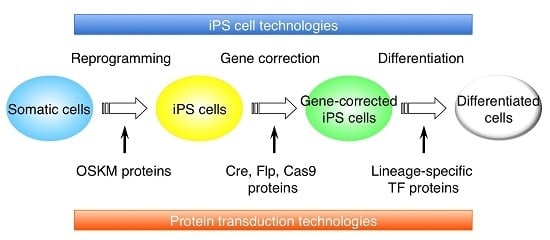
Cell-Penetrating Peptide as a Means of Directing the Differentiation of Induced-Pluripotent Stem Cells
Abstract
:1. Introduction
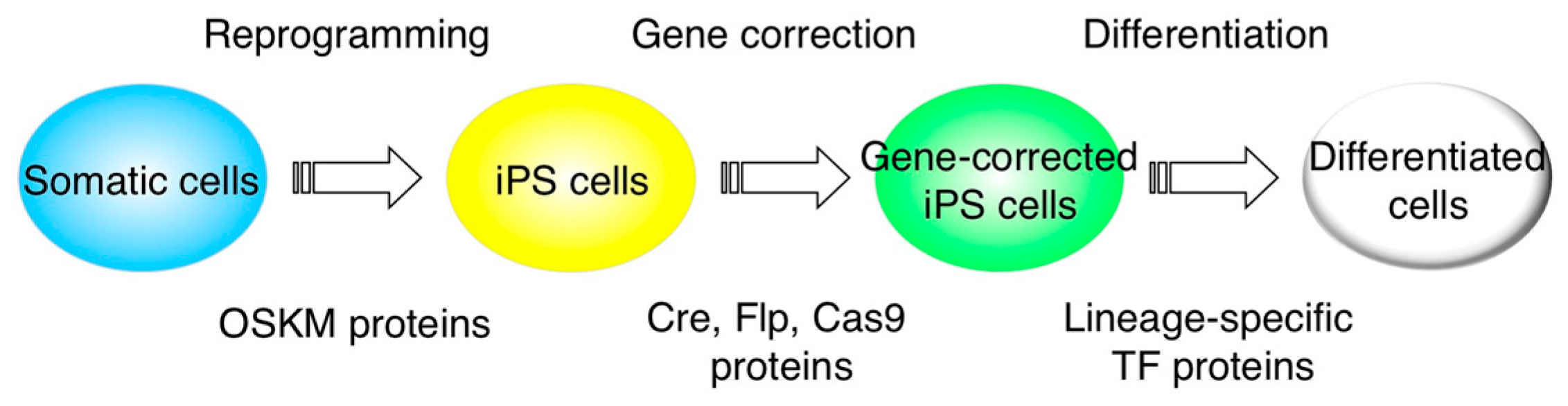
2. CPP-Mediated Protein Transduction
3. Protein Transduction into iPS Cells
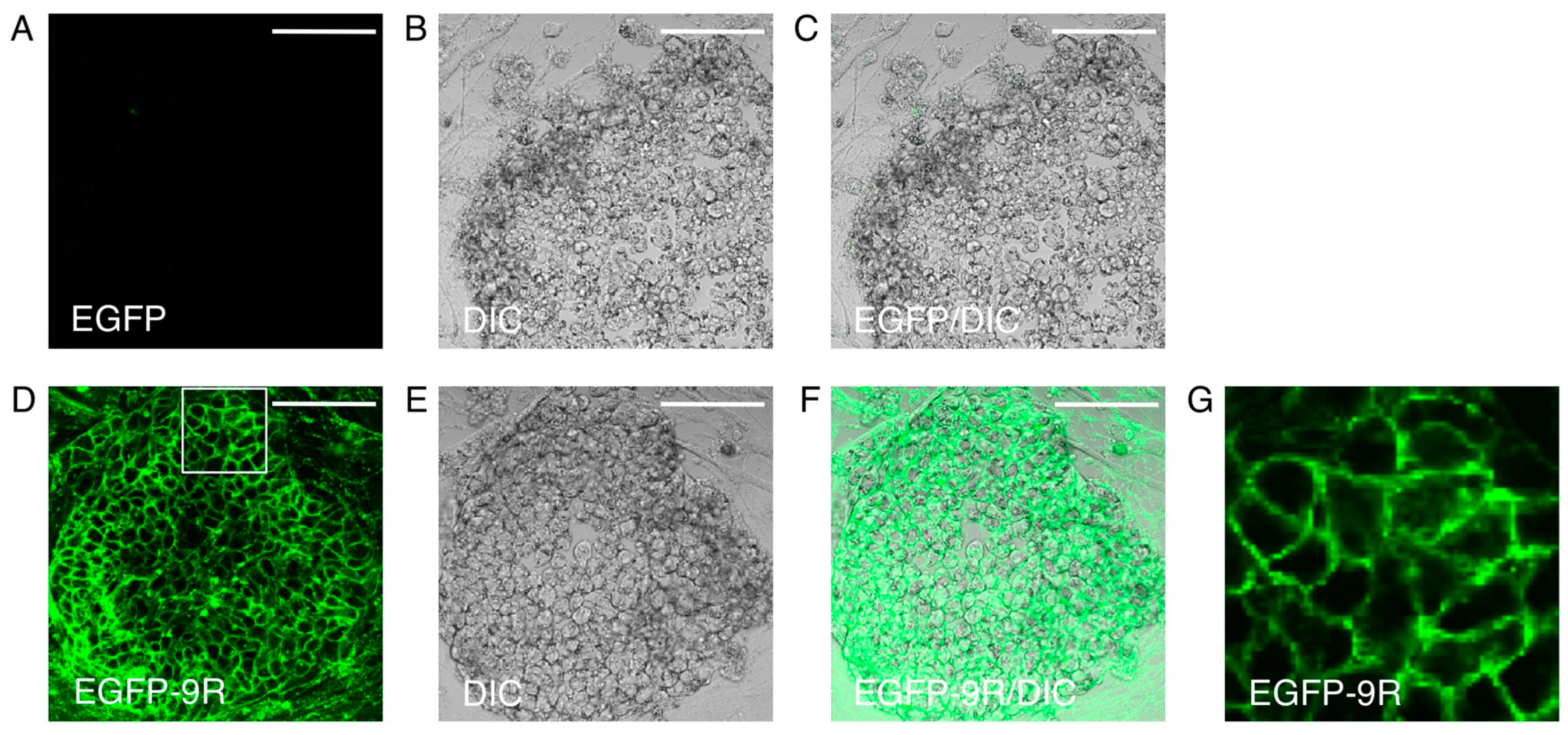
4. iPS Cell Differentiation with Protein Transduction of Specific Transcription Factor

5. Gene Editing with CPP-Mediated Protein Transduction
6. Usage of Protein Transduction in iPS Cell Generation or Direct Conversion
7. Conclusions
| CPPs | Proteins | Supplements | Technologies | Cell Types | References |
|---|---|---|---|---|---|
| Poly-arginine | OSKM | NA | Reprogramming | MEFs | [70] |
| Poly-arginine | OSKM | NA | Reprogramming | HNFs | [71] |
| NA | ES cell-derived extract proteins | Streptolysin O | Reprogramming | Mouse cardiac fibroblasts | [72] |
| Hydrophobic MTDs | OSKMN or OSKML | NA | Partial reprogramming | HDFs | [73] |
| TAT | Cre | NA | Recombination | Mouse ES cells | [60] |
| TAT | Cre | NA | Recombination | Human ES cells | [61] |
| TAT | FLP | dTAT-HA2 peptide | Recombination | Mouse or human ES cells | [62] |
| Poly-arginine | Cas9 and sgRNA | NA | Gene disruption | Human ES cells | [63] |
| NA | Cre or Cas9 | Hypertonic solution and NDSB-201 | Gene editing | Mouse or human ES cells | [22] |
| NA | Cre, TALE or Cas9 | Anionic proteins and cationic lipids | Gene editing | Mouse ES cells | [23] |
| PTDs or Poly-arginine | Pdx1, NeuroD and MafA | NA | Pancreatic differentiation | Mouse ES cells or human iPS cells | [45] |
| TAT | Nkx2.2 | NA | Neural differentiation | Mouse NSCs | [55] |
| TAT | Pax6 | NA | Neural differentiation | Rat NSCs | [57] |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, A.; Dowdy, S.F. Protein transduction domain delivery of therapeutic macromolecules. Curr. Opin. Biotechnol. 2011, 22, 888–893. [Google Scholar] [CrossRef] [PubMed]
- De Duve, C. The origin of eukaryotes: A reappraisal. Nat. Rev. Genet. 2007, 8, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Nakase, I.; Niwa, M.; Takeuchi, T.; Sonomura, K.; Kawabata, N.; Koike, Y.; Takehashi, M.; Tanaka, S.; Ueda, K.; Simpson, J.C.; et al. Cellular uptake of arginine-rich peptides: Roles for macropinocytosis and actin rearrangement. Mol. Ther. 2004, 10, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, I.M.; Wadia, J.S.; Dowdy, S.F. Cationic TAT peptide transduction domain enters cells by macropinocytosis. J. Control. Release 2005, 102, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Swanson, J.A. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 2008, 9, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Green, M.; Loewenstein, P.M. Autonomous functional domains of chemically synthesized human immunodeficiency virus TAT trans-activator protein. Cell 1988, 55, 1179–1188. [Google Scholar] [CrossRef]
