Flavonoid Biosynthesis Genes Putatively Identified in the Aromatic Plant Polygonum minus via Expressed Sequences Tag (EST) Analysis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterisation of the P. minus Standard Library and Normalized Full-Length Enriched cDNA Libraries
2.2. Single-Pass EST Clone Sequencing, Assembly and Annotation
2.3. Transcription Profile Analysis
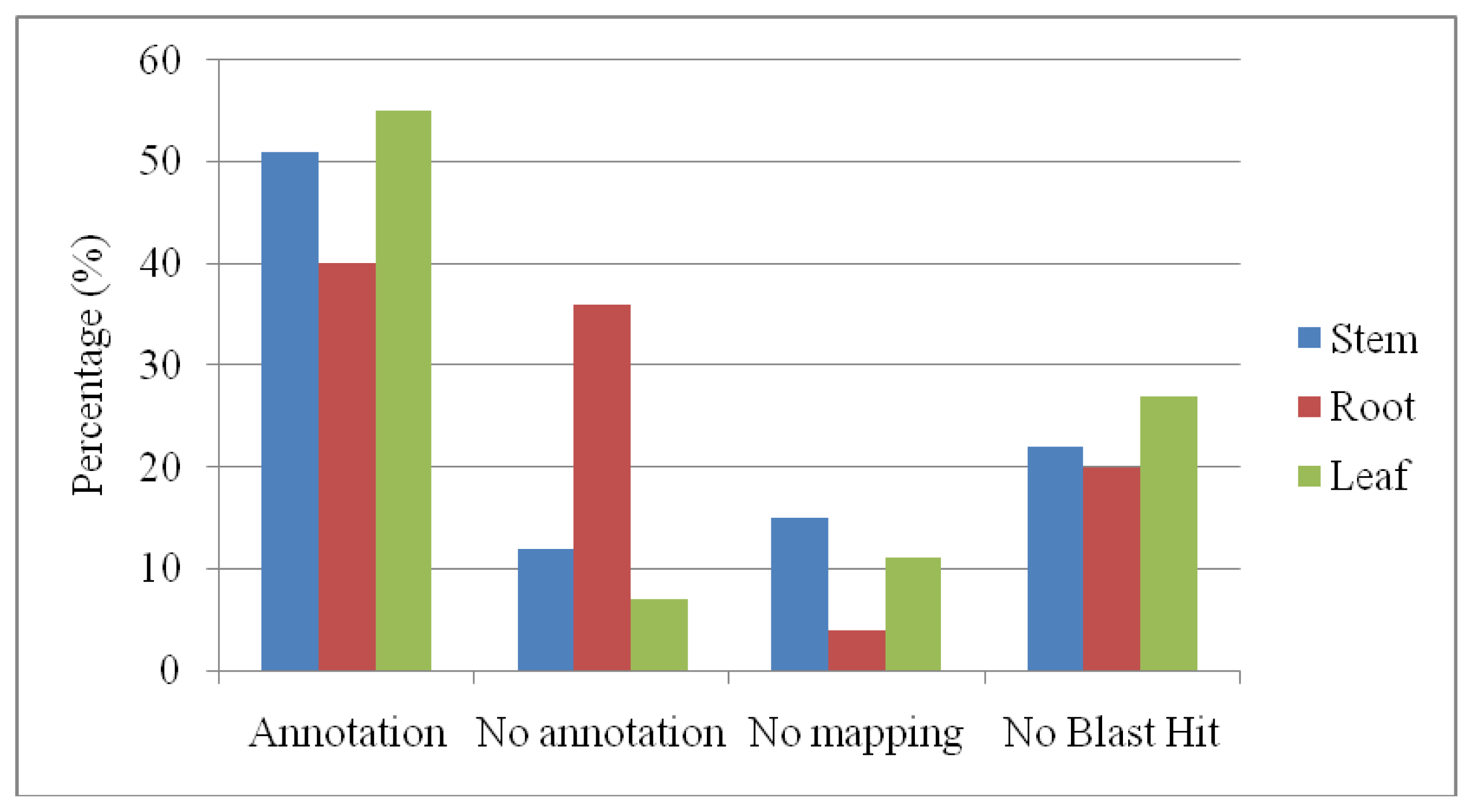
2.4. Gene Ontology Annotations
2.5. KEGG-Based Biochemical Analysis
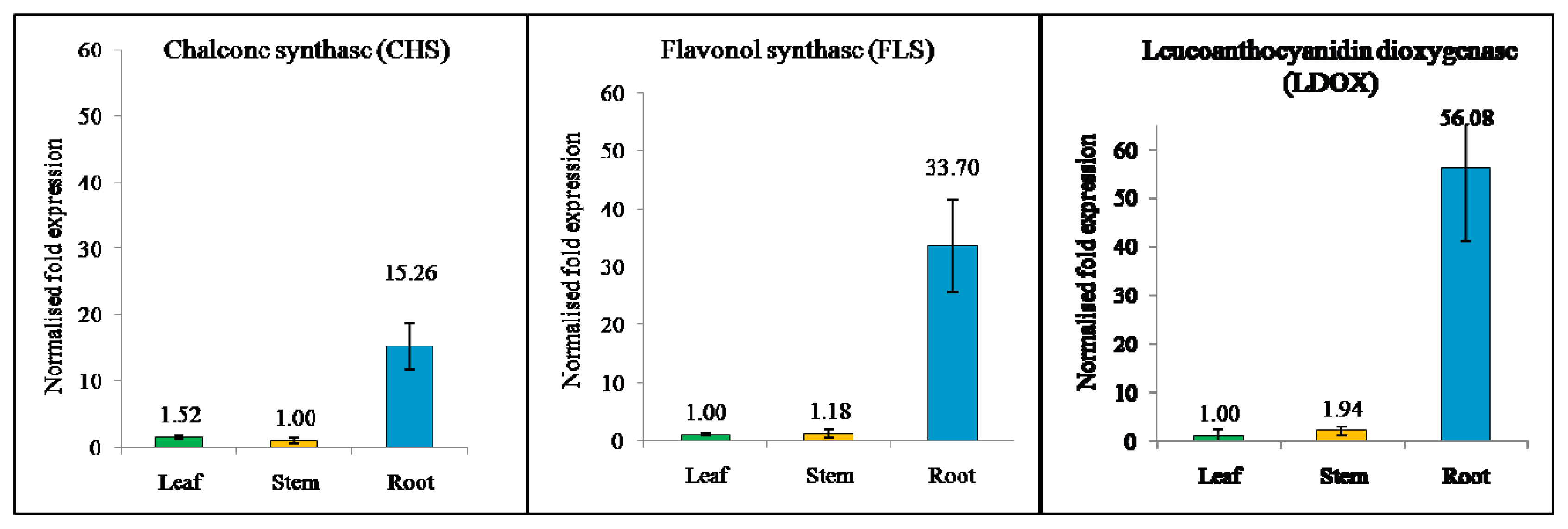
2.6. Expression of Flavonoid Biosynthesis-Related Genes
3. Experimental Section
3.1. Plant Material and Sample Collection
3.2. Construction of Normal cDNA Library from P. minus Leaf
3.3. Construction of Normalized Full-Length Enriched cDNA Libraries from P. minus Root and Stem
3.4. Single-Pass Sequencing
3.5. EST Clustering and Annotations
3.6. qRT-PCR of Selected Flavonoid Related Genes
3.7. Statistical Analysis
4. Conclusions
Supplementary Information
ijms-13-02692-s001.pdfAcknowledgments
References
- Kumar, J.; Gupta, P.K. Molecular approaches for improvement of medicinal and aromatic plants. Plant Biotechnol. Rep 2008, 2, 93–112. [Google Scholar]
- Gómez-Galera, S.; Pelacho, A.M.; Gené, A.; Capell, T.; Christou, P. The genetic manipulation of medicinal and aromatic plants. Plant Cell Rep 2007, 26, 1689–1715. [Google Scholar]
- Azlim Almey, A.A.; Ahmed Jalal Khan, C.; Syed Zahir, I.; Mustapha Suleiman, K.; Aisyah, M.R.; Kamarul Rahim, K. Total phenolic content and primary antioxidant activity of methanolic and ethanolic extracts of aromatic plants’ leaves. J. Food Res. Int 2010, 17, 1077–1084. [Google Scholar]
- Huda-Faujan, N.; Noriham, A.; Norrakiah, A.S.; Babji, A.S. Antioxidative activities of water extracts of some Malaysian herbs. ASEAN Food J 2007, 14, 61–68. [Google Scholar]
- Urones, J.G.; Marcos, I.S.; Pérez, B.G.; Barcala, P.B. Flavonoids from Polygonum minus. Phytochemistry 1990, 29, 3687–3689. [Google Scholar]
- Yaacob, K.B. Kesom oil-A natural source of aliphatic aldehydes. Perfum. Flavor 1987, 12, 27–30. [Google Scholar]
- Baharum, S.N.; Bunawan, H.; Ghani, M.A.; Wan Aida Wan, M.; Noor, N.M. Analysis of the chemical composition of the essential oil of Polygonum minus Huds. Using two-dimensional gas chromatography-time-of-flight mass spectrometry (GC-TOF MS). Molecules 2010, 15, 7006–7015. [Google Scholar]
