Genes Responsive to Low-Intensity Pulsed Ultrasound in MC3T3-E1 Preosteoblast Cells
Abstract
:1. Introduction
2. Results
2.1. Effects of LIPUS on the Cell Growth and Alkaline Phosphatase (ALP) Activity in MC3T3-E1 Cells
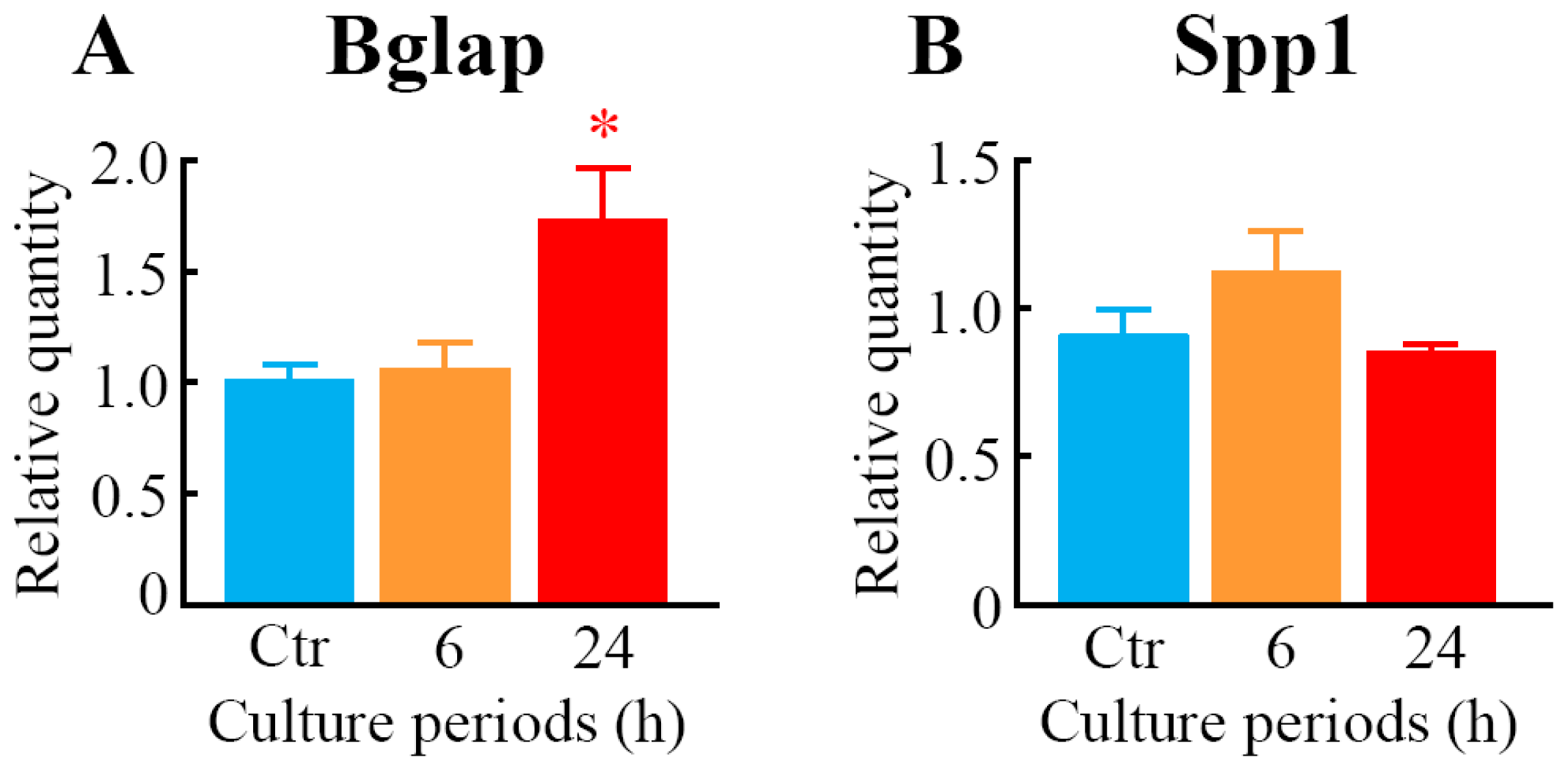
2.2. Effects of LIPUS on the Expression Level of mRNAs for Osteoblast Differentiation Marker Proteins
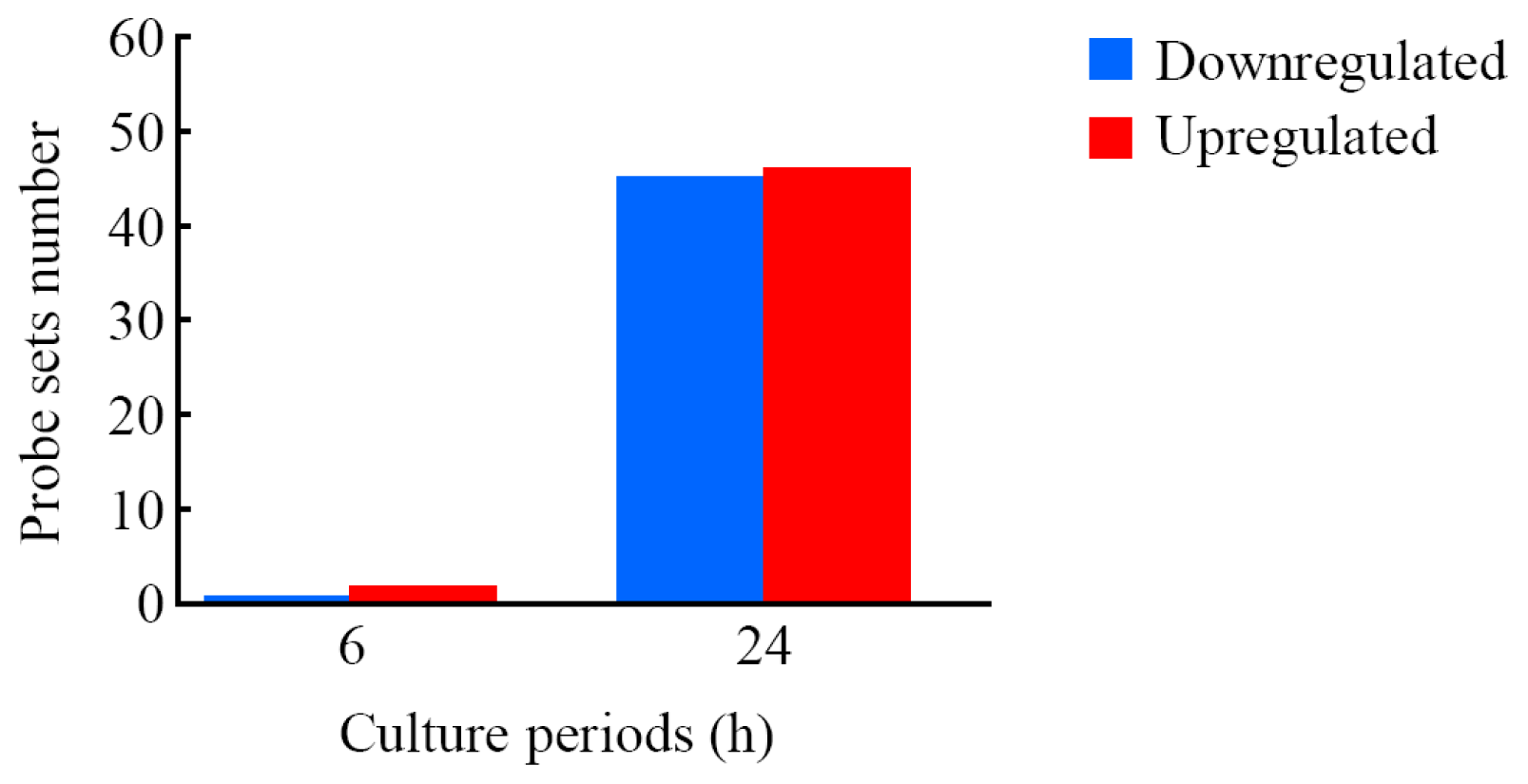
2.3. Genes Responsive to LIPUS
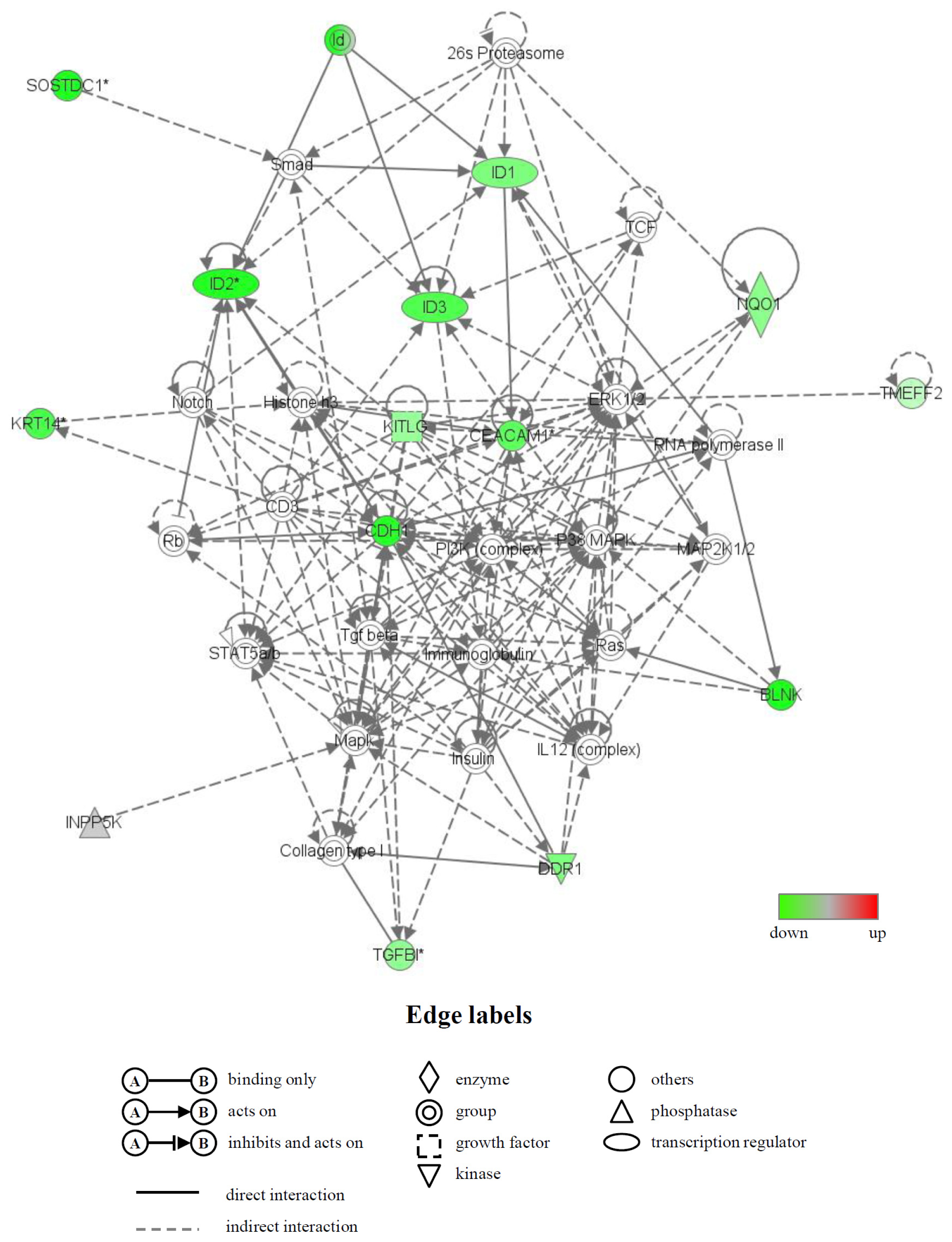
2.4. Identification of Biological Functions and Gene Networks
2.5. Quantitative Analysis of Differentially Expressed Genes
3. Discussion
4. Experimental Section
4.1. Cell Culture
4.2. LIPUS Treatment
4.3. Measurement of Cell Number
4.4. Measurement of ALP Activity
4.5. RNA Isolation
4.6. High-Density Oligonucleotide Microarray and Computational Gene Expression Analyses
4.7. Real-Time Quantitative PCR Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Information
ijms-14-22721-s001.pdf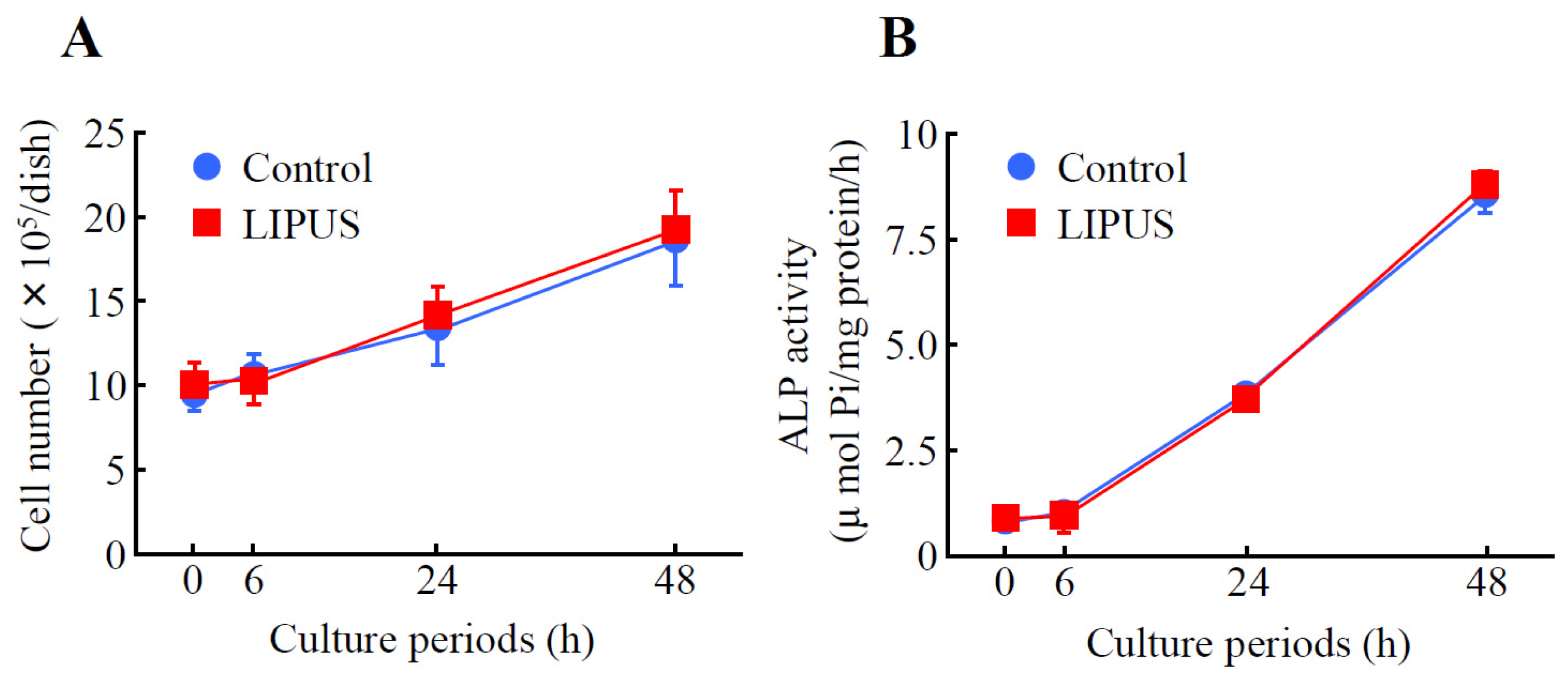

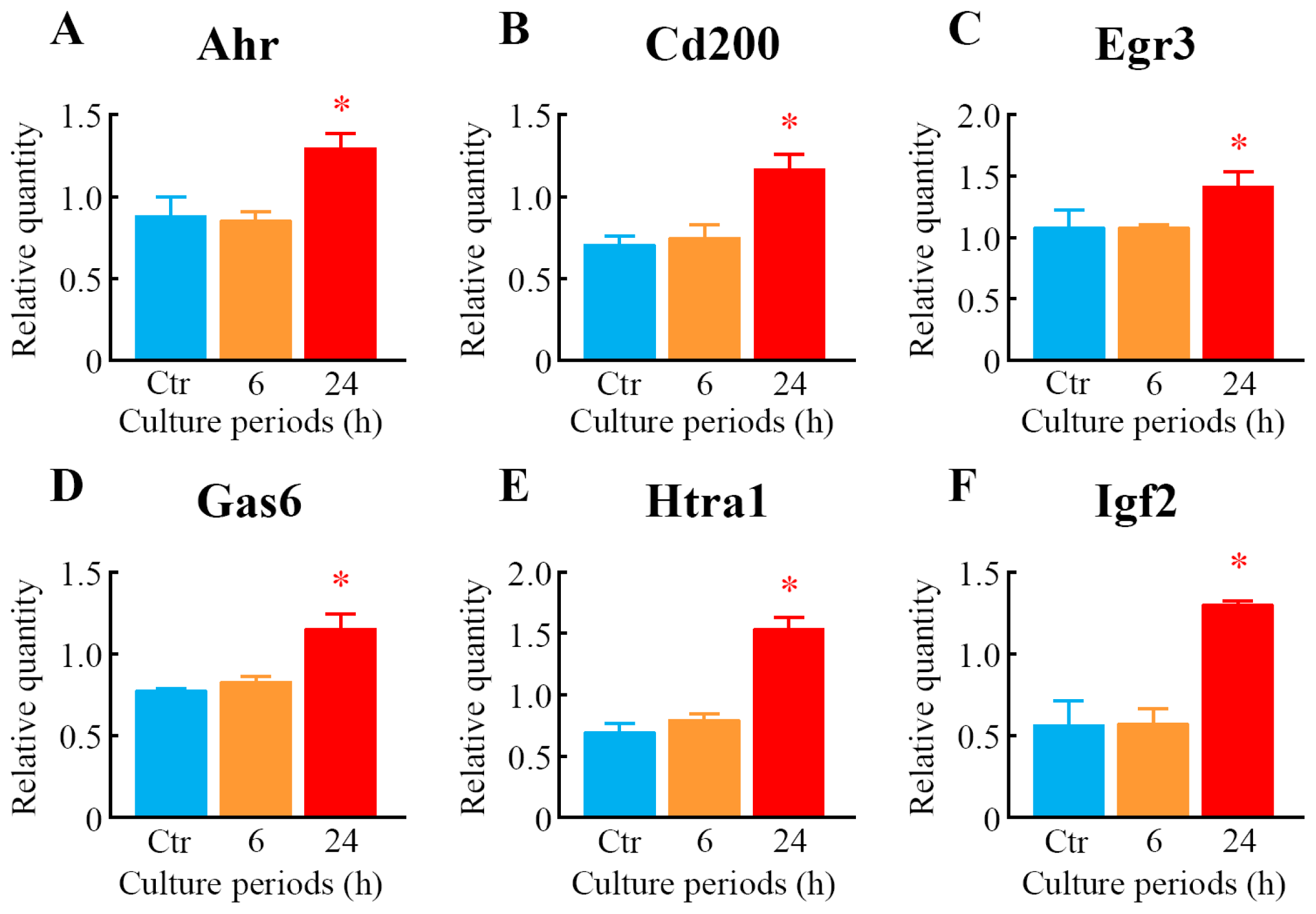
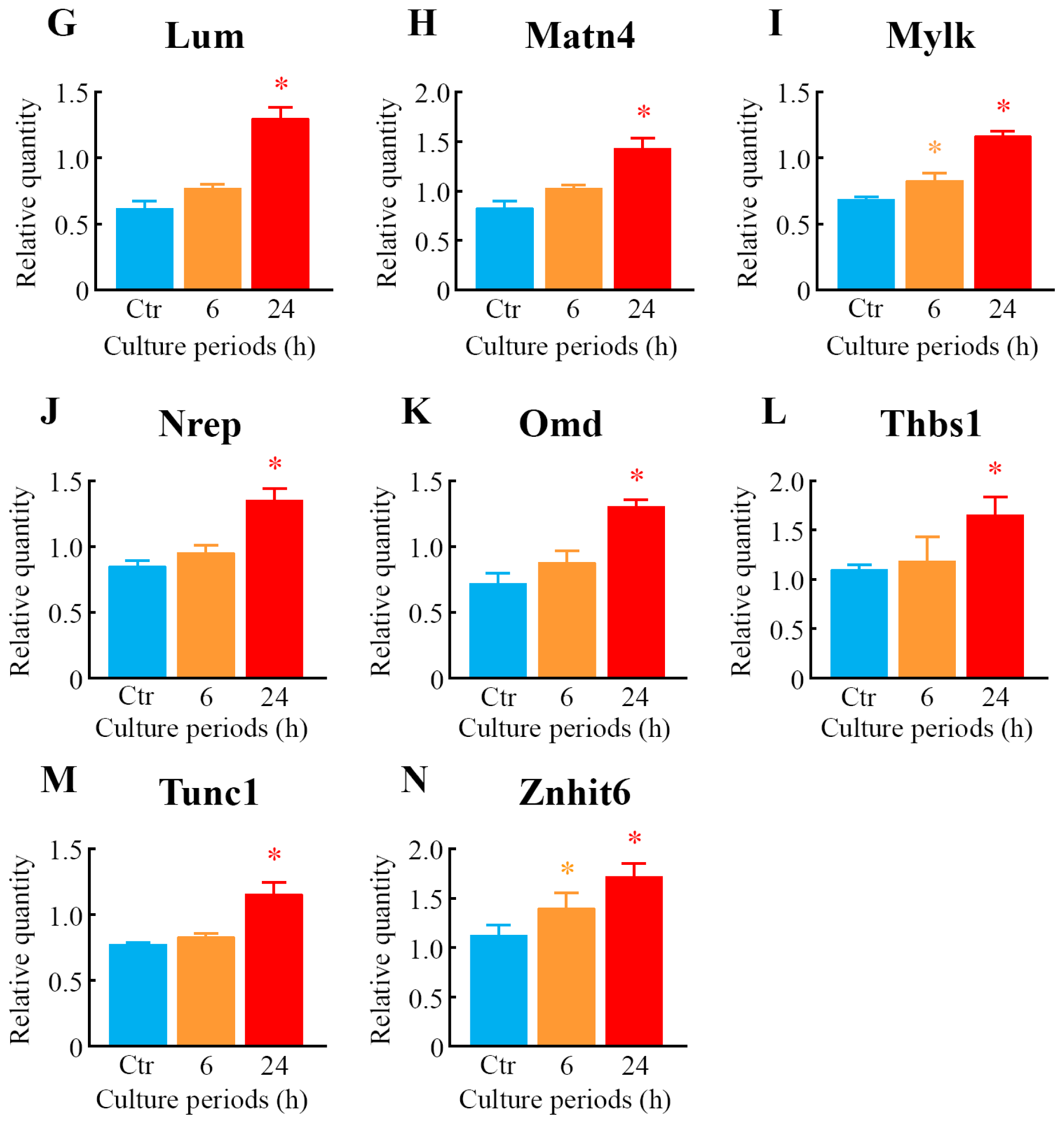


| Probe set ID | Gene symbol | 6 h | 24 h | Gene title | ||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| 1417399_at | Gas6 | 1.04 | 0.13 | 1.53 | 0.05 * | growth arrest specific 6 |
| 1454214_a_at | Znhit6 | 1.11 | 0.02 * | 1.55 | 0.10 * | zinc finger, HIT type 6 |
| 1423854_a_at | Rasl11b | 1.16 | 0.12 | 1.58 | 0.10 * | RAS-like, family 11, member B |
| 1425505_at | Mylk | 0.95 | 0.04 * | 1.58 | 0.05 * | myosin, light polypeptide kinase |
| 1455040_s_at | Nhsl2 | 0.90 | 0.21 | 1.59 | 0.13 * | NHS-like 2 |
| 1456295_at | B230114P17Rik | 1.24 | 0.10 | 1.63 | 0.06 * | RIKEN cDNA B230114P17 gene |
| 1417937_at | Dact1 | 1.01 | 0.08 | 1.67 | 0.11 * | dapper homolog 1, antagonist of beta-catenin (Xenopus) |
| 1421811_at | Thbs1 | 1.11 | 0.12 | 1.68 | 0.26 * | thrombospondin 1 |
| 1426208_x_at | Plagl1 | 1.74 | 0.06 | 1.71 | 0.02 * | pleomorphic adenoma gene-like 1 |
| 1448171_at | Siah2 | 1.01 | 0.76 | 1.71 | 0.18 * | seven in absentia 2 |
| 1448788_at | Cd200 | 1.24 | 0.08 | 1.72 | 0.11 * | CD200 antigen |
| 1433454_at | Abtb2 | 1.29 | 0.12 | 1.73 | 0.12 * | ankyrin repeat and BTB (POZ) domain containing 2 |
| 1441368_at | - | 1.07 | 0.05 | 1.74 | 0.10 * | - |
| 1442157_at | - | 1.14 | 0.06 * | 1.74 | 0.16 * | - |
| 1416225_at | Adh1 | 1.20 | 0.20 | 1.75 | 0.25 * | alcohol dehydrogenase 1 (class I) |
| 1416505_at | Nr4a1 | 1.12 | 0.07 | 1.76 | 0.03 * | nuclear receptor subfamily 4, group A, member 1 |
| 1418464_at | Matn4 | 1.08 | 0.05 | 1.76 | 0.19 * | matrilin 4 |
| 1422631_at | Ahr | 0.98 | 0.04 | 1.76 | 0.09 * | aryl-hydrocarbon receptor |
| 1423261_at | 1500015O10Rik | 1.00 | 0.15 | 1.78 | 0.27 * | RIKEN cDNA 1500015O10 gene |
| 1423607_at | Lum | 0.97 | 0.03 | 1.79 | 0.22 * | lumican |
| 1450839_at | D0H4S114 | 1.07 | 0.06 | 1.80 | 0.17 * | DNA segment, human D4S114 |