- Joliot, A.; Pernelle, C.; Deagostini-Bazin, H.; Prochiantz, A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 1864–1868. [Google Scholar] [CrossRef] [PubMed]
- Futaki, S.; Ohashi, W.; Suzuki, T.; Niwa, M.; Tanaka, S.; Ueda, K.; Harashima, H.; Sugiura, Y. Stearylated arginine-rich peptides: A new class of transfection systems. Bioconjug. Chem. 2001, 12, 1005–1011. [Google Scholar] [CrossRef] [PubMed]
- Matsushita, M.; Tomizawa, K.; Moriwaki, A.; Li, S.T.; Terada, H.; Matsui, H. A high-efficiency protein transduction system demonstrating the role of PKA in long-lasting long-term potentiation. J. Neurosci. 2001, 21, 6000–6007. [Google Scholar] [PubMed]
- Hitsuda, T.; Michiue, H.; Kitamatsu, M.; Fujimura, A.; Wang, F.; Yamamoto, T.; Han, X.J.; Tazawa, H.; Uneda, A.; Ohmori, I.; et al. A protein transduction method using oligo-arginine (3R) for the delivery of transcription factors into cell nuclei. Biomaterials 2012, 33, 4665–4672. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; Rusnati, M.; Presta, M.; Giacca, M. Internalization of HIV-1 TAT requires cell surface heparan sulfate proteoglycans. J. Biol. Chem. 2001, 276, 3254–3261. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Futaki, S.; Niwa, M.; Tanaka, S.; Ueda, K.; Sugiura, Y. Possible existence of common internalization mechanisms among arginine-rich peptides. J. Biol. Chem. 2002, 277, 2437–2443. [Google Scholar] [CrossRef] [PubMed]
- Wadia, J.S.; Stan, R.V.; Dowdy, S.F. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 2004, 10, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Imamura, J.; Suzuki, Y.; Gonda, K.; Roy, C.N.; Gatanaga, H.; Ohuchi, N.; Higuchi, H. Single particle tracking confirms that multivalent Tat protein transduction domain-induced heparan sulfate proteoglycan cross-linkage activates Rac1 for internalization. J. Biol. Chem. 2011, 286, 10581–10592. [Google Scholar] [CrossRef] [PubMed]
- Gump, J.M.; June, R.K.; Dowdy, S.F. Revised role of glycosaminoglycans in TAT protein transduction domain-mediated cellular transduction. J. Biol. Chem. 2010, 285, 1500–1507. [Google Scholar] [CrossRef] [PubMed]
- Hirose, H.; Takeuchi, T.; Osakada, H.; Pujals, S.; Katayama, S.; Nakase, I.; Kobayashi, S.; Haraguchi, T.; Futaki, S. Transient focal membrane deformation induced by arginine-rich peptides leads to their direct penetration into cells. Mol. Ther. 2012, 20, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Katayama, S.; Nakase, I.; Yano, Y.; Murayama, T.; Nakata, Y.; Matsuzaki, K.; Futaki, S. Effects of pyrenebutyrate on the translocation of arginine-rich cell-penetrating peptides through artificial membranes: Recruiting peptides to the membranes, dissipating liquid-ordered phases, and inducing curvature. Biochim. Biophys. Acta 2013, 1828, 2134–2142. [Google Scholar] [CrossRef] [PubMed]
