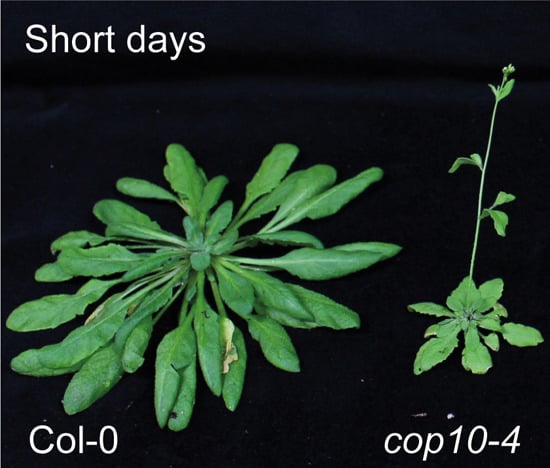
CONSTITUTIVE PHOTOMORPHOGENIC 10 (COP10) Contributes to Floral Repression under Non-Inductive Short Days in Arabidopsis
Abstract
:1. Introduction
2. Results and Discussion
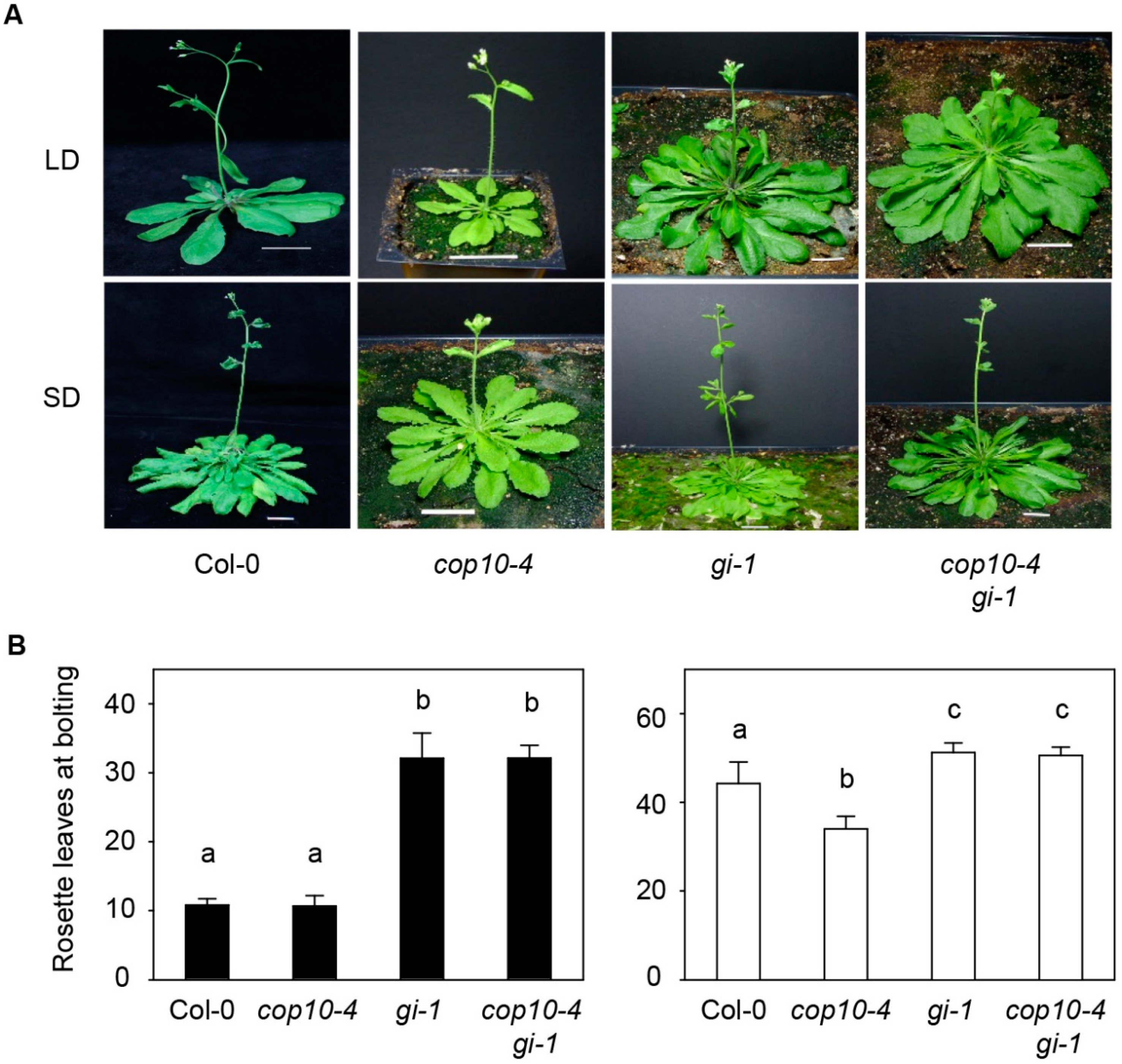
2.1. GIGANTEA (gi-1) Is Epistatic to cop10-4 in the Photoperiodic Pathway of Flowering
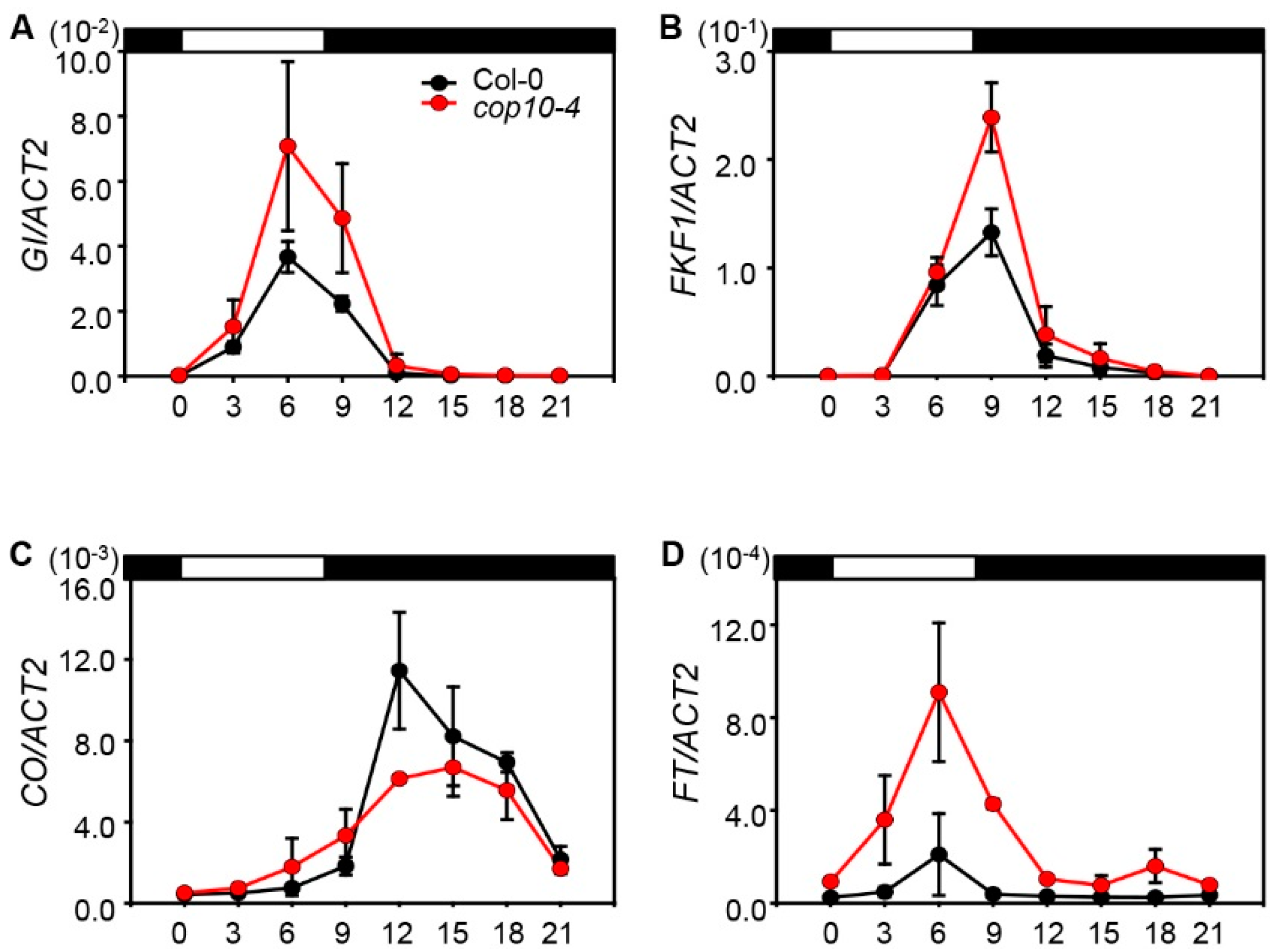
2.2. The cop10 Mutation Alters the Expression of Flowering Time Genes
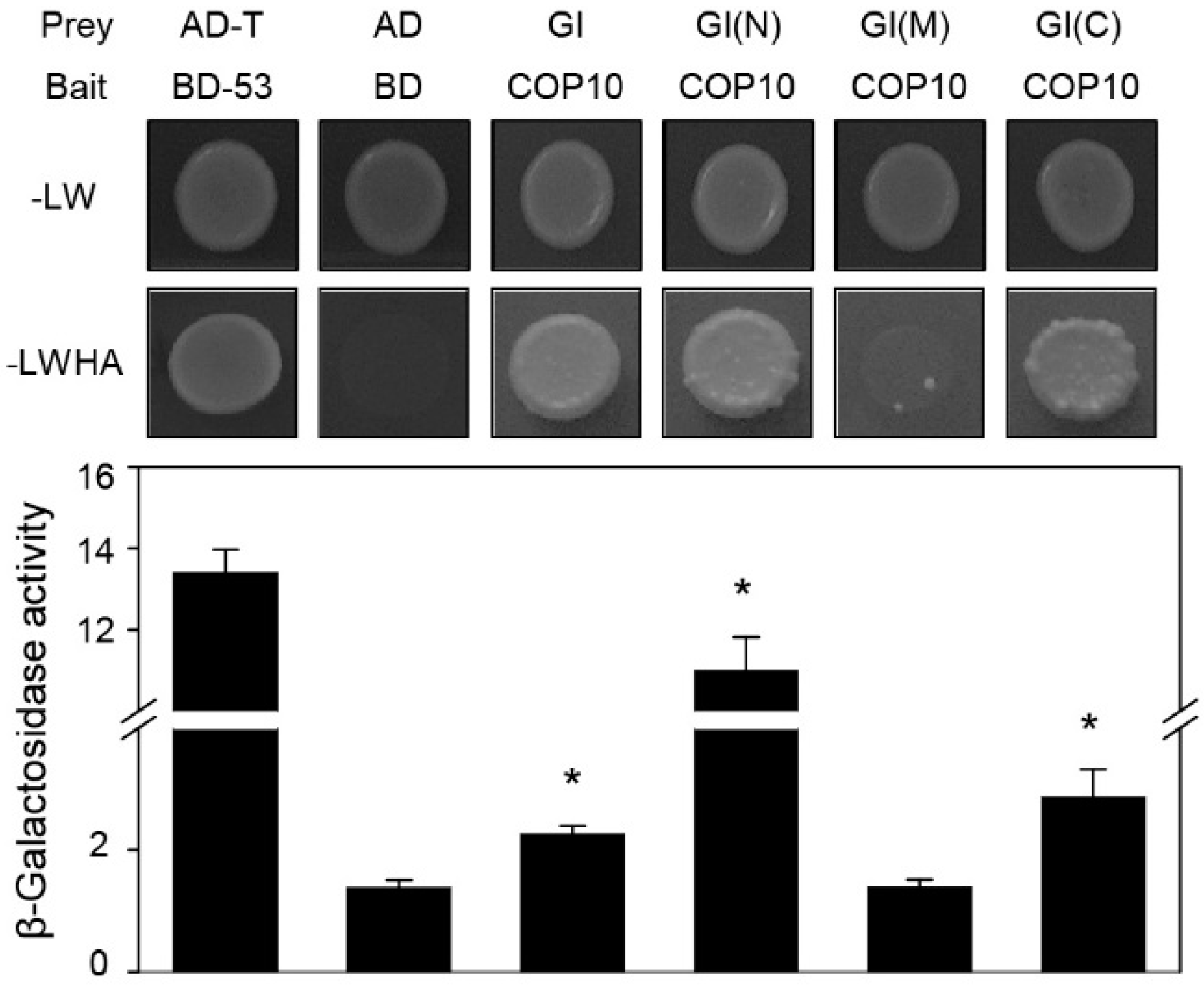
2.3. COP10 Interacts with GI

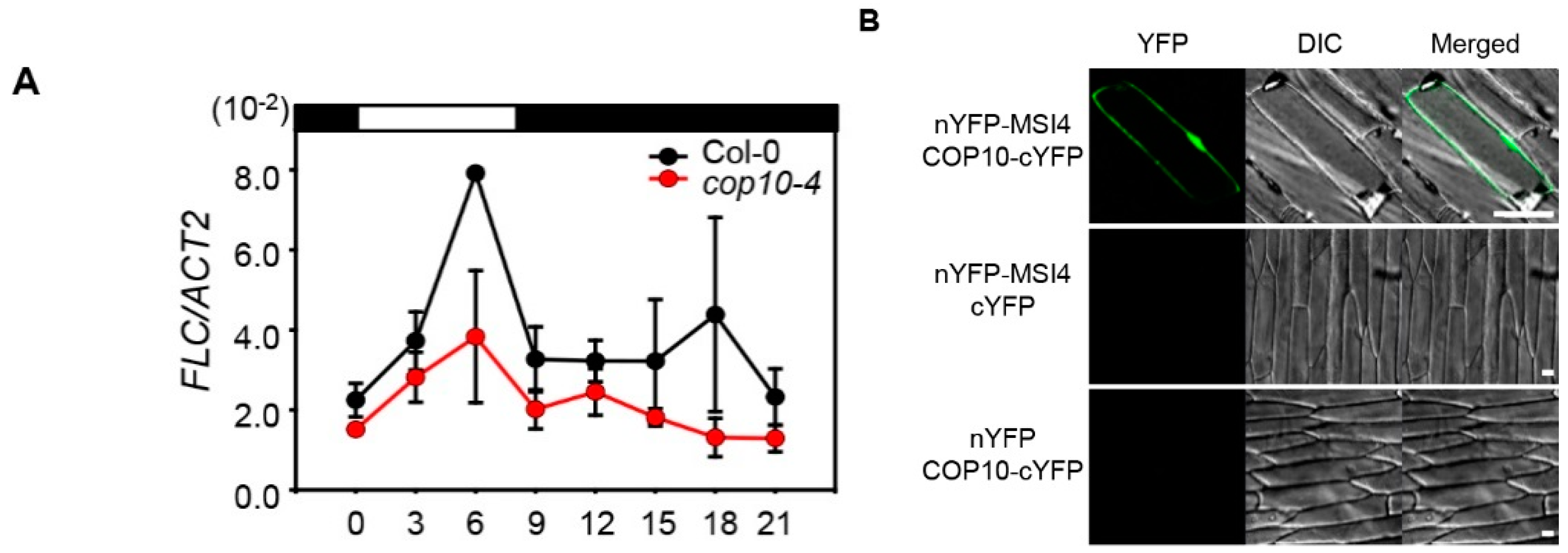
2.4. COP10 Regulates FLOWERING LOCUS C (FLC) Expression through Interaction with MULTICOPY SUPPRESSOR OF IRA1 4 (MSI4) in the Autonomous Pathway

3. Experimental Section
3.1. Plant Materials and Growth Conditions
3.2. Analysis of Flowering Time
3.3. RNA Preparation, Reverse Transcription, and Quantitative Real-Time PCR (qPCR) Analysis
3.4. Yeast Two-Hybrid Assays
3.5. In Vivo Pull-Down Assays
3.6. Bimolecular Fluorescence Complementation Assays in Onion Epidermal Cells
3.7. Accession Numbers
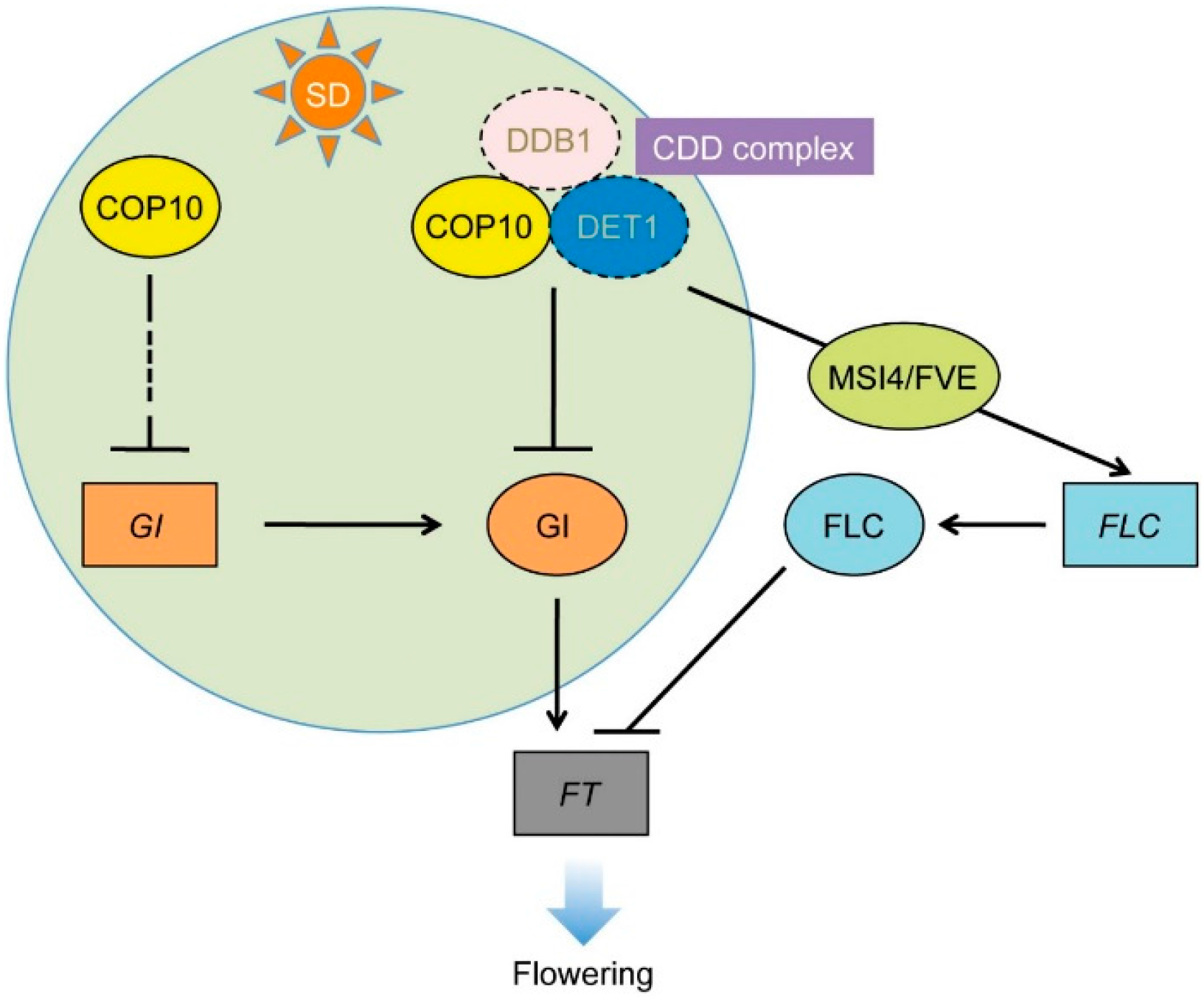
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Golembeski, G.S.; Imaizumi, T. Photoperiodic regulation of florigen function in Arabidopsis thaliana. Arabidopsis Book 2015, 13, e0178. [Google Scholar] [CrossRef] [PubMed]
