Exploiting CRISPR/Cas: Interference Mechanisms and Applications
Abstract
:1. Introduction
2. Three Strategies to Cope with Viruses
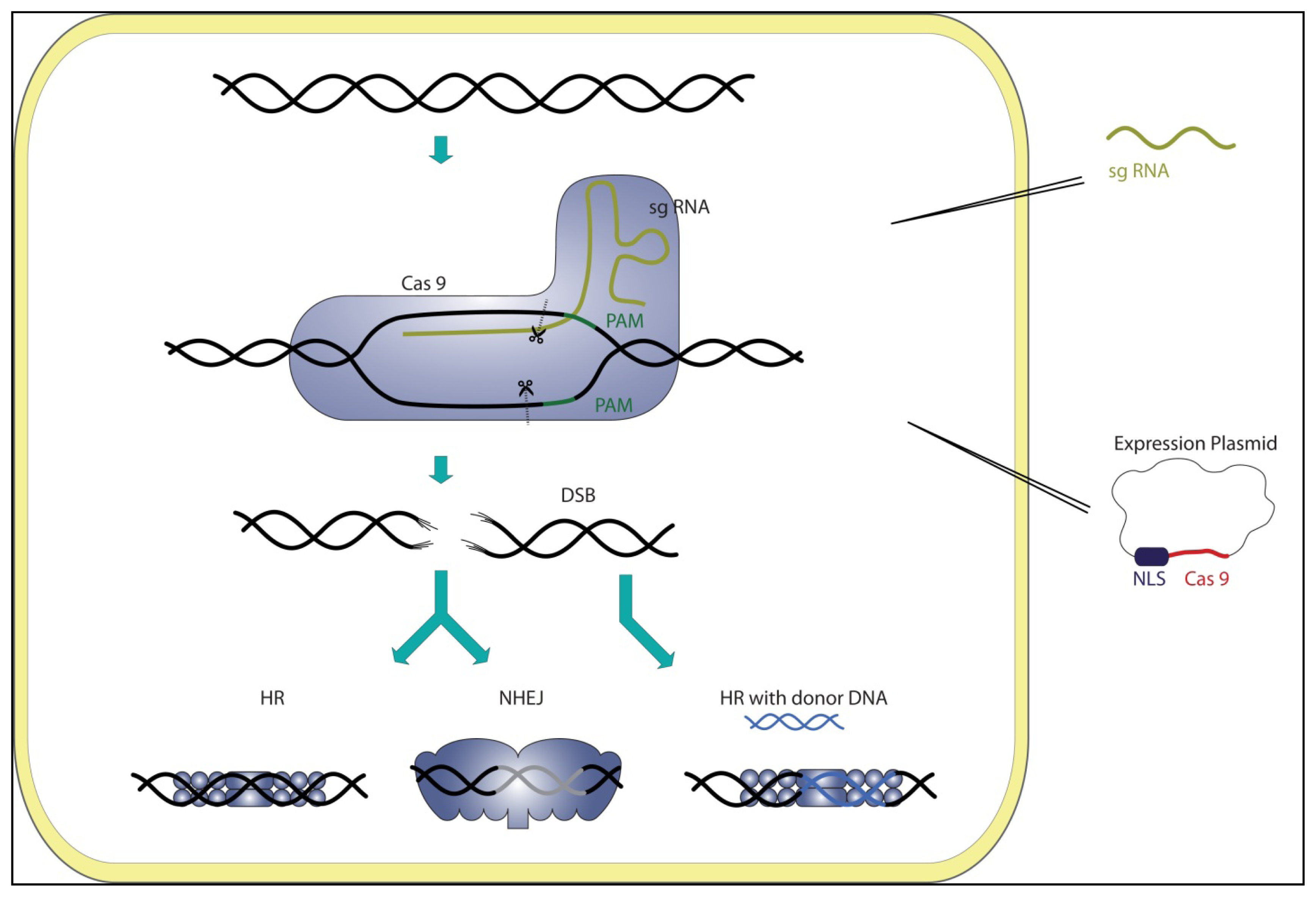
3. CRISPR Systems as Genome Editing Tools
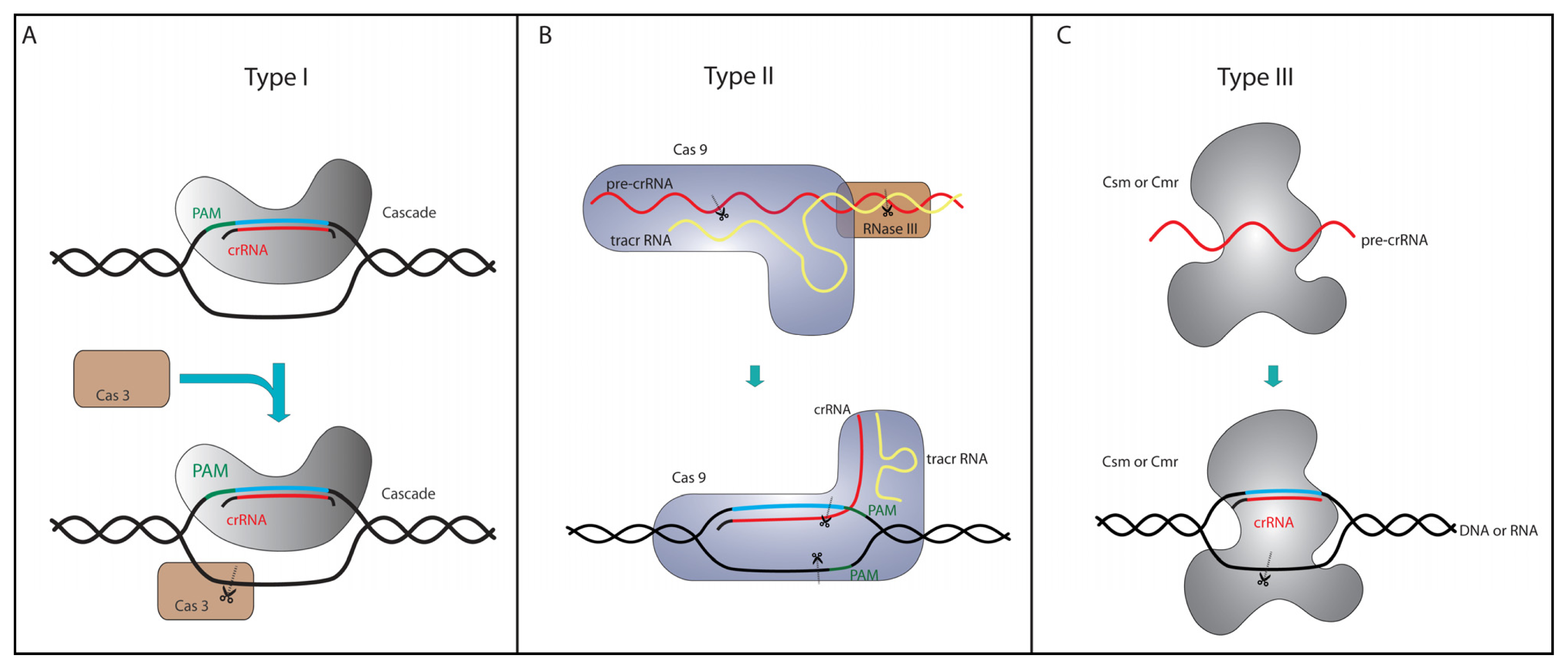
4. Type I and Type III Genome Editing Tools
5. Genome Editing Tools: RGEN, TALEN, ZFN
6. Outlook
Acknowledgments
Conflict of Interest
References
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 2007, 35, W52–W57. [Google Scholar]
- Mojica, F.J.; Diez-Villasenor, C.; Garcia-Martinez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol 2005, 60, 174–182. [Google Scholar]
- Pourcel, C.; Salvignol, G.; Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. [Google Scholar]
- Mojica, F.J.; Diez-Villasenor, C.; Garcia-Martinez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. [Google Scholar]
- Swarts, D.C.; Mosterd, C.; van Passel, M.W.; Brouns, S.J. CRISPR interference directs strand specific spacer acquisition. PLoS One 2012, 7, e35888. [Google Scholar]
- Pul, U.; Wurm, R.; Arslan, Z.; Geissen, R.; Hofmann, N.; Wagner, R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol 2010, 75, 1495–1512. [Google Scholar]
- Yosef, I.; Goren, M.G.; Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 2012, 40, 5569–5576. [Google Scholar]
- Babu, M.; Beloglazova, N.; Flick, R.; Graham, C.; Skarina, T.; Nocek, B.; Gagarinova, A.; Pogoutse, O.; Brown, G.; Binkowski, A.; et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol 2011, 79, 484–502. [Google Scholar]
- Diez-Villasenor, C.; Guzman, N.M.; Almendros, C.; Garcia-Martinez, J.; Mojica, F.J. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol. 2013, 10. [Google Scholar] [CrossRef] [Green Version]
- Hale, C.R.; Majumdar, S.; Elmore, J.; Pfister, N.; Compton, M.; Olson, S.; Resch, A.M.; Glover, C.V., III; Graveley, B.R.; Terns, R.M.; Terns, M.P. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell 2012, 45, 292–302. [Google Scholar]
- Lillestol, R.K.; Shah, S.A.; Brugger, K.; Redder, P.; Phan, H.; Christiansen, J.; Garrett, R.A. CRISPR families of the crenarchaeal genus Sulfolobus: Bidirectional transcription and dynamic properties. Mol. Microbiol 2009, 72, 259–272. [Google Scholar]
- Carte, J.; Wang, R.; Li, H.; Terns, R.M.; Terns, M.P. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 2008, 22, 3489–3496. [Google Scholar]
- Gesner, E.M.; Schellenberg, M.J.; Garside, E.L.; George, M.M.; Macmillan, A.M. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat. Struct. Mol. Biol 2011, 18, 688–692. [Google Scholar]