- Doherty, G.J.; McMahon, H.T. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009, 78, 857–902. [Google Scholar] [CrossRef] [PubMed]
- Tsumuraya, T.; Matsushita, M. COPA and SLC4A4 are required for cellular entry of arginine-rich peptides. PLoS ONE 2014, 9, e86639. [Google Scholar] [CrossRef] [PubMed]
- D’Astolfo, D.S.; Pagliero, R.J.; Pras, A.; Karthaus, W.R.; Clevers, H.; Prasad, V.; Lebbink, R.J.; Rehmann, H.; Geijsen, N. Efficient intracellular delivery of native proteins. Cell 2015, 161, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Zuris, J.A.; Thompson, D.B.; Shu, Y.; Guilinger, J.P.; Bessen, J.L.; Hu, J.H.; Maeder, M.L.; Joung, J.K.; Chen, Z.Y.; Liu, D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Waehler, R.; Russell, S.J.; Curiel, D.T. Engineering targeted viral vectors for gene therapy. Nat. Rev. Genet. 2007, 8, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Kaji, K.; Norrby, K.; Paca, A.; Mileikovsky, M.; Mohseni, P.; Woltjen, K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 2009, 458, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Woltjen, K.; Michael, I.P.; Mohseni, P.; Desai, R.; Mileikovsky, M.; Hämäläinen, R.; Cowling, R.; Wang, W.; Liu, P.; Gertsenstein, M.; et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 2009, 458, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Wu, X.; Li, G.; Han, M.; Zhuang, Y.; Xu, T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 2005, 122, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C. The piggyBac transposon holds promise for human gene therapy. Proc. Natl. Acad. Sci. USA 2006, 103, 14981–14982. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, M.; Yamashita, Y.; Kase, M.; Trifonov, S.; Sugimoto, T. Lineage-specific purification of neural stem/progenitor cells from differentiated mouse induced pluripotent stem cells. Stem Cells Transl. Med. 2013, 2, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Kaitsuka, T.; Tomizawa, K. Generation of functional insulin-producing cells from human pluripotent stem cells via protein transduction methods. Manuscript in preparation. 2015. [Google Scholar]
- Sun, C.; Velazquez, M.A.; Marfy-Smith, S.; Sheth, B.; Cox, A.; Johnston, D.A.; Smyth, N.; Fleming, T.P. Mouse early extra-embryonic lineages activate compensatory endocytosis in response to poor maternal nutrition. Development 2014, 141, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J. Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006, 7, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Dobrowolski, R.; de Robertis, E.M. Endocytic control of growth factor signalling: Multivesicular bodies as signalling organelles. Nat. Rev. Mol. Cell Biol. 2011, 13, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C. The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Wörsdörfer, P.; Maxeiner, S.; Markopoulos, C.; Kirfel, G.; Wulf, V.; Auth, T.; Urschel, S.; von Maltzahn, J.; Willecke, K. Connexin expression and functional analysis of gap junctional communication in mouse embryonic stem cells. Stem Cells 2008, 26, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Knaus, U.G.; Wang, Y.; Reilly, A.M.; Warnock, D.; Jackson, J.H. Structural requirements for PAK activation by Rac GTPases. J. Biol. Chem. 1998, 273, 21512–21518. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.N.; Han, H.J. Laminin regulates mouse embryonic stem cell migration: Involvement of Epac1/Rap1 and Rac1/cdc42. Am. J. Physiol. Cell Physiol. 2010, 298, C1159–C1169. [Google Scholar] [CrossRef] [PubMed]
- Bar-Sagi, D.; Feramisco, J.R. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science 1986, 233, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Commisso, C.; Davidson, S.M.; Soydaner-Azeloglu, R.G.; Parker, S.J.; Kamphorst, J.J.; Hackett, S.; Grabocka, E.; Nofal, M.; Drebin, J.A.; Thompson, C.B.; et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 2013, 497, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Mitsui, K.; Yamanaka, S. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature 2003, 423, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Sasai, Y. Next-generation regenerative medicine: Organogenesis from stem cells in 3D culture. Cell Stem Cell 2013, 12, 520–530. [Google Scholar] [CrossRef] [PubMed]
- Kamao, H.; Mandai, M.; Okamoto, S.; Sakai, N.; Suga, A.; Sugita, S.; Kiryu, J.; Takahashi, M. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Rep. 2014, 2, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, S.; Ito, I.; Nagasaki, T.; Yamamoto, Y.; Konishi, S.; Korogi, Y.; Matsumoto, H.; Muro, S.; Hirai, T.; Funato, M.; et al. Generation of alveolar epithelial spheroids via isolated progenitor cells from human pluripotent stem cells. Stem Cell Rep. 2014, 3, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Pagliuca, F.W.; Millman, J.R.; Gürtler, M.; Segel, M.; van Dervort, A.; Ryu, J.H.; Peterson, Q.P.; Greiner, D.; Melton, D.A. Generation of functional human pancreatic beta cells in vitro. Cell 2014, 159, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Kaitsuka, T.; Noguchi, H.; Shiraki, N.; Kubo, T.; Wei, F.Y.; Hakim, F.; Kume, S.; Tomizawa, K. Generation of functional insulin-producing cells from mouse embryonic stem cells through 804G cell-derived extracellular matrix and protein transduction of transcription factors. Stem Cells Transl. Med. 2014, 3, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Kaneto, H.; Weir, G.C.; Bonner-Weir, S. PDX-1 protein containing its own antennapedia-like protein transduction domain can transduce pancreatic duct and islet cells. Diabetes 2003, 52, 1732–1737. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, H.; Bonner-Weir, S.; Wei, F.Y.; Matsushita, M.; Matsumoto, S. BETA2/NeuroD protein can be transduced into cells due to an arginine- and lysine-rich sequence. Diabetes 2005, 54, 2859–2866. [Google Scholar] [CrossRef] [PubMed]