- Asamizu, E.; Nakamura, Y.; Sato, S.; Tabata, S. A large scale analysis of cDNA in Arabidopsis thaliana: Generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res 2000, 7, 175–180. [Google Scholar]
- Yamamoto, K.; Sasaki, T. Large-scale EST sequencing in rice. Plant Mol. Biol 1997, 35, 135–144. [Google Scholar]
- Sudo, H.; Seki, H.; Sakurai, N.; Suzuki, H.; Shibata, D.; Toyoda, A.; Totoki, Y.; Sakaki, Y.; Lida, O.; Shibata, T.; et al. Expressed sequence tags from rhizomes of Glycyrrhiza uralensis. Plant Biotechnol 2009, 26, 105–107. [Google Scholar]
- Sathiyamoorthy, S.; In, J.-G.; Gayathri, S.; Kim, Y.-J.; Yang, D.C. Generation and gene ontology based analysis of expressed sequence tags (EST) from a Panax ginseng C. A. Meyer roots. Mol. Biol. Rep 2010, 37, 3465–3472. [Google Scholar]
- Senthil, K.; Wasnik, N.; Kim, Y.-J.; Yang, D.C. Generation and analysis of expressed sequence tags from leaf and root of Withania somnifera (Ashwgandha). Mol. Biol. Rep 2010, 37, 893–902. [Google Scholar]
- Park, J.-S.; Kim, J.-B.; Hahn, B.-S.; Kim, K.-H.; Ha, S.-H.; Kim, J.-B.; Kim, Y.-H. EST analysis of genes involved in secondary metabolism in Camellia sinensis (tea), using suppression subtractive hybridization. Plant Sci 2004, 166, 953–961. [Google Scholar]
- López, S.N.; Sierra, M.G.; Gattuso, S.J.; Furlán, R.L.; Zacchino, S.A. An unusual homoisoflavanone and a structurally-related dihydrochalcone from Polygonum ferrugineum (Polygonaceae). Phytochemistry 2006, 67, 2152–2158. [Google Scholar]
- Peng, Z.F.; Strack, D.; Baumert, A.; Subramaniam, R.; Goh, N.K.; Chia, T.F.; Ngin Tan, S.; Chia, L.S. Antioxidant flavonoids from leaves of Polygonum hydropiper L. Phytochemistry 2003, 62, 219–228. [Google Scholar]
- De Fatima Bonaldo, M.; Lennon, G.; Soares, M.B. Normalization and subtraction: Two approaches to facilitate gene discovery. Genome Res 1996, 6, 791–806. [Google Scholar]
- Marques, M.C.; Alonso-Cantabrana, H.; Forment, J.; Arribas, R.; Alamar, S.; Conejero, V.; Perez-Amador, M.A. A new set of ESTs and cDNA clones from full-length and normalized libraries for gene discovery and functional characterization in citrus. BMC Genomics 2009, 10, 428. [Google Scholar]
