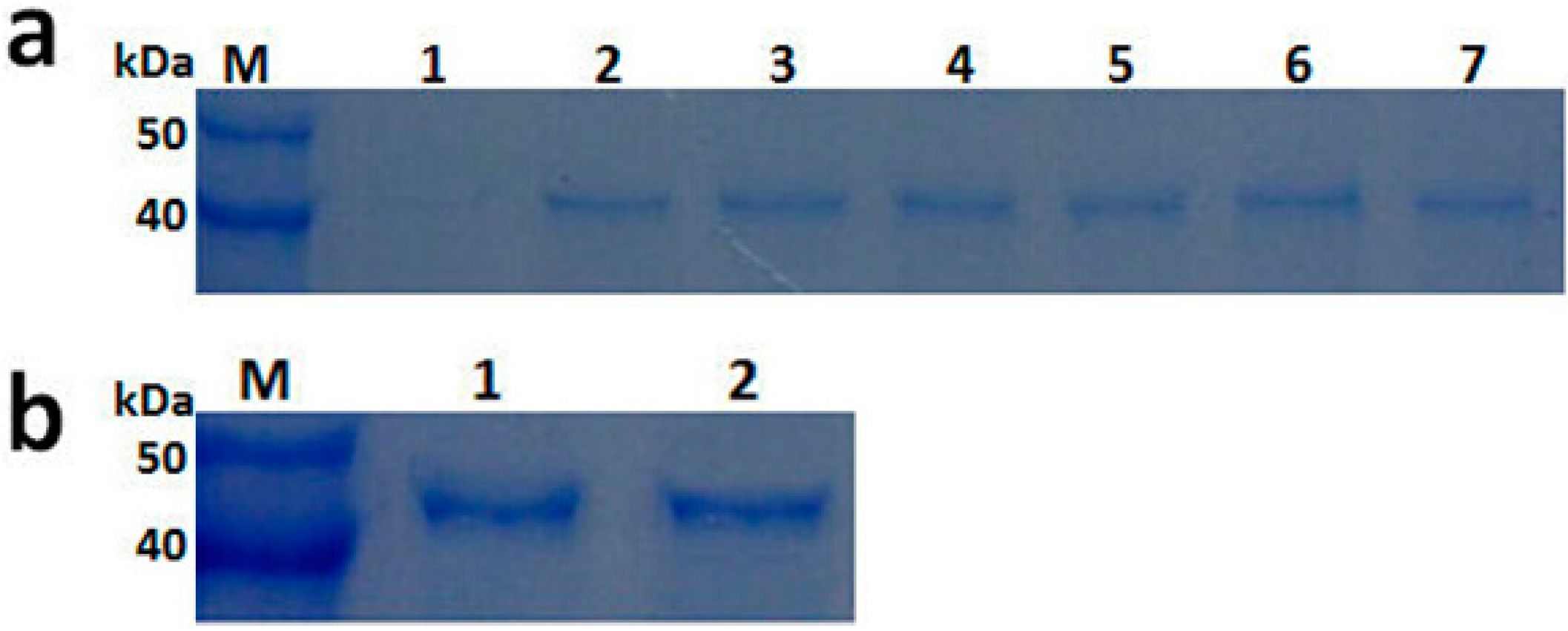
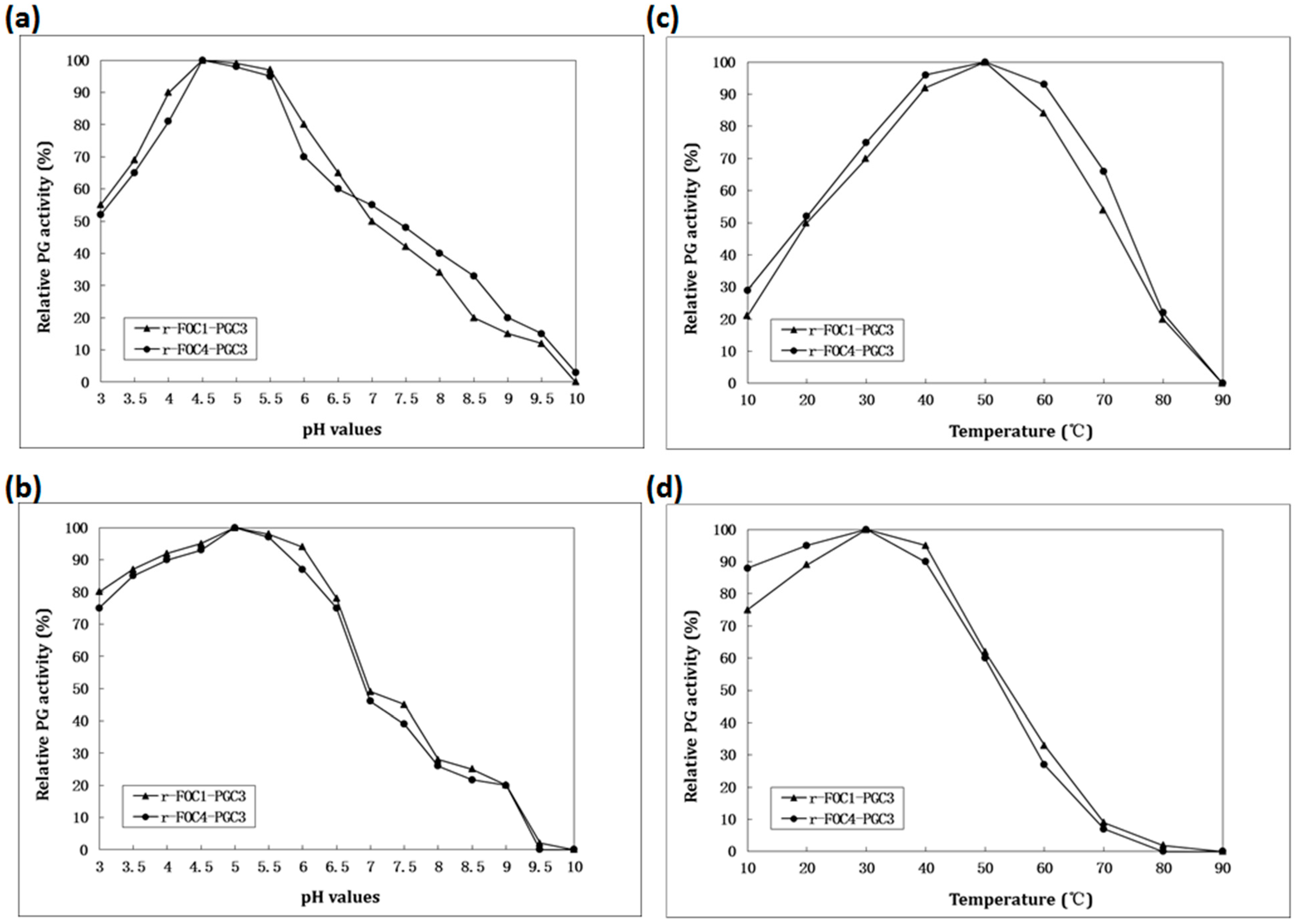
2.4. Biochemical Characterization of PGC3
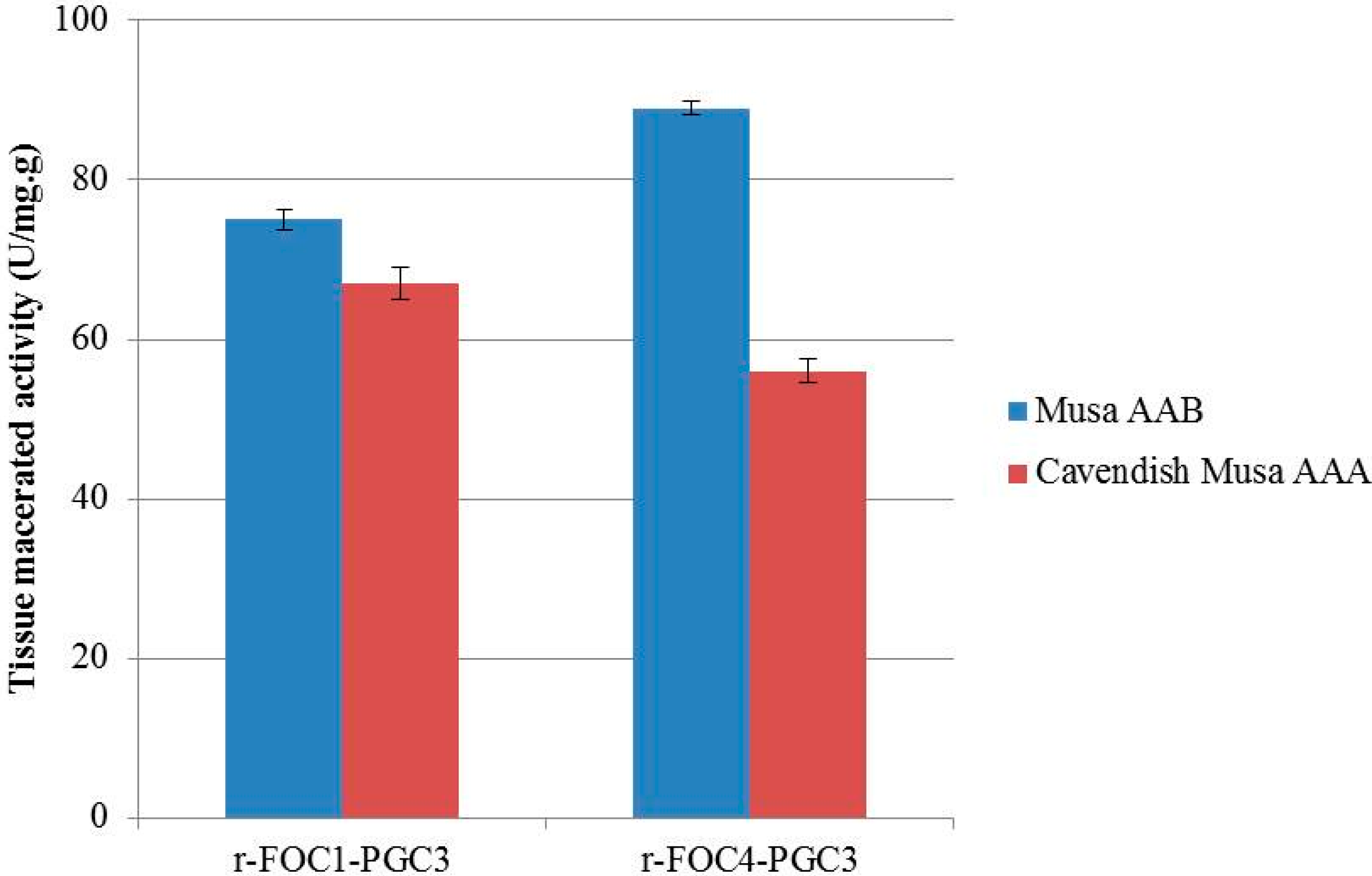
2.5. Active Recombinant PGC3 Causes Tissue Maceration and Necrosis

3. Experimental Section
3.1. Fungal Strains and Growth Condition
3.2. Enzyme Activity and Protein Measurements
3.3. Enzyme Purification
3.4. Enzyme Characterization
3.5. Cloning of Fungal pgc3 Genes
3.6. Expression and Purification of Recombinant Enzymes in P. pastoris
3.7. Tissue Maceration and Necrosis Assayed with Recombinant Enzymes
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Heslop-Harrison, J.S.; Schwarzacher, T. Domestication genomics and the future for banana. Ann. Bot-London 2007, 100, 1073–1084. [Google Scholar] [CrossRef]
- Cheesman, E.E. Pathology of the banana. Nature 1962, 194, 223. [Google Scholar]
- Ploetz, R.C. Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp cubense. Phytopathology 2006, 96, 653–656. [Google Scholar] [CrossRef]
- Butler, D. Fungus threatens top banana. Nature 2013, 504, 195–196. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D. Plant genomics: A bunch of trouble. Science 2008, 322, 1046–1047. [Google Scholar] [CrossRef] [PubMed]
- Walton, J.D. Deconstructing the cell-wall. Plant Physiol. 1994, 104, 1113–1118. [Google Scholar] [PubMed]