- Shahjalal, H.M.; Shiraki, N.; Sakano, D.; Kikawa, K.; Ogaki, S.; Baba, H.; Kume, K.; Kume, S. Generation of insulin-producing beta-like cells from human iPS cells in a defined and completely xeno-free culture system. J. Mol. Cell Biol. 2014, 6, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Rezania, A.; Bruin, J.E.; Arora, P.; Rubin, A.; Batushansky, I.; Asadi, A.; O’Dwyer, S.; Quiskamp, N.; Mojibian, M.; Albrecht, T.; et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 2014, 32, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pak, C.; Han, Y.; Ahlenius, H.; Zhang, Z.; Chanda, S.; Marro, S.; Patzke, C.; Acuna, C.; Covy, J.; et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 2013, 78, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Antonica, F.; Kasprzyk, D.F.; Opitz, R.; Iacovino, M.; Liao, X.H.; Dumitrescu, A.M.; Refetoff, S.; Peremans, K.; Manto, M.; Kyba, M.; et al. Generation of functional thyroid from embryonic stem cells. Nature 2012, 491, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.S.; Shi, X.; Toyama, A.; Arpke, R.W.; Dandapat, A.; Iacovino, M.; Kang, J.; Le, G.; Hagen, H.R.; Garry, D.J.; et al. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell 2013, 12, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Woltjen, K.; Miyake, K.; Hotta, A.; Ikeya, M.; Yamamoto, T.; Nishino, T.; Shoji, E.; Sehara-Fujisawa, A.; Manabe, Y.; et al. Efficient and reproducible myogenic differentiation from human iPS cells: Prospects for modeling Miyoshi Myopathy in vitro. PLoS ONE 2013, 8, e61540. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Tsang, K.S.; Chan, J.C.; Yuan, P.; Fan, R.; Kaneto, H.; Xu, G. The combined expression of Pdx1 and MafA with either Ngn3 or NeuroD improves the differentiation efficiency of mouse embryonic stem cells into insulin-producing cells. Cell Transplant. 2013, 22, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Stock, K.; Nolden, L.; Edenhofer, F.; Quandel, T.; Brüstle, O. Transcription factor-based modulation of neural stem cell differentiation using direct protein transduction. Cell. Mol. Life Sci. 2010, 67, 2439–2449. [Google Scholar] [CrossRef] [PubMed]
- Mie, M.; Kaneko, M.; Henmi, F.; Kobatake, E. Induction of motor neuron differentiation by transduction of Olig2 protein. Biochem. Biophys. Res. Commun. 2012, 427, 531–536. [Google Scholar] [CrossRef] [PubMed]
