Classification of HCV NS5B Polymerase Inhibitors Using Support Vector Machine
Abstract
:1. Introduction
2. Results and Discussion
2.1. Model 1 Built with Global Descriptors
2.2. Model 2 with Global Descriptors and 2D Autocorrelation Descriptors
2.3. Model 3 with Global Descriptors and 3D Autocorrelation Descriptors
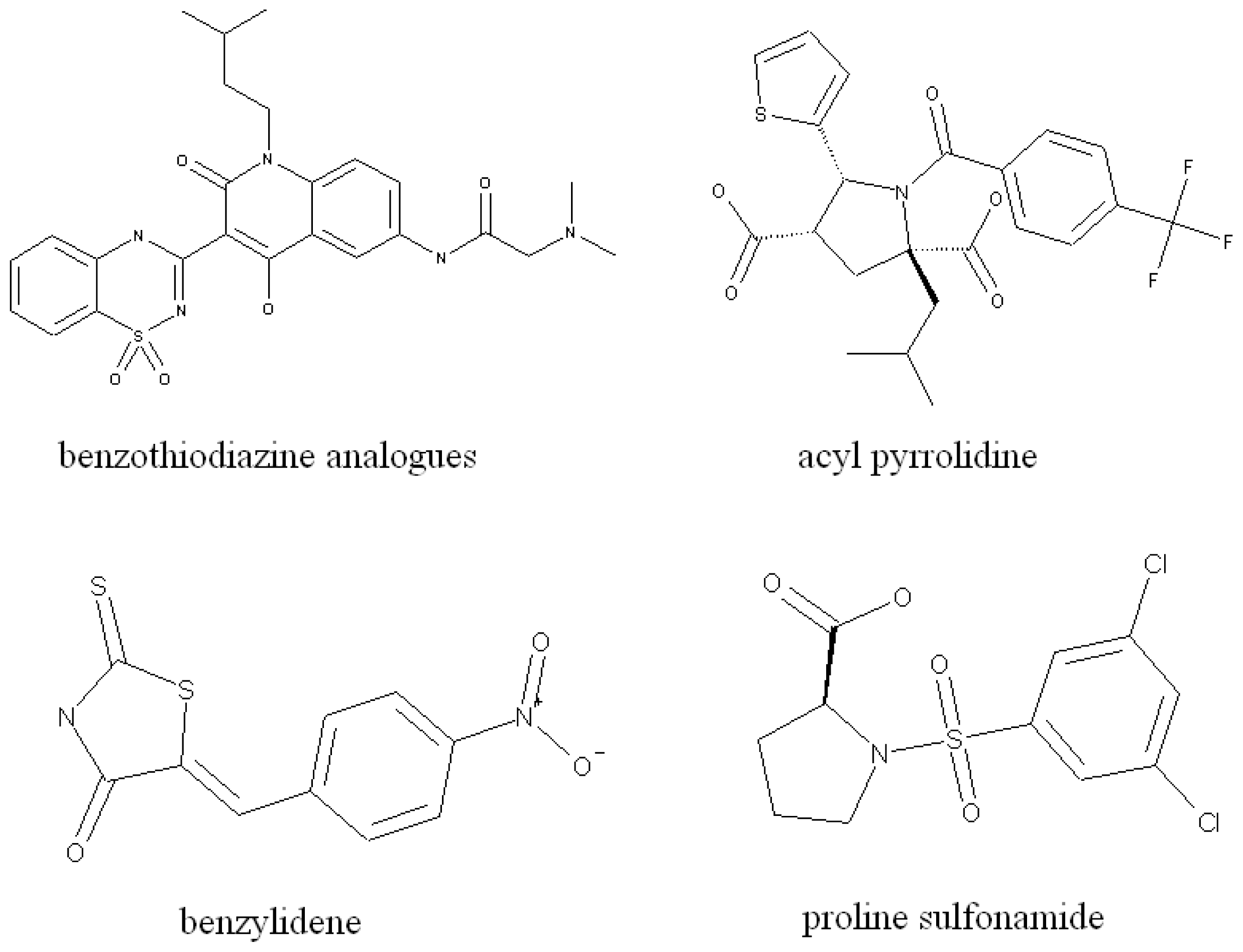
2.4. Relationship between the Selected Molecular Descriptors and Activity
2.5. External Test Set
3. Experimental Section
3.1. Dataset
3.2. Molecular Descriptors
3.3. Descriptors Selection
3.4. Support Vector Machine
3.5. Evaluation of Models
4. Conclusions
Acknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Koch, U.; Attenni, B.; Malancona, S.; Colarusso, S.; Conte, I.; di Filippo, M.; Harper, S.; Pacini, B.; Giomini, C.; Thomas, S.; et al. 2-(2-Thienyl)-5,6-dihydroxy-4-carboxypyrimidines as inhibitors of the hepatitis C virus NS5B polymerase: Discovery, SAR, modeling, and mutagenesis. J. Med. Chem 2006, 49, 1693–1705. [Google Scholar]
- Tedesco, R.; Shaw, A.N.; Bambal, R.; Chai, D.; Concha, N.O.; Darcy, M.G.; Dhanak, D.; Fitch, D.M.; Gates, A.; Gerhardt, W.G.; et al. 3-(1,1-dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinones, potent inhibitors of hepatitis C virus RNA-dependent RNA polymerase. J. Med. Chem 2006, 49, 971–983. [Google Scholar]
- Ontoria, J.M.; Rydberg, E.H.; di Marco, S.; Tomei, L.; Attenni, B.; Malancona, S.; Hernando, J.I.M.; Gennari, N.; Koch, U.; Narjes, F.; et al. Identification and biological evaluation of a series of 1H-benzo[de]isoquinoline-1,3(2H)-diones as hepatitis C virus NS5B polymerase inhibitors. J. Med. Chem 2009, 52, 5217–5227. [Google Scholar]
- Chen, K.X.; Lesburg, C.A.; Vibulbhan, B.; Yang, W. A novel class of highly potent irreversible hepatitis C virus NS5B polymerase inhibitors. J. Med. Chem 2012, 55, 2089–2101. [Google Scholar]
- Krueger, A.C.; Madigan, D.L.; Jiang, W.W.; Kati, W.M.; Liu, D.; Liu, Y.; Maring, C.J.; Masse, S.; McDaniel, K.F.; Middleton, T.; et al. Inhibitors of HCV NS5B polymerase: Synthesis and structure-activity relationships of N-alkyl-4-hydroxyquinolon-3-yl-benzothiadiazine sulfamides. Bioorg. Med. Chem. Lett 2006, 16, 3367–3370. [Google Scholar]
- Ishida, T.; Suzuki, T.; Hirashima, S.; Mizutani, K.; Yoshida, A.; Ando, I.; Ikeda, S.; Adachi, T.; Hashimoto, H. Benzimidazole inhibitors of hepatitis C virus NS5B polymerase: Identification of 2-[(4-diarylmethoxy)phenyl]-benzimidazole. Bioorg. Med. Chem. Lett 2006, 16, 1859–1863. [Google Scholar]