- De Lorenzo, G.; D’Ovidio, R.; Cervone, F. The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu. Rev. Phytopathol. 2001, 39, 313–335. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, V.; Francocci, F.; Ferrari, S.; Volpi, C.; Bellincampi, D.; Galletti, R.; D’Ovidio, R.; de Lorenzo, G.; Cervone, F. Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc. Natl. Acad. Sci. USA 2010, 107, 616–621. [Google Scholar] [CrossRef] [Green Version]
- Scott-Craig, J.S.; Cheng, Y.Q.; Cervone, F.; de Lorenzo, G.; Pitkin, J.W.; Walton, J.D. Targeted mutants of Cochliobolus carbonum lacking the two major extracellular polygalacturonases. Appl. Environ. Microbiol. 1998, 64, 1497–1503. [Google Scholar] [PubMed]
- Garcia-Maceira, F.I.; di Pietro, A.; Roncero, M.I. Cloning and disruption of pgx4 encoding an in planta expressed exopolygalacturonase from Fusarium oxysporum. Mol. Plant Microbe Interact. 2000, 13, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Favaron, F.; Alghisi, P.; Marciano, P.; Magro, P. Polygalacturonase isozymes and oxalic acid produced by Sclerotinia sclerotiorum in soybean hypocotyls as elicitors of glyceollin. Physiol. Mol. Plant Pathol. 1988, 33, 385–395. [Google Scholar] [CrossRef]
- Cervone, F.; de Lorenzo, G.; Pressev, R.; Darvill, A.G.; Albersheim, P. Can Phaseolus PGIP inhibit pectic enzymes from microbes and plants? Phytochemistry 1990, 29, 447–449. [Google Scholar] [CrossRef]