- Bausher, M.; Shatters, R.; Chaparro, J.; Dang, P.; Hunter, W.; Niedz, R. An expressed sequence tag (EST) set from Citrus sinensis L. Osbeck whole seedlings and the implications of further perennial source investigations. Plant Sci 2003, 165, 415–422. [Google Scholar]
- Yazaki, K. ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett 2006, 580, 1183–1191. [Google Scholar]
- Martinoia, E.; Klein, M.; Geisler, M.; Bovet, L.; Forestier, C.; Kolukisaoglu, Ü.; Müller-Röber, B.; Schulz, B. Multifunctionality of plant ABC transporters—more than just detoxifiers. Planta 2002, 214, 345–355. [Google Scholar]
- Moons, A. Transcriptional profiling of the gene family in rice roots in response to plant growth regulators, redox perturbations and weak organic acid stresses. Planta 2008, 229, 53–71. [Google Scholar]
- Lee, M.; Lee, K.; Lee, J.; Noh, E.W.; Lee, Y. AtPDR12 contributes to lead resistance in Arabidopsis. Plant Physiol 2005, 138, 827–836. [Google Scholar]
- Rizhsky, L.; Liang, H.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 2002, 130, 1143–1151. [Google Scholar]
- Molina, A.; Segura, A.; García-Olmedo, F. Lipid transfer proteins (nsLTPs) from barley and maize leaves are potent inhibitors of bacterial and fungal plant pathogens. FEBS Lett 1993, 316, 119–122. [Google Scholar] [Green Version]
- Brandle, J.E.; Richman, A.; Swanson, A.K.; Chapman, B.P. Leaf ESTs from Stevia rebaudiana: A resource for gene discovery in diterpene synthesis. Plant Mol. Biol 2002, 50, 613–622. [Google Scholar]
- Jeter, C.R.; Roux, S.J. Plant responses to extracellular nucleotides: Cellular processes and biological effects. Purinergic Sig 2006, 2, 443–449. [Google Scholar]
- Pelletier, M.K.; Murrell, J.R.; Shirley, B.W. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in arabidopsis: Further evidence for differential regulation of “early” and “late” genes. Plant Physiol 1997, 113, 1427–1445. [Google Scholar]
- Cook, N.C.; Samman, S. Flavonoids—Chemistry, metabolism, cardioprotective effects, and dietary sources. J. Nut. Biochem 1996, 7, 66–76. [Google Scholar]
- Yonekura-Sakakibara, K.; Saito, K. Functional genomics for plant natural product biosynthesis. Nat. Prod. Rep 2009, 26, 1466–1487. [Google Scholar]