- Das, D.; Hong, J.; Chen, S.H.; Wang, G.; Beigelman, L.; Seiwert, S.D.; Buckman, B.O. Recent advances in drug discovery of benzothiadiazine and related analogs as HCV NS5B polymerase inhibitors. Bioorg. Med. Chem 2011, 19, 4690–4703. [Google Scholar]
- Pfefferkorn, J.A.; Greene, M.L.; Nugent, R.A.; Gross, R.J.; Mitchell, M.A.; Finzel, B.C.; Harris, M.S.; Wells, P.A.; Shelly, J.A.; Anstadt, R.A.; et al. Inhibitors of HCV NS5B polymerase. Part 1: Evaluation of the southern region of (2Z)-2-(benzoylamino)-3-(5-phenyl-2-furyl)acrylic acid. Bioorg. Med. Chem. Lett 2005, 15, 2481–2486. [Google Scholar]
- Donner, P.L.; Xie, Q.; Pratt, J.K.; Maring, C.J.; Kati, W.; Jiang, W.; Liu, Y.; Koev, G.; Masse, S.; Montgomery, D.; et al. Des-A-ring benzothiadiazines: Inhibitors of HCV genotype 1 NS5B RNA-dependent RNA polymerase. Bioorg. Med. Chem. Lett 2008, 18, 2735–2738. [Google Scholar]
- Powdrill, M.H.; Bernatchez, J.A.; Gotte, M. Inhibitors of the hepatitis C virus RNA-dependent RNA polymerase NS5B. Viruses 2010, 2, 2169–2195. [Google Scholar]
- Wang, Z.; Chen, Y.; Liang, H.; Bender, A.; Glen, R.C.; Yan, A. P-glycoprotein substrate models using support vector machines based on a comprehensive data set. J. Chem. Inf. Model 2011, 51, 1447–1456. [Google Scholar]
- Subramaniam, S.; Mehrotra, M.; Gupta, D. Support vector machine based classification model for screening Plasmodium falciparum proliferation Inhibitors and non-inhibitors. Biomed. Eng. Comput. Biol 2011, 3, 13–24. [Google Scholar]
- Liew, C.Y.; Ma, X.H.; Liu, X.; Yap, C.W. SVM model for virtual screening of Lck inhibitors. J. Chem. Inf. Model 2009, 49, 877–885. [Google Scholar]
- Armutlu, P.; Ozdemir, M.E.; Uney-Yuksektepe, F.; Kavakli, I.H.; Turkay, M. Classification of drug molecules considering their IC50 values using mixed-integer linear programming based hyper-boxes method. BMC Bioinforma 2008, 9. [Google Scholar] [CrossRef]
- Hao, M.; Li, Y.; Wang, Y.; Zhang, S. A classification study of respiratory syncytial virus (RSV) inhibitors by variable selection with random forest. Int. J. Mol. Sci 2011, 12, 1259–1280. [Google Scholar]
- Lv, W.; Xue, Y. Prediction of hepatitis c virus non-structural proteins 5B polymerase inhibitors using machine learning methods. Acta Phys. Chim. Sin 2011, 27, 1407–1416. [Google Scholar]
