Multi-Parametric Profiling Network Based on Gene Expression and Phenotype Data: A Novel Approach to Developmental Neurotoxicity Testing
Abstract
:1. Introduction
2. Results and Discussion
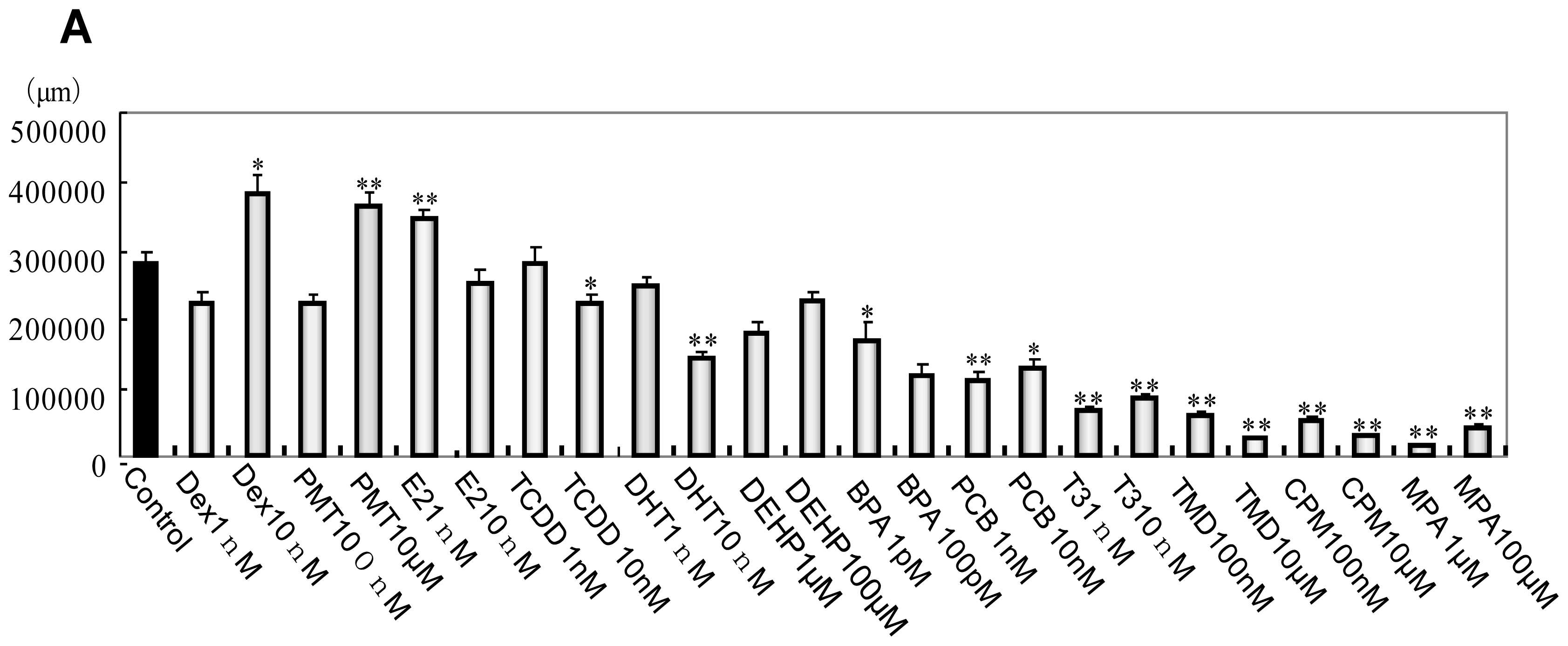
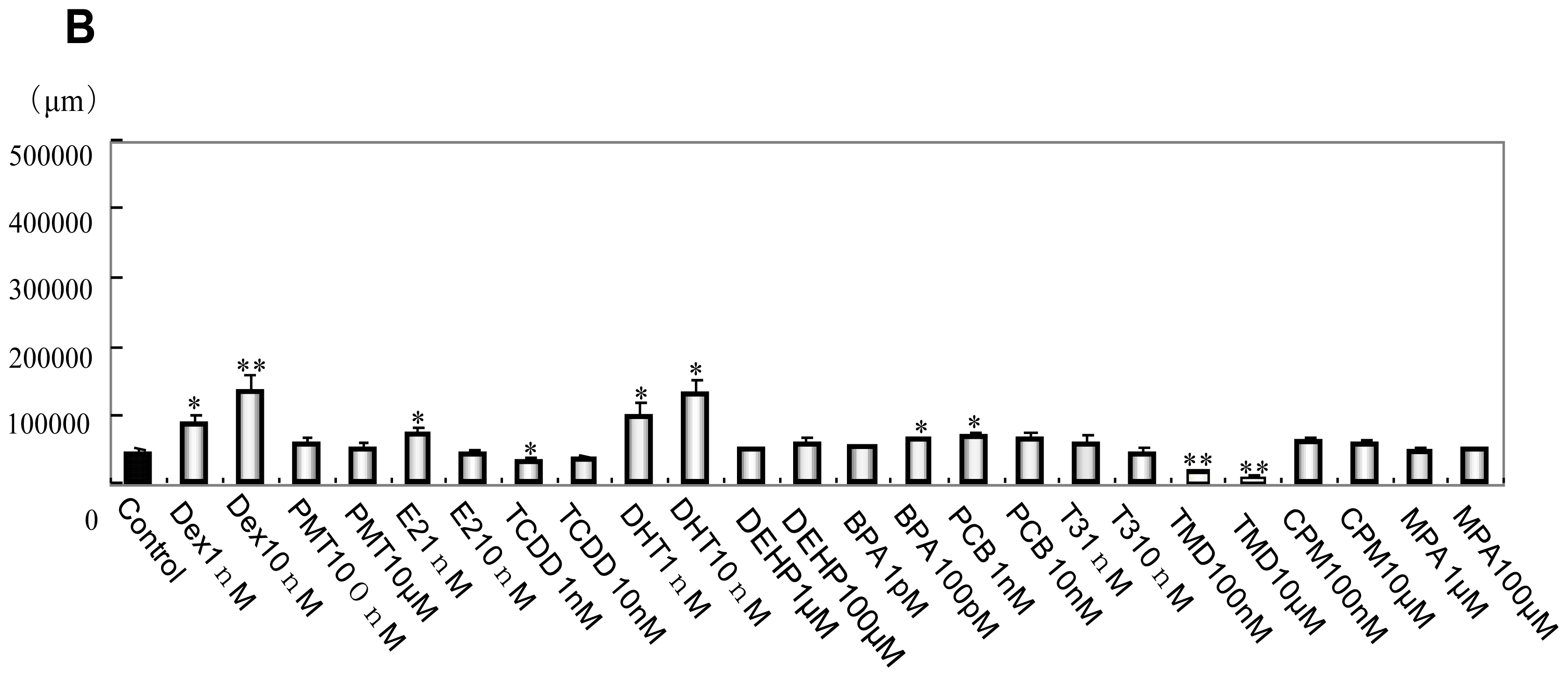
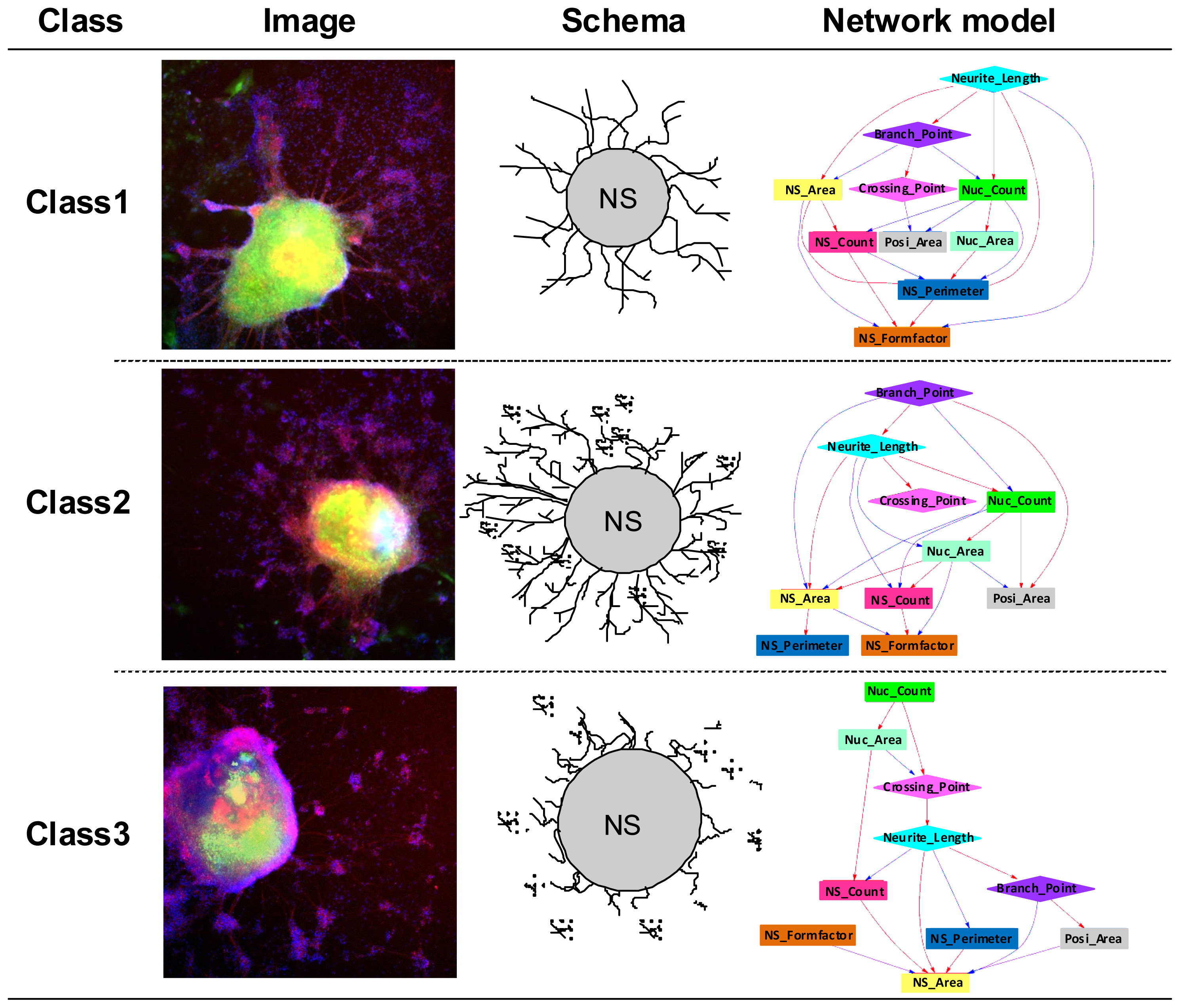
2.1. Phenotype Profiling Based on the Morphology of Differentiated Neuronal Cells by High-Content Image