- Kardailsky, I.; Shukla, V.K.; Ahn, J.H.; Dagenais, N.; Christensen, S.K.; Nguyen, J.T.; Chory, J.; Harrison, M.J.; Weigel, D. Activation tagging of the floral inducer FT. Science 1999, 286, 1962–1965. [Google Scholar] [CrossRef] [PubMed]
- Park, D.H.; Somers, D.E.; Kim, Y.S.; Choy, Y.H.; Lim, H.K.; Soh, M.S.; Kim, H.J.; Kay, S.A.; Nam, H.G. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 1999, 285, 1579–1582. [Google Scholar] [CrossRef] [PubMed]
- Suárez-López, P.; Wheatley, K.; Robson, F.; Onouchi, H.; Valverde, F.; Coupland, G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 2001, 410, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Valverde, F.; Mouradov, A.; Soppe, W.; Ravenscroft, D.; Samach, A.; Coupland, G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Marchal, V.; Panigrahi, K.C.; Wenkel, S.; Soppe, W.; Deng, X.W.; Valverde, F.; Coupland, G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008, 27, 1277–1288. [Google Scholar] [CrossRef] [PubMed]
- Yanovsky, M.J.; Kay, S.A. Molecular basis of seasonal time measurement in Arabidopsis. Nature 2002, 419, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.J.; Zhang, Y.C.; Li, Q.H.; Sang, Y.; Mao, J.; Lian, H.L.; Wang, L.; Yang, H.Q. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 2008, 20, 292–306. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi, T.; Schultz, T.F.; Harmon, F.G.; Ho, L.A.; Kay, S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 2005, 309, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Kay, S.A.; Imaizumi, T. Photoperiodic flowering occurs under internal and external coincidence. Plant Signal. Behav. 