- Spitere, K.; Toulouse, A.; O’Sullivan, D.B.; Sullivan, A.M. TAT-PAX6 protein transduction in neural progenitor cells: A novel approach for generation of dopaminergic neurones in vitro. Brain Res. 2008, 1208, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Hidema, S.; Tonomura, Y.; Date, S.; Nishimori, K. Effects of protein transduction with intact myogenic transcription factors tagged with HIV-1 Tat-PTD (T-PTD) on myogenic differentiation of mouse primary cells. J. Biosci. Bioeng. 2012, 113, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Noda, T.; Fujino, T.; Mie, M.; Kobatake, E. Transduction of MyoD protein into myoblasts induces myogenic differentiation without addition of protein transduction domain. Biochem. Biophys. Res. Commun. 2009, 382, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Peitz, M.; Pfannkuche, K.; Rajewsky, K.; Edenhofer, F. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: A tool for efficient genetic engineering of mammalian genomes. Proc. Natl. Acad. Sci. USA 2002, 99, 4489–4494. [Google Scholar] [CrossRef] [PubMed]
- Nolden, L.; Edenhofer, F.; Haupt, S.; Koch, P.; Wunderlich, F.T.; Siemen, H.; Brüstle, O. Site-specific recombination in human embryonic stem cells induced by cell-permeant Cre recombinase. Nat. Methods 2006, 3, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Patsch, C.; Peitz, M.; Otte, D.M.; Kesseler, D.; Jungverdorben, J.; Wunderlich, F.T.; Brüstle, O.; Zimmer, A.; Edenhofer, F. Engineering cell-permeant FLP recombinase for tightly controlled inducible and reversible overexpression in embryonic stem cells. Stem Cells 2010, 28, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, S.; Kwaku Dad, A.B.; Beloor, J.; Gopalappa, R.; Lee, S.K.; Kim, H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014, 24, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, A.; Meade, B.R.; Chang, Y.C.; Fredrickson, C.T.; Willert, K.; Puri, N.; Dowdy, S.F. Efficient siRNA delivery into primary cells by a peptide transduction domain-dsRNA binding domain fusion protein. Nat. Biotechnol. 2009, 27, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Wernig, M.; Meissner, A.; Foreman, R.; Brambrink, T.; Ku, M.; Hochedlinger, K.; Bernstein, B.E.; Jaenisch, R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 2007, 448, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Kadari, A.; Lu, M.; Li, M.; Sekaran, T.; Thummer, R.P.; Guyette, N.; Chu, V.; Edenhofer, F. Excision of viral reprogramming cassettes by Cre protein transduction enables rapid, robust and efficient derivation of transgene-free human induced pluripotent stem cells. Stem Cell Res. Ther. 2014, 5, 47. [Google Scholar] [CrossRef] [PubMed]