- ADRIANA. Code, Molecular Networks GmbH: Erlangen, Germany. Available online: http://www.molecular-networks.com accessed on 22 March 2012.
- Gasteiger, J.; Jochum, C. An algorithm for the perception of synthetically important rings. J. Chem. Inf. Comput. Sci 1979, 19, 43–48. [Google Scholar]
- Todeschini, R.; Consonni, V. Handbook of Molecular Descriptors; Wiley-VCH: Weinheim, Germany, 2000; Volume 11. [Google Scholar]
- Pfefferkorn, J.A.; Nugent, R.; Gross, R.J.; Greene, M.; Mitchell, M.A.; Reding, M.T.; Funk, L.A.; Anderson, R.; Wells, P.A.; Shelly, J.A.; et al. Inhibitors of HCV NS5B polymerase. Part 2: Evaluation of the northern region of (2Z)-2-benzoylamino-3-(4-phenoxy-phenyl)-acrylic acid. Bioorg. Med. Chem. Lett 2005, 15, 2812–2818. [Google Scholar]
- Zhou, Y.; Webber, S.E.; Murphy, D.E.; Li, L.S.; Dragovich, P.S.; Tran, C.V.; Sun, Z.; Ruebsam, F.; Shah, A.M.; Tsan, M.; et al. Novel HCV NS5B polymerase inhibitors derived from 4-(1′,1′-dioxo- 1′,4′-dihydro-1′lambda6-benzo[1′,2′,4′]thiadiazin-3′-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 1: Exploration of 7′-substitution of benzothiadiazine. Bioorg. Med. Chem. Lett 2008, 18, 1413–1418. [Google Scholar]
- Burton, G.; Ku, T.W.; Carr, T.J.; Kiesow, T.; Sarisky, R.T.; Lin-Goerke, J.; Baker, A.; Earnshaw, D.L.; Hofmann, G.A.; Keenan, R.M.; et al. Identification of small molecule inhibitors of the hepatitis C virus RNA-dependent RNA polymerase from a pyrrolidine combinatorial mixture. Bioorg. Med. Chem. Lett 2005, 15, 1553–1556. [Google Scholar]
- De Vicente, J.; Hendricks, R.T.; Smith, D.B.; Fell, J.B.; Fischer, J.; Spencer, S.R.; Stengel, P.J.; Mohr, P.; Robinson, J.E.; Blake, J.F.; et al. Non-nucleoside inhibitors of HCV polymerase NS5B. Part 2: Synthesis and structure-activity relationships of benzothiazine-substituted quinolinediones. Bioorg. Med. Chem. Lett 2009, 19, 3642–3646. [Google Scholar]
- De Vicente, J.; Hendricks, R.T.; Smith, D.B.; Fell, J.B.; Fischer, J.; Spencer, S.R.; Stengel, P.J.; Mohr, P.; Robinson, J.E.; Blake, J.F.; et al. Non-nucleoside inhibitors of HCV polymerase NS5B. Part 3: Synthesis and optimization studies of benzothiazine-substituted tetramic acids. Bioorg. Med. Chem. Lett 2009, 19, 5648–5651. [Google Scholar]