2008, 3, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Kröber, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Castillejo, C.; Pelaz, S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 2008, 18, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Hepworth, S.R.; Valverde, F.; Ravenscroft, D.; Mouradov, A.; Coupland, G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 2002, 21, 4327–4337. [Google Scholar] [CrossRef] [PubMed]
- He, Y. Chromatin regulation of flowering. Trends Plant Sci. 2012, 17, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Jiang, D.; Yang, W.; Jacob, Y.; Michaels, S.D.; He, Y. Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet. 2011, 7, e1002366. [Google Scholar] [CrossRef] [PubMed]
- Pazhouhandeh, M.; Molinier, J.; Berr, A.; Genschik, P. MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 3430–3435. [Google Scholar] [CrossRef] [PubMed]
- Chory, J.; Peto, C.; Feinbaum, R.; Pratt, L.; Ausubel, F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 1989, 58, 991–999. [Google Scholar] [CrossRef]
- Kang, M.Y.; Yoo, S.C.; Kwon, H.Y.; Lee, B.D.; Cho, J.N.; Noh, Y.S.; Paek, N.C. Negative regulatory roles of DE-ETIOLATED1 in flowering time in Arabidopsis. Sci. Rep. 2015, 5, 9728. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.W.; Rubio, V.; Lee, N.Y.; Bai, S.; Lee, S.Y.; Kim, S.S.; Liu, L.; Zhang, Y.; Irigoyen, M.L.; Sullivan, J.A.; et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 2008, 32, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Sullivan, J.A.; Wang, H.; Yang, J.; Shen, Y.; Rubio, V.; Ma, L.; Hoecker, U.; Deng, X.W. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003, 17, 2642–2647. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, G.; Yanagawa, Y.; Kwok, S.F.; Matsui, M.; Deng, X.W. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 2002, 16, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Shen, Y.; Tang, X.; Yu, L.; Wang, J.; Guo, L.; Zhang, Y.; Zhang, H.; Feng, S.; Strickland, E.; et al. Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 2006, 18, 1991–2004. [Google Scholar] [CrossRef] [PubMed]
- Lau, O.S.; Huang, X.; Charron, J.B.; Lee, J.H.; Li, G.; Deng, X.W. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell 2011, 43, 703–712. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R. The photomorphogenic protein, DE-ETIOLATED 1, is a critical transcriptional corepressor in the central loop of the Arabidopsis circadian clock. Mol. Cell 2011, 43, 693–694. [Google Scholar] [CrossRef] [PubMed]
- Castle, L.A.; Meinke, D.W. A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell 1994, 6, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Wei, N.; Kwok, S.F.; von Arnim, A.G.; Lee, A.; McNellis, T.W.; Piekos, B.; Deng, X.W. Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 1994, 6, 629–643. [Google Scholar] [CrossRef] [PubMed]