- Haurwitz, R.E.; Jinek, M.; Wiedenheft, B.; Zhou, K.; Doudna, J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 2010, 329, 1355–1358. [Google Scholar]
- Richter, H.; Zoephel, J.; Schermuly, J.; Maticzka, D.; Backofen, R.; Randau, L. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res 2012, 40, 9887–9896. [Google Scholar]
- Sashital, D.G.; Jinek, M.; Doudna, J.A. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat. Struct. Mol. Biol 2011, 18, 680–687. [Google Scholar]
- Jore, M.M.; Lundgren, M.; van Duijn, E.; Bultema, J.B.; Westra, E.R.; Waghmare, S.P.; Wiedenheft, B.; Pul, U.; Wurm, R.; Wagner, R.; et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol 2011, 18, 529–536. [Google Scholar]
- Brouns, S.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.; Snijders, A.P.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008, 321, 960–964. [Google Scholar]
- Wiedenheft, B.; Lander, G.C.; Zhou, K.; Jore, M.M.; Brouns, S.J.; van der Oost, J.; Doudna, J.A.; Nogales, E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 2011, 477, 486–489. [Google Scholar]
- Wiedenheft, B.; van Duijn, E.; Bultema, J.B.; Waghmare, S.P.; Zhou, K.; Barendregt, A.; Westphal, W.; Heck, A.J.; Boekema, E.J.; Dickman, M.J.; Doudna, J.A. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 10092–10097. [Google Scholar]
- Bondy-Denomy, J.; Pawluk, A.; Maxwell, K.L.; Davidson, A.R. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 2013, 493, 429–432. [Google Scholar]
- Haft, D.H.; Selengut, J.; Mongodin, E.F.; Nelson, K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol 2005, 1, e60. [Google Scholar]
- Makarova, K.S.; Aravind, L.; Wolf, Y.I.; Koonin, E.V. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct 2011, 6. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol 2011, 9, 467–477. [Google Scholar]
- Beloglazova, N.; Petit, P.; Flick, R.; Brown, G.; Savchenko, A.; Yakunin, A.F. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J 2011, 30, 4616–4627. [Google Scholar]
- Mulepati, S.; Bailey, S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3). J. Biol. Chem 2011, 286, 31896–31903. [Google Scholar]
- Sinkunas, T.; Gasiunas, G.; Fremaux, C.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 2011, 30, 1335–1342. [Google Scholar]
- Westra, E.R.; Nilges, B.; van Erp, P.B.; van der Oost, J.; Dame, R.T.; Brouns, S.J. Cascade-mediated binding and bending of negatively supercoiled DNA. RNA Biol 2012, 9, 1134–1138. [Google Scholar]