- Redmer, T.; Diecke, S.; Grigoryan, T.; Quiroga-Negreira, A.; Birchmeier, W.; Besser, D. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011, 12, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Gao, W.Q. Concise Reviews: Patient-Derived Stem Cell Research for Monogenic Disorders. Stem Cells 2015. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Nakano, T.; Hotta, A. Genetic correction using engineered nucleases for gene therapy applications. Dev. Growth Differ. 2014, 56, 63–77. [Google Scholar] [PubMed]
- Zhou, H.; Wu, S.; Joo, J.Y.; Zhu, S.; Han, D.W.; Lin, T.; Trauger, S.; Bien, G.; Yao, S.; Zhu, Y.; et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 2009, 4, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, C.H.; Moon, J.I.; Chung, Y.G.; Chang, M.Y.; Han, B.S.; Ko, S.; Yang, E.; Cha, K.Y.; Lanza, R.; et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 2009, 4, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Lee, C.S.; Kwon, Y.W.; Paek, J.S.; Lee, S.H.; Hur, J.; Lee, E.J.; Roh, T.Y.; Chu, I.S.; Leem, S.H.; et al. Induction of pluripotent stem cells from adult somatic cells by protein-based reprogramming without genetic manipulation. Blood 2010, 116, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Kim, J.; Kang, J.; Jo, D. Partial somatic to stem cell transformations induced by cell-permeable reprogramming factors. Sci. Rep. 2014, 4, 4361. [Google Scholar] [CrossRef] [PubMed]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; He, Z.; Ji, S.; Sun, H.; Xiang, D.; Liu, C.; Hu, Y.; Wang, X.; Hui, L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 2011, 475, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, S.; Suzuki, A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 2011, 475, 390–393. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Zuchero, J.B.; Ahlenius, H.; Marro, S.; Ng, Y.H.; Vierbuchen, T.; Hawkins, J.S.; Geissler, R.; Barres, B.A.; Wernig, M. Generation of oligodendroglial cells by direct lineage conversion. Nat. Biotechnol. 2013, 31, 434–439. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaitsuka, T.; Tomizawa, K. Cell-Penetrating Peptide as a Means of Directing the Differentiation of Induced-Pluripotent Stem Cells. Int. J. Mol. Sci. 2015, 16, 26667-26676. https://doi.org/10.3390/ijms161125986
Kaitsuka T, Tomizawa K. Cell-Penetrating Peptide as a Means of Directing the Differentiation of Induced-Pluripotent Stem Cells. International Journal of Molecular Sciences. 2015; 16(11):26667-26676. https://doi.org/10.3390/ijms161125986
Chicago/Turabian StyleKaitsuka, Taku, and Kazuhito Tomizawa. 2015. "Cell-Penetrating Peptide as a Means of Directing the Differentiation of Induced-Pluripotent Stem Cells" International Journal of Molecular Sciences 16, no. 11: 26667-26676. https://doi.org/10.3390/ijms161125986
APA StyleKaitsuka, T., & Tomizawa, K. (2015). Cell-Penetrating Peptide as a Means of Directing the Differentiation of Induced-Pluripotent Stem Cells. International Journal of Molecular Sciences, 16(11), 26667-26676. https://doi.org/10.3390/ijms161125986