- Kwok, S.F.; Piekos, B.; Misera, S.; Deng, X.W. A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 1996, 110, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.P.; Schuerman, P.; Woeste, K.; Brandstatter, I.; Kieber, J.J. Isolation and characterization of Arabidopsis mutants defective in the induction of ethylene biosynthesis by cytokinin. Genetics 1998, 149, 417–427. [Google Scholar] [PubMed]
- Yanagawa, Y.; Sullivan, J.A.; Komatsu, S.; Gusmaroli, G.; Suzuki, G.; Yin, J.; Ishibashi, T.; Saijo, Y.; Rubio, V.; Kimura, S.; et al. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 2004, 18, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, T.; Wright, L.; Fujiwara, S.; Cremer, F.; Lee, K.; Onouchi, H.; Mouradov, A.; Fowler, S.; Kamada, H.; Putterill, J.; et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 2005, 17, 2255–2270. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Kay, S.A. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 11698–11703. [Google Scholar] [CrossRef] [PubMed]
- Simpson, G.G. The autonomous pathway: Epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Curr. Opin. Plant Boil. 2004, 7, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Ausin, I.; Alonso-Blanco, C.; Jarillo, J.A.; Ruiz-Garcia, L.; Martinez-Zapater, J.M. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 2004, 36, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Benvenuto, G.; Formiggini, F.; Laflamme, P.; Malakhov, M.; Bowler, C. The photomorphogenesis regulator DET1 binds the amino-terminal tail of histone H2B in a nucleosome context. Curr. Biol. 2002, 12, 1529–1534. [Google Scholar] [CrossRef]
- Schroeder, D.F.; Gahrtz, M.; Maxwell, B.B.; Cook, R.K.; Kan, J.M.; Alonso, J.M.; Ecker, J.R.; Chory, J. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 2002, 12, 1462–1472. [Google Scholar] [CrossRef]
- Jarillo, J.A.; Piñeiro, M.; Cubas, P.; Martinez-Zapater, J.M. Chromatin remodeling in plant development. Int. J. Dev. Biol. 2009, 53, 1581–1596. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lim, J.; Yeom, M.; Kim, H.; Kim, J.; Wang, L.; Kim, W.Y.; Somers, D.E.; Nam, H.G. ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Rep. 2013, 3, 671–677. [Google Scholar] [CrossRef] [PubMed]
- James, P.; Halladay, J.; Craig, E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1996, 144, 1425–1436. [Google Scholar] [PubMed]
- Gietz, D.; St Jean, A.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef]
- Kim, W.Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Qiang, Z.; et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 1352. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Kim, D.; Kim, Y.S.; Hörtensteiner, S.; Paek, N.C. Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett. 2014, 588, 3830–3837. [Google Scholar] [CrossRef] [PubMed]