| 1436329_at | Egr3 | 1.02 | 0.12 | 1.81 | 0.23 * | early growth response 3 |
| 1438870_at | Fbn1 | 1.11 | 0.11 | 1.82 | 0.30 * | fibrillin 1 |
| 1447927_at | Gbp10 | 1.12 | 0.38 | 1.82 | 0.13 * | guanylate-binding protein 10 |
| 1418252_at | Padi2 | 1.08 | 0.15 | 1.86 | 0.23 * | peptidyl arginine deiminase, type II |
| 1418745_at | Omd | 1.05 | 0.05 | 1.87 | 0.18 * | osteomodulin |
| 1438251_x_at | Htra1 | 0.89 | 0.04 | 2.02 | 0.16 * | HtrA serine peptidase 1 |
| 1448152_at | Igf2 | 0.95 | 0.08 | 2.04 | 0.69 | insulin-like growth factor 2 |
| 1416749_at | Htra1 | 1.00 | 0.14 | 2.06 | 0.06 * | HtrA serine peptidase 1 |
| 1420448_at | Rhox2a | 1.20 | 0.41 | 2.08 | 0.66 | reproductive homeobox 2A |
| 1427149_at | Plekha6 | 0.87 | 0.33 | 2.10 | 0.77 | pleckstrin homology domain containing, family A member 6 |
| 1460049_s_at | 1500015O10Rik | 1.05 | 0.10 | 2.12 | 0.17 * | RIKEN cDNA 1500015O10 gene |
| 1427053_at | Abi3bp | 1.16 | 0.23 | 2.19 | 0.84 | ABI gene family, member 3 (NESH) binding protein |
| 1416371_at | Apod | 1.00 | 0.04 | 2.26 | 0.31 * | apolipoprotein D |
| 1443746_x_at | Dmp1 | 1.10 | 0.16 | 2.29 | 0.19 * | dentin matrix protein 1 |
| 1426081_a_at | Dio2 | 1.03 | 0.17 | 2.38 | 0.55 * | deiodinase, iodothyronine, type II |
| 1427054_s_at | Abi3bp | 1.01 | 0.11 | 2.38 | 0.32 * | ABI gene family, member 3 (NESH) binding protein |
| 1436996_x_at | Lyz1 | 0.86 | 0.51 | 2.41 | 0.76 | lysozyme 1 |
| 1448929_at | F13a1 | 1.56 | 0.18 | 2.42 | 0.16 * | coagulation factor XIII, A1 subunit |
| 1443745_s_at | Dmp1 | 1.11 | 0.08 | 2.44 | 0.35 * | dentin matrix protein 1 |
| 1426139_a_at | Ccrl1 | 1.09 | 0.04 | 2.47 | 0.94 * | chemokine (C-C motif) receptor-like 1 |
| 1417256_at | Mmp13 | 0.83 | 0.13 | 2.65 | 0.46 * | matrix metallopeptidase 13 |
| 1448326_a_at | Crabp1 | 1.18 | 0.08 | 2.68 | 0.50 * | cellular retinoic acid binding protein I |
| 1418370_at | Tnnc1 | 0.92 | 0.09 | 2.82 | 0.08 * | troponin C, cardiac/slow skeletal |
| 1418937_at | Dio2 | 0.97 | 0.04 | 3.05 | 0.36 * | deiodinase, iodothyronine, type II |
| 1427338_at | Crocc | 2.48 | 1.53 | 2.26 | 1.41 | ciliary rootlet coiled-coil, rootletin |
| Probe set ID | Gene symbol | 6 h | 24 h | Gene title | ||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| 1451780_at | Blnk | 0.96 | 0.22 | 0.37 | 0.08 * | B-cell linker |
| 1422537_a_at | Id2 | 1.05 | 0.29 | 0.40 | 0.04 * | inhibitor of DNA binding 2 |
| 1423935_x_at | Krt14 | 0.96 | 0.06 | 0.43 | 0.01 * | keratin 14 |
| 1425538_x_at | Ceacam1 | 1.05 | 0.39 | 0.45 | 0.07 * | carcinoembryonic antigen-related cell adhesion molecule 1 |
| 1425789_s_at | Anxa8 | 0.89 | 0.01 * | 0.46 | 0.04 * | annexin A8 |
| 1448261_at | Cdh1 | 0.86 | 0.08 | 0.46 | 0.05 * | cadherin 1 |
| 1460347_at | Krt14 | 0.97 | 0.07 | 0.46 | 0.06 * | keratin 14 |
| 1435176_a_at | Id2 | 0.91 | 0.17 | 0.47 | 0.11 * | inhibitor of DNA binding 2 |