- Schacht, T.; Unger, C.; Pich, A.; Wydra, K. Endo- and exopolygalacturonases of Ralstonia solanacearum are inhibited by polygalacturonase-inhibiting protein (PGIP) activity in tomato stem extracts. Plant Physiol. Biochem. 2011, 49, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Y.; Wang, Z.Z. Isolation and characterization of an exopolygalacturonase from Fusarium oxysporum f. sp. cubense race 1 and race 4. BMC Biochem. 2011, 12, 51. [Google Scholar]
- Tomassini, A.; Sella, L.; Raiola, A.; Ovidio, R.D.; Favaron, F. Characterization and expression of Fusarium graminearum endo-polygalacturonases in vitro and during wheat infection. Plant Pathol. 2009, 58, 556–564. [Google Scholar] [CrossRef]
- Di Pietro, A.; Roncero, M.I.G. Endopolygalacturonase from Fusarium oxysporum f. sp. lycopersici: Purification, characterization, and production during infection of tomato plants. Phytopathology 1996, 86, 1324–1330. [Google Scholar]
- Di Pietro, A.; Roncero, M.I. Purification and characterization of an exo-polygalacturonase from the tomato vascular wilt pathogen Fusarium oxysporum f. sp. lycopersici. Phytopathology 1996, 145, 295–299. [Google Scholar]
- Maceira, G.; Pietro, I.F.; Roncero, M.I. Purification and characterization of a novel exopolygalacturonase from Fusarium oxysporum f. sp. lycopersici. FEMS Microbiol. Lett. 1997, 154, 37–43. [Google Scholar] [CrossRef]
- Yadav, S.; Anand, G.; Dubey, A.K.; Yadav, D. Purification and characterization of an exo-polygalacturonase secreted by Rhizopus oryzae MTCC 1987 and its role in retting of Crotalaria juncea fibre. Biologia 2012, 67, 1069–1074. [Google Scholar] [CrossRef]