- Citovsky, V.; Lee, L.Y.; Vyas, S.; Glick, E.; Chen, M.H.; Vainstein, A.; Gafni, Y.; Gelvin, S.B.; Tzfira, T. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 2006, 362, 1120–1131. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, M.-Y.; Kwon, H.-Y.; Kim, N.-Y.; Sakuraba, Y.; Paek, N.-C. CONSTITUTIVE PHOTOMORPHOGENIC 10 (COP10) Contributes to Floral Repression under Non-Inductive Short Days in Arabidopsis. Int. J. Mol. Sci. 2015, 16, 26493-26505. https://doi.org/10.3390/ijms161125969
Kang M-Y, Kwon H-Y, Kim N-Y, Sakuraba Y, Paek N-C. CONSTITUTIVE PHOTOMORPHOGENIC 10 (COP10) Contributes to Floral Repression under Non-Inductive Short Days in Arabidopsis. International Journal of Molecular Sciences. 2015; 16(11):26493-26505. https://doi.org/10.3390/ijms161125969
Chicago/Turabian StyleKang, Min-Young, Hye-Young Kwon, Na-Yun Kim, Yasuhito Sakuraba, and Nam-Chon Paek. 2015. "CONSTITUTIVE PHOTOMORPHOGENIC 10 (COP10) Contributes to Floral Repression under Non-Inductive Short Days in Arabidopsis" International Journal of Molecular Sciences 16, no. 11: 26493-26505. https://doi.org/10.3390/ijms161125969
APA StyleKang, M.-Y., Kwon, H.-Y., Kim, N.-Y., Sakuraba, Y., & Paek, N.-C. (2015). CONSTITUTIVE PHOTOMORPHOGENIC 10 (COP10) Contributes to Floral Repression under Non-Inductive Short Days in Arabidopsis. International Journal of Molecular Sciences, 16(11), 26493-26505. https://doi.org/10.3390/ijms161125969