| 1460684_at | Tm7sf2 | 1.00 | 0.09 | 0.47 | 0.08 * | transmembrane 7 superfamily member 2 |
| 1416630_at | Id3 | 0.94 | 0.04 | 0.48 | 0.07 * | inhibitor of DNA binding 3 |
| 1418595_at | Plin4 | 1.12 | 0.51 | 0.50 | 0.16 * | perilipin 4 |
| 1460250_at | Sostdc1 | 1.02 | 0.05 | 0.50 | 0.08 * | sclerostin domain containing 1 |
| 1425895_a_at | Id1 | 0.98 | 0.13 | 0.51 | 0.16 * | inhibitor of DNA binding 1 |
| 1439382_x_at | Ddr1 | 0.96 | 0.20 | 0.51 | 0.21 | discoidin domain receptor family, member 1 |
| 1428738_a_at | D14Ertd449e | 1.00 | 0.28 | 0.52 | 0.09 * | DNA segment, Chr 14, ERATO Doi 449, expressed |
| 1436520_at | Ahnak2 | 1.05 | 0.20 | 0.52 | 0.08 * | AHNAK nucleoprotein 2 |
| 1452656_at | Zdhhc2 | 0.98 | 0.24 | 0.53 | 0.12 * | zinc finger, DHHC domain containing 2 |
| 1424162_at | Trim29 | 0.80 | 0.34 | 0.54 | 0.16 * | tripartite motif-containing 29 |
| 1434046_at | AA467197 | 0.92 | 0.01 * | 0.54 | 0.05 * | expressed sequence AA467197 |
| 1417732_at | Anxa8 | 0.90 | 0.08 | 0.55 | 0.06 * | annexin A8 |
| 1438152_at | - | 1.35 | 0.57 | 0.55 | 0.13 * | - |
| 1449060_at | Kif2c | 0.82 | 0.08 | 0.55 | 0.10 * | kinesin family member 2C |
| 1454702_at | 4930503L19Rik | 0.87 | 0.05 * | 0.55 | 0.03 * | RIKEN cDNA 4930503L19 gene |
| 1426152_a_at | Kitl | 1.01 | 0.02 | 0.56 | 0.09 * | kit ligand |
| 1458236_at | - | 1.06 | 0.05 | 0.56 | 0.05 * | - |
| 1415871_at | Tgfbi | 0.90 | 0.06 | 0.57 | 0.04 * | transforming growth factor, beta induced |
| 1427357_at | Cda | 0.94 | 0.13 | 0.57 | 0.05 * | cytidine deaminase |
| 1449340_at | Sostdc1 | 0.97 | 0.07 | 0.57 | 0.08 * | sclerostin domain containing 1 |
| 1419489_at | Fam19a5 | 0.96 | 0.06 | 0.58 | 0.03 * | family with sequence similarity 19, member A5 |
| 1422123_s_at | Ceacam1 | 0.87 | 0.26 | 0.58 | 0.06 * | carcinoembryonic antigen-related cell adhesion molecule 1 |
| 1456412_a_at | Inpp5k | 0.71 | 0.21 | 0.58 | 0.04 * | inositol polyphosphate 5-phosphatase K |
| 1432202_a_at | Poc1a | 0.78 | 0.14 | 0.58 | 0.08 * | POC1 centriolar protein homolog A (Chlamydomonas) |
| 1458385_at | Hspa4l | 0.95 | 0.06 | 0.58 | 0.04 * | heat shock protein 4 like |
| 1419073_at | Tmeff2 | 0.98 | 0.17 | 0.59 | 0.07 * | transmembrane protein with EGF-like and two follistatin-like domains 2 |
| 1440924_at | Kif20b | 0.99 | 0.06 | 0.61 | 0.02 * | kinesin family member 20B |
| 1417751_at | Stk10 | 0.95 | 0.16 | 0.62 | 0.03 * | serine/threonine kinase 10 |
| 1423627_at | Nqo1 | 1.02 | 0.02 | 0.62 | 0.05 * | NAD(P)H dehydrogenase, quinone 1 |
| 1452654_at | Zdhhc2 | 0.98 | 0.12 | 0.62 | 0.06 * | zinc finger, DHHC domain containing 2 |
| 1430193_at | Casc5 | 0.85 | 0.10 | 0.62 | 0.03 * | cancer susceptibility candidate 5 |
| 1430617_at | Oip5 | 0.92 | 0.12 | 0.62 | 0.04 * | Opa interacting protein 5 |
| 1417823_at | Gcat | 0.86 | 0.14 | 0.63 | 0.03 * | glycine C-acetyltransferase (2-amino-3-ketobutyrate-coenzyme A ligase) |