- Harker, C.L.; Ellis, T.H.N.; Coen, E.S. Identification and genetic regulation of the chalcone synthase multigene family in pea. Plant Cell 1990, 2, 185–194. [Google Scholar]
- Mahajan, M.; Ahuja, P.S.; Yadav, S.K. Post-transcriptional silencing of flavonol synthase mrna in tobacco leads to fruits with arrested seed set. PLoS one 2011, 6, e28315. [Google Scholar]
- Jaganath, I.B.; Crozier, A. Dietary Flavonoids and Phenolic Compounds. In Plant Phenolics and Human Health; John Wiley & Sons, Inc: Hoboken, NJ, USA, 2009; pp. 1–49. [Google Scholar]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071–108. [Google Scholar]
- Liu, X.; Chen, M.; Li, M.; Yang, C.; Fu, Y.; Zhang, Q.; Zeng, L.; Liao, Z. The anthocyanidin synthase gene from sweetpotato [Ipomoea batatas (L.) Lam]: Cloning, characterization and tissue expression analysis. Afr. J. Biotechnol 2010, 9, 3748–3752. [Google Scholar]
- Koes, R.E.; Quattrocchio, F.; Mol, J.N.M. The flavonoid biosynthetic pathway in plants: Function and evolution. BioEssays 1994, 16, 123–132. [Google Scholar]
- Buer, C.S.; Muday, G.K.; Djordjevic, M.A. Implications of long-distance flavonoid movement in Arabidopsis thaliana. Plant Signal. Behav 2008, 3, 415–417. [Google Scholar]
- Lopez-Gomez, R.; Gomez Lim, M.A. Method for extracting intact RNA from fruits rich in polysaccharides using ripe mango mesocarp. HortScience 1992, 27, 440–442. [Google Scholar]
| Features | Standard leaf library | Normalized stem library | Normalized root library |
|---|---|---|---|
| Total number of clones sequenced | 3260 | 2016 | 2016 |
| Number of high-quality sequences | 1977 | 1398 | 1767 |
| Average length of high-quality ESTs (bp) | 630 bp | 600 bp | 600 bp |
| Number of consensus/contigs | 392 contigs | 92 consensus | 130 consensus |
| Number of singletons | 922 | 1179 | 1481 |
| Number of unigene transcripts (UT) | 1314 a | 1271 a | 1611 a |
| Redundant ESTs | 663 (33.5) b | 127 (9) b | 156 (8.8) b |
| Functional analysis: | |||
| UT with GO match | 731 (55.6) c | 657 (51.6) c | 636 (39.4) c |
| Molecular function | 1392 | 1036 | 832 |
| Biological process | 1779 | 1482 | 1321 |
| Cellular component | 816 | 692 | 918 |
| UTs without GO match | 583 (44.4) c | 614 (48.4) c | 975 (60.6) c |
| (a) | |||||