- Maller, A.; da Silva, T.M.; de Lima Damasio, A.R.; Hirata, I.Y.; Jorge, J.A.; Terenzi, H.F.; de Moraes, M.D.L.T. Functional properties of a manganese-activated exo-polygalacturonase produced by a thermotolerant fungus Aspergillus niveus. Folia Microbiol. 2013, 58, 615–621. [Google Scholar] [CrossRef]
- Li, C.; Shao, J.; Wang, Y.; Li, W.; Guo, D.; Yan, B.; Xia, Y.; Peng, M. Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f. sp. cubense. BMC Genomics 2013, 14, 851. [Google Scholar] [CrossRef]
- Somogyi, M. Notes on sugar determination. J. Biol. Chem. 1952, 195, 19–23. [Google Scholar] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Z.; Wang, Z. Isolation and Heterologous Expression of a Polygalacturonase Produced by Fusarium oxysporum f. sp. cubense Race 1 and 4. Int. J. Mol. Sci. 2015, 16, 7595-7607. https://doi.org/10.3390/ijms16047595
Dong Z, Wang Z. Isolation and Heterologous Expression of a Polygalacturonase Produced by Fusarium oxysporum f. sp. cubense Race 1 and 4. International Journal of Molecular Sciences. 2015; 16(4):7595-7607. https://doi.org/10.3390/ijms16047595
Chicago/Turabian StyleDong, Zhangyong, and Zhenzhong Wang. 2015. "Isolation and Heterologous Expression of a Polygalacturonase Produced by Fusarium oxysporum f. sp. cubense Race 1 and 4" International Journal of Molecular Sciences 16, no. 4: 7595-7607. https://doi.org/10.3390/ijms16047595