| 1423569_at | Gatm | 1.08 | 0.04 | 0.64 | 0.02 * | glycine amidinotransferase (l-arginine: glycine amidinotransferase) |
| 1456250_x_at | Tgfbi | 0.94 | 0.03 | 0.64 | 0.02 * | transforming growth factor, beta induced |
| 1435554_at | Tmcc3 | 0.93 | 0.08 | 0.64 | 0.02 * | transmembrane and coiled coil domains 3 |
| 1452968_at | Cthrc1 | 0.99 | 0.02 | 0.64 | 0.02 * | collagen triple helix repeat containing 1 |
| 1436654_at | Gen1 | 1.00 | 0.13 | 0.65 | 0.01 * | Gen homolog 1, endonuclease (Drosophila) |
| 1456569_x_at | Gsn | 0.58 | 0.08* | 5.62 | 8.87 | gelsolin |
| 1436622_at | Iqsec2 | 0.62 | 0.01* | 0.98 | 0.21 | IQ motif and Sec7 domain 2 |
Acknowledgments
Conflicts of Interest
References
- Riesz, P.; Kondo, T. Free radical formation induced by ultrasound and its biological implications. Free Radic. Biol. Med 1992, 13, 247–270. [Google Scholar]
- Feril, L.B., Jr.; Kondo, T. Biological effects of low intensity ultrasound: The mechanism involved, and its implications on therapy and on biosafety of ultrasound. J. Radiat. Res 2004, 45, 479–489. [Google Scholar]
- Pounder, N.M.; Harrison, A.J. Low intensity pulsed ultrasound for fracture healing: A review of the clinical evidence and the associated biological mechanism of action. Ultrasonics 2008, 48, 330–338. [Google Scholar]
- Martinez de Albornoz, P.; Khanna, A.; Longo, U.G.; Forriol, F.; Maffulli, N. The evidence of low-intensity pulsed ultrasound for in vitro, animal and human fracture healing. Br. Med. Bull 2011, 100, 39–57. [Google Scholar]
- Riboh, J.C.; Leversedge, F.J. The use of low-intensity pulsed ultrasound bone stimulators for fractures of the hand and upper extremity. J. Hand. Surg. Am 2012, 37, 1456–1461. [Google Scholar]
- Bashardoust Tajali, S.; Houghton, P.; MacDermid, J.C.; Grewal, R. Effects of low-intensity pulsed ultrasound therapy on fracture healing: A systematic review and meta-analysis. Am. J. Phys. Med. Rehabil 2012, 91, 349–367. [Google Scholar]
- Duarte, L.R. The stimulation of bone growth by ultrasound. Arch. Orthop. Trauma Surg 1983, 101, 153–159. [Google Scholar]
- Shimazaki, A.; Inui, K.; Azuma, Y.; Nishimura, N.; Yamano, Y. Low-intensity pulsed ultrasound accelerates bone maturation in distraction osteogenesis in rabbits. J. Bone Joint Surg. Br 2000, 82, 1077–1082. [Google Scholar]
- Azuma, Y.; Ito, M.; Harada, Y.; Takagi, H.; Ohta, T.; Jingushi, S. Low-intensity pulsed ultrasound accelerates rat femoral fracture healing by acting on the various cellular reactions in the fracture callus. J. Bone Miner. Res 2001, 16, 671–680. [Google Scholar]
- Machen, M.S.; Tis, J.E.; Inoue, N.; Meffert, R.H.; Chao, E.Y.; McHale, K.A. The effect of low intensity pulsed ultrasound on regenerate bone in a less-than-rigid biomechanical environment. Biomed. Mater. Eng 2002, 12, 239–247. [Google Scholar]
- Uglow, M.G.; Peat, R.A.; Hile, M.S.; Bilston, L.E.; Smith, E.J.; Little, D.G. Low-intensity ultrasound stimulation in distraction osteogenesis in rabbits. Clin. Orthop. Relat. Res 2003, 303–312. [Google Scholar]