- Dragovich, P.S.; Blazel, J.K.; Ellis, D.A.; Han, Q.; Kamran, R.; Kissinger, C.R.; LeBrun, L.A.; Li, L.S.; Murphy, D.E.; Noble, M.; et al. Novel HCV NS5B polymerase inhibitors derived from 4-(1′,1′-dioxo-1′,4′-dihydro-1′lambda(6)-benzo[1′,2′,4′]thiadiazin-3′-yl)-5-hydroxy-2H-pyridazin- 3-ones. Part 5: Exploration of pyridazinones containing 6-amino-substituents. Bioorg. Med. Chem. Lett 2008, 18, 5635–5639. [Google Scholar]
- Gopalsamy, A.; Chopra, R.; Lim, K.; Ciszewski, G.; Shi, M.; Curran, K.J.; Sukits, S.F.; Svenson, K.; Bard, J.; Ellingboe, J.W.; et al. Discovery of proline sulfonamides as potent and selective hepatitis C virus NS5b polymerase inhibitors. Evidence for a new NS5b polymerase binding site. J. Med. Chem 2006, 49, 3052–3055. [Google Scholar]
- Li, L.S.; Zhou, Y.; Murphy, D.E.; Stankovic, N.; Zhao, J.; Dragovich, P.S.; Bertolini, T.; Sun, Z.; Ayida, B.; Tran, C.V.; et al. Novel HCV NS5B polymerase inhibitors derived from 4-(1′,1′-dioxo- 1′,4′-dihydro-1′lambda(6)-benzo[1′,2′,4′]thiadiazin-3′-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 3: Further optimization of the 2-, 6-, and 7′-substituents and initial pharmacokinetic assessments. Bioorg. Med. Chem. Lett 2008, 18, 3446–3455. [Google Scholar]
- Powers, J.P.; Piper, D.E.; Li, Y.; Mayorga, V.; Anzola, J.; Chen, J.M.; Jaen, J.C.; Lee, G.; Liu, J.; Peterson, M.G.; et al. SAR and mode of action of novel non-nucleoside inhibitors of hepatitis C NS5b RNA polymerase. J. Med. Chem 2006, 49, 1034–1046. [Google Scholar]
- Ruebsam, F.; Tran, C.V.; Li, L.S.; Kim, S.H.; Xiang, A.X.; Zhou, Y.; Blazel, J.K.; Sun, Z.; Dragovich, P.S.; Zhao, J.; et al. 5,6-Dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase. Bioorg. Med. Chem. Lett 2009, 19, 451–458. [Google Scholar]
- Ruebsam, F.; Webber, S.E.; Tran, M.T.; Tran, C.V.; Murphy, D.E.; Zhao, J.; Dragovich, P.S.; Kim, S.H.; Li, L.S.; Zhou, Y.; et al. Pyrrolo[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase. Bioorg. Med. Chem. Lett 2008, 18, 3616–3621. [Google Scholar]
- Tedesco, R.; Chai, D.; Darcy, M.G.; Dhanak, D.; Fitch, D.M.; Gates, A.; Johnston, V.K.; Keenan, R.M.; Lin-Goerke, J.; Sarisky, R.T.; et al. Synthesis and biological activity of heteroaryl 3-(1,1-dioxo-2H-(1,2,4)-benzothiadizin-3-yl)-4-hydroxy-2(1H)-quinolinone derivatives as hepatitis C virus NS5B polymerase inhibitors. Bioorg. Med. Chem. Lett 2009, 19, 4354–4358. [Google Scholar]
- MOE (The Molecular Operating Environment), version 2010; Chemical Computing Group Inc: Montreal, Canada, 2010.
- SONNIA Software, version 4; Molecular Networks GmbH: Erlangen, Germany, 2010.