|---|---|---|---|---|---|
| UT ID | EST Count | Description | Organism Source | Identity | Accession |
| root_cn10 | 2 | Multidrug/pheromone exporter, MDR family, ABC transporter family | Populus trichocarpa | 50 | JG732266 |
| root_cn11 | 2 | Hypothetical protein LOC100280328 (putative ABC transporter) | Zea mays | 86 | JG732267 |
| root_cn8 | 2 | Os03g0641200 (Amino acid/polyamine transporter I family protein) | Oryza sativa Japonica Group | 66 | JG732264 |
| root_cn1 | 2 | Dipeptidyl-peptidase, putative | Ricinus communis | 84 | JG732257 |
| root_cn2 | 2 | Calcium-binding EF hand family protein | Arabidopsis thaliana | 49 | JG732258 |
| root_cn3 | 2 | Cation-transporting ATPase plant, putative | Ricinus communis | 78 | JG732259 |
| root_cn4 | 2 | Unnamed protein product | Vitis vinifera | 84 | JG732260 |
| root_cn7 | 2 | Ormdl, putative | Ricinus communis | 89 | JG732263 |
| root_cn12 | 2 | Small glutamine-rich tetratricopeptide repeat-containing protein A, putative (co-chaperon) | Ricinus communis | 70 | JG732268 |
| root_cn14 | 2 | Predicted protein | Populus trichocarpa | 48 | JG732270 |
| (b) | |||||
|---|---|---|---|---|---|
| UT ID | EST Count | Description | Organism Source | Identity | Accession |
| stem_cn65 | 4 | Photosystem II protein K | - | 62 | JG700241 |
| stem_cn33 | 3 | Predicted protein | Populus trichocarpa | 99 | JG700209 |
| stem_cn42 | 3 | Predicted: similar to small nuclear ribonucleoprotein E | Vitis vinifera | 94 | JG700218 |
| stem_cn83 | 3 | Os12g0207300 (Clathrin coat assembly protein AP17) | Oryza sativa Japonica Group | 98 | JG700257 |
| stem_cn39 | 2 | Transcription elongation factor, putative (TF) | Ricinus communis | 91 | JG700215 |
| stem_cn21 | 2 | GATA transcription factor, putative (TF) | Ricinus communis | 81 | JG700197 |
| stem_cn91 | 2 | GRAS family transcription factor (TF) | Populus trichocarpa | 67 | JG700265 |
| stem_cn35 | 2 | ARF domain class transcription factor (TF) | Malus x domestica | 48 | JG700211 |
| stem_cn1 | 2 | Dehydration-induced protein | Aegiceras corniculatum | 50 | JG700177 |
| S005.A07 | 1 | WRKY Protein (TF) | Rheum australe | 43 | JG701357 |
| S017.A08 | 1 | MYBR domain class transcription factor (TF) | Malus x domestica | 71 | JG700745 |
| (c) | |||||
|---|---|---|---|---|---|