- Qin, L.; Fok, P.; Lu, H.; Shi, S.; Leng, Y.; Leung, K. Low intensity pulsed ultrasound increases the matrix hardness of the healing tissues at bone-tendon insertion-a partial patellectomy model in rabbits. Clin. Biomech 2006, 21, 387–394. [Google Scholar]
- Lu, H.; Qin, L.; Fok, P.; Cheung, W.; Lee, K.; Guo, X.; Wong, W.; Leung, K. Low-intensity pulsed ultrasound accelerates bone-tendon junction healing: A partial patellectomy model in rabbits. Am. J. Sports Med 2006, 34, 1287–1296. [Google Scholar]
- Iwai, T.; Harada, Y.; Imura, K.; Iwabuchi, S.; Murai, J.; Hiramatsu, K.; Myoui, A.; Yoshikawa, H.; Tsumaki, N. Low-intensity pulsed ultrasound increases bone ingrowth into porous hydroxyapatite ceramic. J. Bone Miner. Metab 2007, 25, 392–399. [Google Scholar]
- Walsh, W.R.; Langdown, A.J.; Auld, J.W.; Stephens, P.; Yu, Y.; Vizesi, F.; Bruce, W.J.; Pounder, N. Effect of low intensity pulsed ultrasound on healing of an ulna defect filled with a bone graft substitute. J. Biomed. Mater. Res. B 2008, 86, 74–81. [Google Scholar]
- Lu, C.C.; Liu, Y.C.; Cheng, Y.M.; Chih, T.T.; Tien, Y.C. Augmentation of tendon-bone interface healing with low-intensity pulsed ultrasound. Orthopedics 2009, 32, 173. [Google Scholar]
- Shakouri, K.; Eftekharsadat, B.; Oskuie, M.R.; Soleimanpour, J.; Tarzamni, M.K.; Salekzamani, Y.; Hoshyar, Y.; Nezami, N. Effect of low-intensity pulsed ultrasound on fracture callus mineral density and flexural strength in rabbit tibial fresh fracture. J. Orthop. Sci 2010, 15, 240–244. [Google Scholar]
- Xie, L.K.; Wangrangsimakul, K.; Suttapreyasri, S.; Cheung, L.K.; Nuntanaranont, T. A preliminary study of the effect of low intensity pulsed ultrasound on new bone formation during mandibular distraction osteogenesis in rabbits. Int. J. Oral Maxillofac. Surg 2011, 40, 730–736. [Google Scholar]
- Hantes, M.E.; Mavrodontidis, A.N.; Zalavras, C.G.; Karantanas, A.H.; Karachalios, T.; Malizos, K.N. Low-intensity transosseous ultrasound accelerates osteotomy healing in a sheep fracture model. J. Bone Joint Surg. Am 2004, 86-A, 2275–2282. [Google Scholar]
- Walsh, W.R.; Stephens, P.; Vizesi, F.; Bruce, W.; Huckle, J.; Yu, Y. Effects of low-intensity pulsed ultrasound on tendon-bone healing in an intra-articular sheep knee model. Arthroscopy 2007, 23, 197–204. [Google Scholar]
- Lovric, V.; Ledger, M.; Goldberg, J.; Harper, W.; Bertollo, N.; Pelletier, M.H.; Oliver, R.A.; Yu, Y.; Walsh, W.R. The effects of low-intensity pulsed ultrasound on tendon-bone healing in a transosseous-equivalent sheep rotator cuff model. Knee Surg. Sports Traumatol. Arthrosc 2013, 21, 466–475. [Google Scholar]
- Heckman, J.D.; Ryaby, J.P.; McCabe, J.; Frey, J.J.; Kilcoyne, R.F. Acceleration of tibial fracture-healing by non-invasive, low-intensity pulsed ultrasound. J. Bone Joint Surg. Am 1994, 76, 26–34. [Google Scholar]