- Sinkunas, T.; Gasiunas, G.; Waghmare, S.P.; Dickman, M.J.; Barrangou, R.; Horvath, P.; Siksnys, V. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J 2013, 32, 385–394. [Google Scholar]
- Semenova, E.; Jore, M.M.; Datsenko, K.A.; Semenova, A.; Westra, E.R.; Wanner, B.; van der Oost, J.; Brouns, S.J.; Severinov, K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. USA 2011, 108, 10098–10103. [Google Scholar]
- Marraffini, L.A.; Sontheimer, E.J. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 2010, 463, 568–571. [Google Scholar]
- Sashital, D.G.; Wiedenheft, B.; Doudna, J.A. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell 2012, 46, 606–615. [Google Scholar]
- Nam, K.H.; Huang, Q.; Ke, A. Nucleic acid binding surface and dimer interface revealed by CRISPR-associated CasB protein structures. FEBS Lett 2012, 586, 3956–3961. [Google Scholar]
- Mulepati, S.; Orr, A.; Bailey, S. Crystal structure of the largest subunit of a bacterial RNA-guided immune complex and its role in DNA target binding. J. Biol. Chem 2012, 287, 22445–22449. [Google Scholar]
- Lintner, N.G.; Kerou, M.; Brumfield, S.K.; Graham, S.; Liu, H.; Naismith, J.H.; Sdano, M.; Peng, N.; She, Q.; Copie, V.; et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J. Biol. Chem 2011, 286, 21643–21656. [Google Scholar]
- Plagens, A.; Tjaden, B.; Hagemann, A.; Randau, L.; Hensel, R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J. Bacteriol 2012, 194, 2491–2500. [Google Scholar]
- Fischer, S.; Maier, L.K.; Stoll, B.; Brendel, J.; Fischer, E.; Pfeiffer, F.; Dyall-Smith, M.; Marchfelder, A. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J. Biol. Chem 2012, 287, 33351–33363. [Google Scholar]
- Peng, W.; Li, H.; Hallstrom, S.; Peng, N.; Liang, Y.X.; She, Q. Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol. 2013, 10. [Google Scholar] [CrossRef]
- Sapranauskas, R.; Gasiunas, G.; Fremaux, C.; Barrangou, R.; Horvath, P.; Siksnys, V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 2011, 39, 9275–9582. [Google Scholar]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar]
- Chylinski, K.; Le Rhun, A.; Charpentier, E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013, 10. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar]
- Garneau, J.E.; Dupuis, M.E.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadan, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 2008, 322, 1843–1845. [Google Scholar]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genetics 2010, 11, 181–190. [Google Scholar]