| UT ID | EST count | Description | Organism Source | Identity | Accession |
| leaf_cn134 | 420 | Putative ribulose-1,5-biphosphate carboxylase small subunit precursor | Rumex obtusifolius | 85 | JG745178 |
| leaf_cn312 | 140 | Lipid transfer protein | Dianthus caryophyllus | 52 | JG745356 |
| leaf_cn185 | 110 | Chlorophyll a-b binding protein 3C-like | Solanum tuberosum | 94 | JG745226 |
| leaf_cn4 | 99 | Hairpin-induced protein | Rheum australe | 69 | JG745048 |
| leaf_cn278 | 63 | S-adenosylmethionine synthase | - | 93 | JG745322 |
| leaf_cn184 | 60 | AF220527_1 chlorophyll a/b binding protein precursor | Euphorbia esula | 91 | JG745229 |
| leaf_cn20 | 55 | 14 kDa proline-rich protein DC2.15 precursor, putative | Ricinus communis | 77 | JG745064 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Roslan, N.D.; Yusop, J.M.; Baharum, S.N.; Othman, R.; Mohamed-Hussein, Z.-A.; Ismail, I.; Noor, N.M.; Zainal, Z. Flavonoid Biosynthesis Genes Putatively Identified in the Aromatic Plant Polygonum minus via Expressed Sequences Tag (EST) Analysis. Int. J. Mol. Sci. 2012, 13, 2692-2706. https://doi.org/10.3390/ijms13032692
Roslan ND, Yusop JM, Baharum SN, Othman R, Mohamed-Hussein Z-A, Ismail I, Noor NM, Zainal Z. Flavonoid Biosynthesis Genes Putatively Identified in the Aromatic Plant Polygonum minus via Expressed Sequences Tag (EST) Analysis. International Journal of Molecular Sciences. 2012; 13(3):2692-2706. https://doi.org/10.3390/ijms13032692
Chicago/Turabian StyleRoslan, Nur Diyana, Jastina Mat Yusop, Syarul Nataqain Baharum, Roohaida Othman, Zeti-Azura Mohamed-Hussein, Ismanizan Ismail, Normah Mohd Noor, and Zamri Zainal. 2012. "Flavonoid Biosynthesis Genes Putatively Identified in the Aromatic Plant Polygonum minus via Expressed Sequences Tag (EST) Analysis" International Journal of Molecular Sciences 13, no. 3: 2692-2706. https://doi.org/10.3390/ijms13032692
APA StyleRoslan, N. D., Yusop, J. M., Baharum, S. N., Othman, R., Mohamed-Hussein, Z.-A., Ismail, I., Noor, N. M., & Zainal, Z. (2012). Flavonoid Biosynthesis Genes Putatively Identified in the Aromatic Plant Polygonum minus via Expressed Sequences Tag (EST) Analysis. International Journal of Molecular Sciences, 13(3), 2692-2706. https://doi.org/10.3390/ijms13032692