- Kristiansen, T.K.; Ryaby, J.P.; McCabe, J.; Frey, J.J.; Roe, L.R. Accelerated healing of distal radial fractures with the use of specific, low-intensity ultrasound. A multicenter, prospective, randomized, double-blind, placebo-controlled study. J. Bone Joint Surg. Am 1997, 79, 961–973. [Google Scholar]
- Naruse, K.; Miyauchi, A.; Itoman, M.; Mikuni-Takagaki, Y. Distinct anabolic response of osteoblast to low-intensity pulsed ultrasound. J. Bone Miner. Res 2003, 18, 360–369. [Google Scholar]
- Bandow, K.; Nishikawa, Y.; Ohnishi, T.; Kakimoto, K.; Soejima, K.; Iwabuchi, S.; Kuroe, K.; Matsuguchi, T. Low-intensity pulsed ultrasound (LIPUS) induces RANKL, MCP-1, and MIP-1beta expression in osteoblasts through the angiotensin II type 1 receptor. J. Cell. Physiol 2007, 211, 392–398. [Google Scholar]
- Takayama, T.; Suzuki, N.; Ikeda, K.; Shimada, T.; Suzuki, A.; Maeno, M.; Otsuka, K.; Ito, K. Low-intensity pulsed ultrasound stimulates osteogenic differentiation in ROS 17/2.8 cells. Life Sci 2007, 80, 965–971. [Google Scholar]
- Unsworth, J.; Kaneez, S.; Harris, S.; Ridgway, J.; Fenwick, S.; Chenery, D.; Harrison, A. Pulsed low intensity ultrasound enhances mineralisation in preosteoblast cells. Ultrasound Med. Biol 2007, 33, 1468–1474. [Google Scholar]
- Leskinen, J.J.; Karjalainen, H.M.; Olkku, A.; Hynynen, K.; Mahonen, A.; Lammi, M.J. Genome-wide microarray analysis of MG-63 osteoblastic cells exposed to ultrasound. Biorheology 2008, 45, 345–354. [Google Scholar]
- Lu, H.; Qin, L.; Lee, K.; Cheung, W.; Chan, K.; Leung, K. Identification of genes responsive to low-intensity pulsed ultrasound stimulations. Biochem. Biophys. Res. Commun 2009, 378, 569–573. [Google Scholar]
- Watabe, H.; Furuhama, T.; Tani-Ishii, N.; Mikuni-Takagaki, Y. Mechanotransduction activates α5β1 integrin and PI3K/Akt signaling pathways in mandibular osteoblasts. Exp. Cell Res 2011, 317, 2642–2649. [Google Scholar]
- Kitamura, K.; Suzuki, N.; Sato, Y.; Nemoto, T.; Ikegame, M.; Shimizu, N.; Kondo, T.; Furusawa, Y.; Wada, S.; Hattori, A. Osteoblast activity in the goldfish scale responds sensitively to mechanical stress. Comp. Biochem. Physiol. A 2010, 156, 357–363. [Google Scholar]
- Naruse, K.; Mikuni-Takagaki, Y.; Azuma, Y.; Ito, M.; Oota, T.; Kameyama, K.; Itoman, M. Anabolic response of mouse bone-marrow-derived stromal cell clone ST2 cells to low-intensity pulsed ultrasound. Biochem. Biophys. Res. Commun 2000, 268, 216–220. [Google Scholar]
- Sena, K.; Angle, S.R.; Kanaji, A.; Aher, C.; Karwo, D.G.; Sumner, D.R.; Virdi, A.S. Low-intensity pulsed ultrasound (LIPUS) and cell-to-cell communication in bone marrow stromal cells. Ultrasonics 2011, 51, 639–644. [Google Scholar]
- Mukai, S.; Ito, H.; Nakagawa, Y.; Akiyama, H.; Miyamoto, M.; Nakamura, T. Transforming growth factor-beta1 mediates the effects of low-intensity pulsed ultrasound in chondrocytes. Ultrasound Med. Biol 2005, 31, 1713–1721. [Google Scholar]