- Zhang, J.; Rouillon, C.; Kerou, M.; Reeks, J.; Brugger, K.; Graham, S.; Reimann, J.; Cannone, G.; Liu, H.; Albers, S.V.; et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell 2012, 45, 303–313. [Google Scholar]
- Shao, Y.; Cocozaki, A.I.; Ramia, N.F.; Terns, R.M.; Terns, M.P.; Li, H. Structure of the Cmr2–Cmr3 subcomplex of the Cmr RNA silencing complex. Structure 2013, 21, 376–384. [Google Scholar]
- Osawa, T.; Inanaga, H.; Numata, T. Crystal structure of the Cmr2-Cmr3 subcomplex in the CRISPR-Cas RNA silencing effector complex. J. Mol. Biol. 2013, in press. [Google Scholar]
- Hale, C.R.; Zhao, P.; Olson, S.; Duff, M.O.; Graveley, B.R.; Wells, L.; Terns, R.M.; Terns, M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 2009, 139, 945–956. [Google Scholar]
- Deng, L.; Garrett, R.A.; Shah, S.A.; Peng, X.; She, Q. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol 2013, 87, 1088–1099. [Google Scholar]
- Vergnaud, G.; Li, Y.; Gorge, O.; Cui, Y.; Song, Y.; Zhou, D.; Grissa, I.; Dentovskaya, S.V.; Platonov, M.E.; Rakin, A.; et al. Analysis of the three Yersinia pestis CRISPR loci provides new tools for phylogenetic studies and possibly for the investigation of ancient DNA. Adv. Exp. Med. Biol 2007, 603, 327–338. [Google Scholar]
- Dale, J.W.; Brittain, D.; Cataldi, A.A.; Cousins, D.; Crawford, J.T.; Driscoll, J.; Heersma, H.; Lillebaek, T.; Quitugua, T.; Rastogi, N.; et al. Spacer oligonucleotide typing of bacteria of the Mycobacterium tuberculosis complex: Recommendations for standardised nomenclature. Int. J. Tuberc. Lung Dis 2001, 5, 216–219. [Google Scholar]
- Qi, L.; Haurwitz, R.E.; Shao, W.; Doudna, J.A.; Arkin, A.P. RNA processing enables predictable programming of gene expression. Nat. Biotechnol 2012, 30, 1002–1006. [Google Scholar]
- Lee, H.Y.; Haurwitz, R.E.; Apffel, A.; Zhou, K.; Smart, B.; Wenger, C.D.; Laderman, S.; Bruhn, L.; Doudna, J.A. RNA-protein analysis using a conditional CRISPR nuclease. Proc. Natl. Acad. Sci. USA 2013, 110, 5416–5421. [Google Scholar]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; Zhang, F. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol 2013, 31, 230–232. [Google Scholar]
- Chang, N.; Sun, C.; Gao, L.; Zhu, D.; Xu, X.; Zhu, X.; Xiong, J.W.; Xi, J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 2013, 23, 465–472. [Google Scholar]
- Dicarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 2013, 41, 4336–4343. [Google Scholar]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol 2013, 31, 227–229. [Google Scholar]