- Sadowski, J.; Gasteiger, J. From atoms and bonds to three-dimensional atomic coordinates: Automatic model builders. J. Chem. Rev 1993, 93, 2567–2581. [Google Scholar]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem 2002, 45, 2615–2623. [Google Scholar]
- Rodgers, J.L.; Nicewander, W.A. Thirteen ways to look at the correlation coefficient. Am. Stat 1988, 42, 59–66. [Google Scholar]
- Cortes, C.; Vapnik, V.N. Support-vector networks. Mach. Learn 1995, 20, 273–297. [Google Scholar]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machine. 2001. Available online: http://www.csie.ntu.edu.tw/~cjlin/libsvm accessed on 22 March 2012.
| Activity | InertiaZ | HAcc | NAtoms | NViolationsRo5 | LogS | InertiaX | Span | HDon | HDon_N | NRotBond | RComplexity | Dipole | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| InertiaZ | 0.540 | 1 | |||||||||||
| HAcc | 0.492 | 0.794 | 1 | ||||||||||
| NAtoms | 0.457 | 0.812 | 0.841 | 1 | |||||||||
| NViolationsRo5 | 0.452 | 0.770 | 0.734 | 0.607 | 1 | ||||||||
| LogS | −0.439 | −0.735 | −0.404 | −0.607 | −0.475 | 1 | |||||||
| InertiaX | 0.426 | 0.830 | 0.720 | 0.822 | 0.673 | −0.721 | 1 | ||||||
| Span | 0.403 | 0.824 | 0.677 | 0.760 | 0.501 | −0.625 | 0.667 | 1 | |||||
| HDon | 0.397 | 0.629 | 0.724 | 0.655 | 0.515 | −0.241 | 0.569 | 0.523 | 1 | ||||
| HDon_N | 0.364 | 0.653 | 0.604 | 0.503 | 0.553 | −0.314 | 0.460 | 0.576 | 0.792 | 1 | |||
| NRotBond | 0.323 | 0.686 | 0.681 | 0.759 | 0.615 | −0.431 | 0.702 | 0.658 | 0.461 | 0.348 | 1 | ||
| RComplexity | 0.298 | 0.280 | 0.426 | 0.497 | −0.007 | −0.246 | 0.266 | 0.483 | 0.366 | 0.259 | 0.077 | 1 | |
| Dipole | 0.296 | 0.303 | 0.416 | 0.464 | 0.067 | −0.306 | 0.322 | 0.534 | 0.288 | 0.237 | 0.179 | 0.794 | 1 |
| Eccentric | 0.168 | 0.275 | 0.139 | −0.039 | 0.189 | −0.018 | −0.241 | 0.267 | 0.089 | 0.374 | −0.045 | 0.009 | −0.008 |
| Activity | Description of Selected Descriptors | |
|---|---|---|
| Span | 0.403 | Radius of the smallest sphere centered at the center of mass which completely encloses all atoms in the molecule |
| NRotBond | 0.323 | Number of open-chain, single rotatable bonds |
| LogS | −0.439 | Solubility of the molecule in water in [log units] |
| InertiaZ | 0.540 | Principal component of the inertia tensor in z-direction |
| InertiaX | 0.426 | Principal component of the inertia tensor in x-direction |
| 2DACorr_TotChg_11 | −0.277 | The eleventh component of 2D autocorrelation coefficients for σ and π charges, where the distance d = 10 |
| 2DACorr_TotChg_1 | 0.523 | The first component of 2D autocorrelation coefficients for σ and π charges, where the distance d = 0 |
| 2DACorr_SigChg_4 | −0.452 | The fourth component of 2D autocorrelation coefficients for σ charge, where the distance d = 3 |
| 2DACorr_SigChg_3 | 0.272 | The third component of 2D autocorrelation coefficients for σ charge, where the distance d = 2 |
| 2DACorr_SigChg_2 | −0.249 | The second component of 2D autocorrelation coefficients for σ charge, where the distance d = 1 |
| 2DACorr_PiChg_10 | 0.326 | The tenth component of 2D autocorrelation coefficients for π charges, where the distance d = 9 |
| 2DACorr_LpEN_8 | 0.305 | The eighth component of 2D autocorrelation coefficient for lone pair electronegativities, where the distance d = 7 |
| 2DACorr_LpEN_6 | 0.582 | The sixth component of 2D autocorrelation coefficient for lone pair electronegativities, where the distance d = 5 |
| 2DACorr_LpEN_4 | 0.198 | The fourth component of 2D autocorrelation coefficient for lone pair electronegativities, where the distance d = 3 |
| 2DACorr_LpEN_10 | 0.166 | The tenth component of 2D autocorrelation coefficient for lone pair electronegativities, where the distance d = 9 |