- Takeuchi, R.; Ryo, A.; Komitsu, N.; Mikuni-Takagaki, Y.; Fukui, A.; Takagi, Y.; Shiraishi, T.; Morishita, S.; Yamazaki, Y.; Kumagai, K.; et al. Low-intensity pulsed ultrasound activates the phosphatidylinositol 3 kinase/Akt pathway and stimulates the growth of chondrocytes in three-dimensional cultures: a basic science study. Arthritis Res. Ther 2008, 10, R77. [Google Scholar]
- Zhou, S.; Schmelz, A.; Seufferlein, T.; Li, Y.; Zhao, J.; Bachem, M.G. Molecular mechanisms of low intensity pulsed ultrasound in human skin fibroblasts. J. Biol. Chem 2004, 279, 54463–54469. [Google Scholar]
- Kobayashi, Y.; Sakai, D.; Iwashina, T.; Iwabuchi, S.; Mochida, J. Low-intensity pulsed ultrasound stimulates cell proliferation, proteoglycan synthesis and expression of growth factor-related genes in human nucleus pulposus cell line. Eur. Cell Mater 2009, 17, 15–22. [Google Scholar]
- Li, L.; Yang, Z.; Zhang, H.; Chen, W.; Chen, M.; Zhu, Z. Low-intensity pulsed ultrasound regulates proliferation and differentiation of osteoblasts through osteocytes. Biochem. Biophys. Res. Commun 2012, 418, 296–300. [Google Scholar]
- Tabuchi, Y.; Takasaki, I.; Zhao, Q.L.; Wada, S.; Hori, T.; Feril, L.B., Jr.; Tachibana, K.; Nomura, T.; Kondo, T. Genetic networks responsive to low-intensity pulsed ultrasound in human lymphoma U937 cells. Cancer Lett 2008, 270, 286–294. [Google Scholar]
- Ahmed, K.; Furusawa, Y.; Tabuchi, Y.; Emam, H.F.; Piao, J.L.; Hassan, M.A.; Yamamoto, T.; Kondo, T.; Kadowaki, M. Chemical inducers of heat shock proteins derived from medicinal plants and cytoprotective genes response. Int. J. Hyperthermia 2012, 28, 1–8. [Google Scholar]
- Tabuchi, Y.; Wada, S.; Furusawa, Y.; Ohtsuka, K.; Kondo, T. Gene networks related to the cell death elicited by hyperthermia in human oral squamous cell carcinoma HSC-3 cells. Int. J. Mol. Med 2012, 29, 380–386. [Google Scholar]
- Ito, A.; Aoyama, T.; Yamaguchi, S.; Zhang, X.; Akiyama, H.; Kuroki, H. Low-intensity pulsed ultrasound inhibits messenger RNA expression of matrix metalloproteinase-13 induced by interleukin-1β in chondrocytes in an intensity-dependent manner. Ultrasound Med. Biol 2012, 38, 1726–1733. [Google Scholar]
- Al-Daghreer, S.; Doschak, M.; Sloan, A.J.; Major, P.W.; Heo, G.; Scurtescu, C.; Tsui, Y.Y.; El-Bialy, T. Long term effect of low intensity pulsed ultrasound on a human tooth slice organ culture. Arch. Oral Biol 2012, 57, 760–768. [Google Scholar]
- Feng, J.Q.; Scott, G.; Guo, D.; Jiang, B.; Harris, M.; Ward, T.; Ray, M.; Bonewald, L.F.; Harris, S.E.; Mishina, Y. Generation of a conditional null allele for Dmp1 in mouse. Genesis 2008, 46, 87–91. [Google Scholar]
- Carta, L.; Pereira, L.; Arteaga-Solis, E.; Lee-Arteaga, S.Y.; Lenart, B.; Starcher, B.; Merkel, C.A.; Sukoyan, M.; Kerkis, A.; Hazeki, N.; et al. Fibrillins 1 and 2 perform partially overlapping functions during aortic development. J. Biol. Chem 2006, 281, 8016–8023. [Google Scholar]