- Jiang, W.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol 2013, 31, 233–239. [Google Scholar]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. ELife 2013, 2, e00471. [Google Scholar]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar]
- Gratz, S.J.; Cummings, A.M.; Nguyen, J.N.; Hamm, D.C.; Donohue, L.K.; Harrison, M.M.; Wildonger, J.; O’Connor-Giles, K.M. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 2013. [Google Scholar] [CrossRef]
- Segal, D.J.; Meckler, J.F. Genome engineering at the dawn of the golden age. Ann. Rev. Genomics Hum. Genet. 2013. [Google Scholar] [CrossRef]
- Qiu, P.; Shandilya, H.; D’Alessio, J.M.; O’Connor, K.; Durocher, J.; Gerard, G.F. Mutation detection using Surveyor nuclease. BioTechniques 2004, 36, 702–707. [Google Scholar]
- Horvath, P.; Barrangou, R. RNA-guided genome editing a la carte. Cell Res. 2013. [Google Scholar] [CrossRef]
- Kim, Y.; Kweon, J.; Kim, A.; Chon, J.K.; Yoo, J.Y.; Kim, H.J.; Kim, S.; Lee, C.; Jeong, E.; Chung, E.; et al. A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol 2013, 31, 251–258. [Google Scholar]
- Ramalingam, S.; Annaluru, N.; Chandrasegaran, S. A CRISPR way to engineer the human genome. Genome Biol. 2013, 14. [Google Scholar] [CrossRef]
- Mercer, A.C.; Gaj, T.; Fuller, R.P.; Barbas, C.F., III. Chimeric TALE recombinases with programmable DNA sequence specificity. Nucleic Acids Res. 2012, 40, 11163–11172. [Google Scholar]
- Gaj, T.; Mercer, A.C.; Gersbach, C.A.; Gordley, R.M.; Barbas, C.F., III. Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proc. Natl. Acad. Sci. USA 2011, 108, 498–503. [Google Scholar]
- Yant, S.R.; Huang, Y.; Akache, B.; Kay, M.A. Site-directed transposon integration in human cells. Nucleic Acids Res 2007, 35, e50. [Google Scholar]
- Xiao, A.; Wang, Z.; Hu, Y.; Wu, Y.; Luo, Z.; Yang, Z.; Zu, Y.; Li, W.; Huang, P.; Tong, X.; et al. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 2013, in press. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Richter, H.; Randau, L.; Plagens, A. Exploiting CRISPR/Cas: Interference Mechanisms and Applications. Int. J. Mol. Sci. 2013, 14, 14518-14531. https://doi.org/10.3390/ijms140714518
Richter H, Randau L, Plagens A. Exploiting CRISPR/Cas: Interference Mechanisms and Applications. International Journal of Molecular Sciences. 2013; 14(7):14518-14531. https://doi.org/10.3390/ijms140714518
Chicago/Turabian StyleRichter, Hagen, Lennart Randau, and André Plagens. 2013. "Exploiting CRISPR/Cas: Interference Mechanisms and Applications" International Journal of Molecular Sciences 14, no. 7: 14518-14531. https://doi.org/10.3390/ijms140714518