| 2DACorr_Ident_11 | 0.421 | The eleventh component of 2D autocorrelation coefficient for identity, where the distance d = 10 |
| Activity | Description of Selected Descriptors | |
|---|---|---|
| HDon | 0.397 | Number of hydrogen bonding donors derived from the sum of N-H and O-H groups in the molecule |
| HAcc_N | 0.431 | Number of hydrogen bonding acceptors derived from the nitrogen atoms in the molecule |
| HAcc_O | 0.417 | Number of hydrogen bonding acceptors derived from the oxygen atoms in the molecule |
| LogS | −0.439 | Solubility of the molecule in water in [log units] |
| NRotBond | 0.323 | Number of open-chain, single rotatable bonds |
| InertiaX | 0.426 | Principal component of the inertia tensor in x-direction |
| InertiaZ | 0.540 | Principal component of the inertia tensor in z-direction |
| Span | 0.403 | Radius of the smallest sphere centered at the center of mass which completely encloses all atoms in the molecule |
| Eccentric | 0.168 | Molecular eccentricity [19] |
| 3DACorr_SigChg_2 | −0.210 | 3D autocorrelation weighted by σ atom charges, where d is in the range of 2–3 Å |
| 3DACorr_SigChg_6 | −0.364 | 3D autocorrelation weighted by σ atom charges, where d is in the range of 6–7 Å |
| 3DACorr_SigChg_7 | 0.345 | 3D autocorrelation weighted by σ atom charges, where d is in the range of 7–8 Å |
| 3DACorr_PiChg_4 | −0.165 | 3D autocorrelation weighted by π atom charges, where d is in the range of 4–5 Å |
| 3DACorr_PiChg_10 | 0.166 | 3D autocorrelation weighted by π atom charges, where d is in the range of 10–11 Å |
| 3DACorr_TotChg_1 | −0.514 | 3D autocorrelation weighted by total atom charges (sum of σ, π charges), where d is in the range of 1–2 Å |
| 3DACorr_TotChg_7 | 0.348 | 3D autocorrelation weighted by total atom charges (sum of σ, π charges), where d is in the range of 7–8 Å |
| 3DACorr_PiEN_7 | 0.436 | 3D autocorrelation weighted by π atom electronegativities, where d is in the range of 7–8 Å |
| 3DACorr_LpEN_5 | 0.413 | 3D autocorrelation weighted by lone pair electronegativities, where d is in the range of 5–6 Å |
| 3DACorr_LpEN_12 | 0.350 | 3D autocorrelation weighted by lone pair electronegativities, where d is in the range of 12–13 Å |
| Model | Number of Descriptors | Number of Compounds | Training Set | Test Set | |||
|---|---|---|---|---|---|---|---|
| Training Set/Test Set | Accuracy | SE b | SP c | Accuracy | MCC d | ||
| Model 1 | 13 | 266/102 | 87.97% | 97.92% | 61.11% | 78.43% | 0.625 |
| Model 2 | 16 | 266/102 | 95.49% | 100% | 77.78% | 88.24% | 0.789 |
| Model 3 | 19 | 266/102 | 95.11% | 100% | 64.81% | 81.37% | 0.681 |
| Model | Number of Descriptors | Number of Compounds | SE b | SP c | Accuracy | MCC d |
|---|---|---|---|---|---|---|
| Model 1 | 13 | 80 | 92.11% | 80.95% | 86.25% | 0.732 |
| Model 2 | 16 | 80 | 92.11% | 69.05% | 80.00% | 0.623 |
| Model 3 | 19 | 80 | 65.79% | 54.76% | 60.00% | 0.206 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, M.; Wang, K.; Yan, A.; Yu, C. Classification of HCV NS5B Polymerase Inhibitors Using Support Vector Machine. Int. J. Mol. Sci. 2012, 13, 4033-4047. https://doi.org/10.3390/ijms13044033
Wang M, Wang K, Yan A, Yu C. Classification of HCV NS5B Polymerase Inhibitors Using Support Vector Machine. International Journal of Molecular Sciences. 2012; 13(4):4033-4047. https://doi.org/10.3390/ijms13044033
Chicago/Turabian StyleWang, Maolin, Kai Wang, Aixia Yan, and Changyuan Yu. 2012. "Classification of HCV NS5B Polymerase Inhibitors Using Support Vector Machine" International Journal of Molecular Sciences 13, no. 4: 4033-4047. https://doi.org/10.3390/ijms13044033