- Eggenschwiler, J.; Ludwig, T.; Fisher, P.; Leighton, P.A.; Tilghman, S.M.; Efstratiadis, A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes Dev 1997, 11, 3128–3142. [Google Scholar]
- Jepsen, K.J.; Wu, F.; Peragallo, J.H.; Paul, J.; Roberts, L.; Ezura, Y.; Oldberg, A.; Birk, D.E.; Chakravarti, S. A syndrome of joint laxity and impaired tendon integrity in lumican- and fibromodulin-deficient mice. J. Biol. Chem 2002, 277, 35532–35540. [Google Scholar]
- Stickens, D.; Behonick, D.J.; Ortega, N.; Heyer, B.; Hartenstein, B.; Yu, Y.; Fosang, A.J.; Schorpp-Kistner, M.; Angel, P.; Werb, Z. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development 2004, 131, 5883–5895. [Google Scholar]
- Crawford, S.E.; Stellmach, V.; Murphy-Ullrich, J.E.; Ribeiro, S.M.; Lawler, J.; Hynes, R.O.; Boivin, G.P.; Bouck, N. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell 1998, 93, 1159–1170. [Google Scholar]
- Kalajzic, I.; Staal, A.; Yang, W.P.; Wu, Y.; Johnson, S.E.; Feyen, J.H.; Krueger, W.; Maye, P.; Yu, F.; Zhao, Y.; et al. Expression profile of osteoblast lineage at defined stages of differentiation. J. Biol. Chem 2005, 280, 24618–24626. [Google Scholar]
- Cui, W.; Cuartas, E.; Ke, J.; Zhang, Q.; Einarsson, H.B.; Sedgwick, J.D.; Li, J.; Vignery, A. CD200 and its receptor, CD200R, modulate bone mass via the differentiation of osteoclasts. Proc. Natl. Acad. Sci. USA 2007, 104, 14436–14441. [Google Scholar]
- Eapen, A.; Ramachandran, A.; Pratap, J.; George, A. Activation of the ERK1/2 mitogen-activated protein kinase cascade by dentin matrix protein 1 promotes osteoblast differentiation. Cells Tissues Organs 2011, 194, 255–260. [Google Scholar]
- Miyazono, K.; Miyazawa, K. Id: A target of BMP signaling. Sci. STKE 2002, 2002, pe40. [Google Scholar]
- Peng, Y.; Kang, Q.; Luo, Q.; Jiang, W.; Si, W.; Liu, B.A.; Luu, H.H.; Park, J.K.; Li, X.; Luo, J.; et al. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J. Biol. Chem 2004, 279, 32941–32949. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tabuchi, Y.; Sugahara, Y.; Ikegame, M.; Suzuki, N.; Kitamura, K.-i.; Kondo, T. Genes Responsive to Low-Intensity Pulsed Ultrasound in MC3T3-E1 Preosteoblast Cells. Int. J. Mol. Sci. 2013, 14, 22721-22740. https://doi.org/10.3390/ijms141122721
Tabuchi Y, Sugahara Y, Ikegame M, Suzuki N, Kitamura K-i, Kondo T. Genes Responsive to Low-Intensity Pulsed Ultrasound in MC3T3-E1 Preosteoblast Cells. International Journal of Molecular Sciences. 2013; 14(11):22721-22740. https://doi.org/10.3390/ijms141122721
Chicago/Turabian StyleTabuchi, Yoshiaki, Yuuki Sugahara, Mika Ikegame, Nobuo Suzuki, Kei-ichiro Kitamura, and Takashi Kondo. 2013. "Genes Responsive to Low-Intensity Pulsed Ultrasound in MC3T3-E1 Preosteoblast Cells" International Journal of Molecular Sciences 14, no. 11: 22721-22740. https://doi.org/10.3390/ijms141122721