Analysis and Generation of Phenotypic Networks
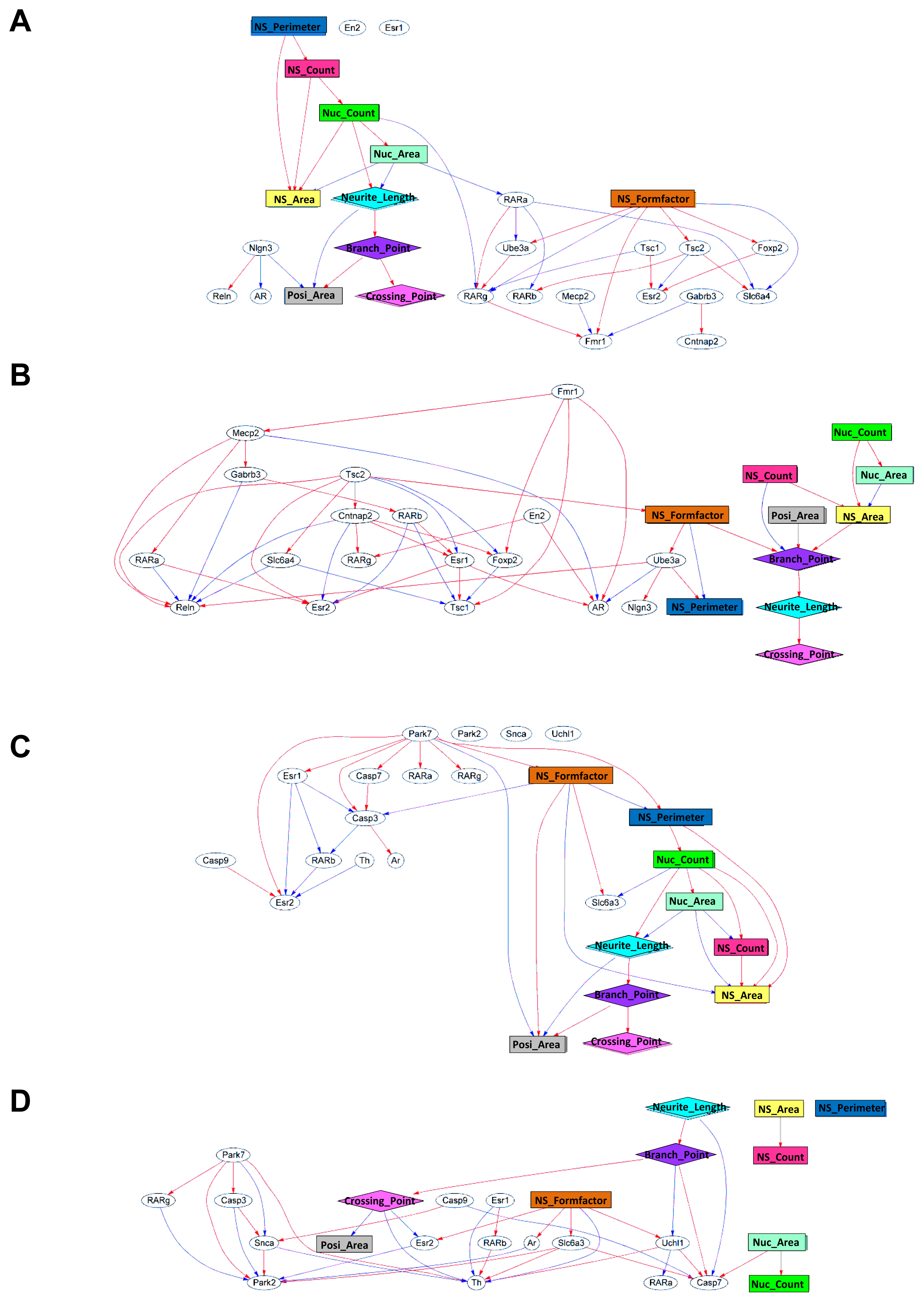
2.2. Generation of a Comprehensive Network Based on Gene Expression and Phenotype Profiling by a Bayesian Network Model
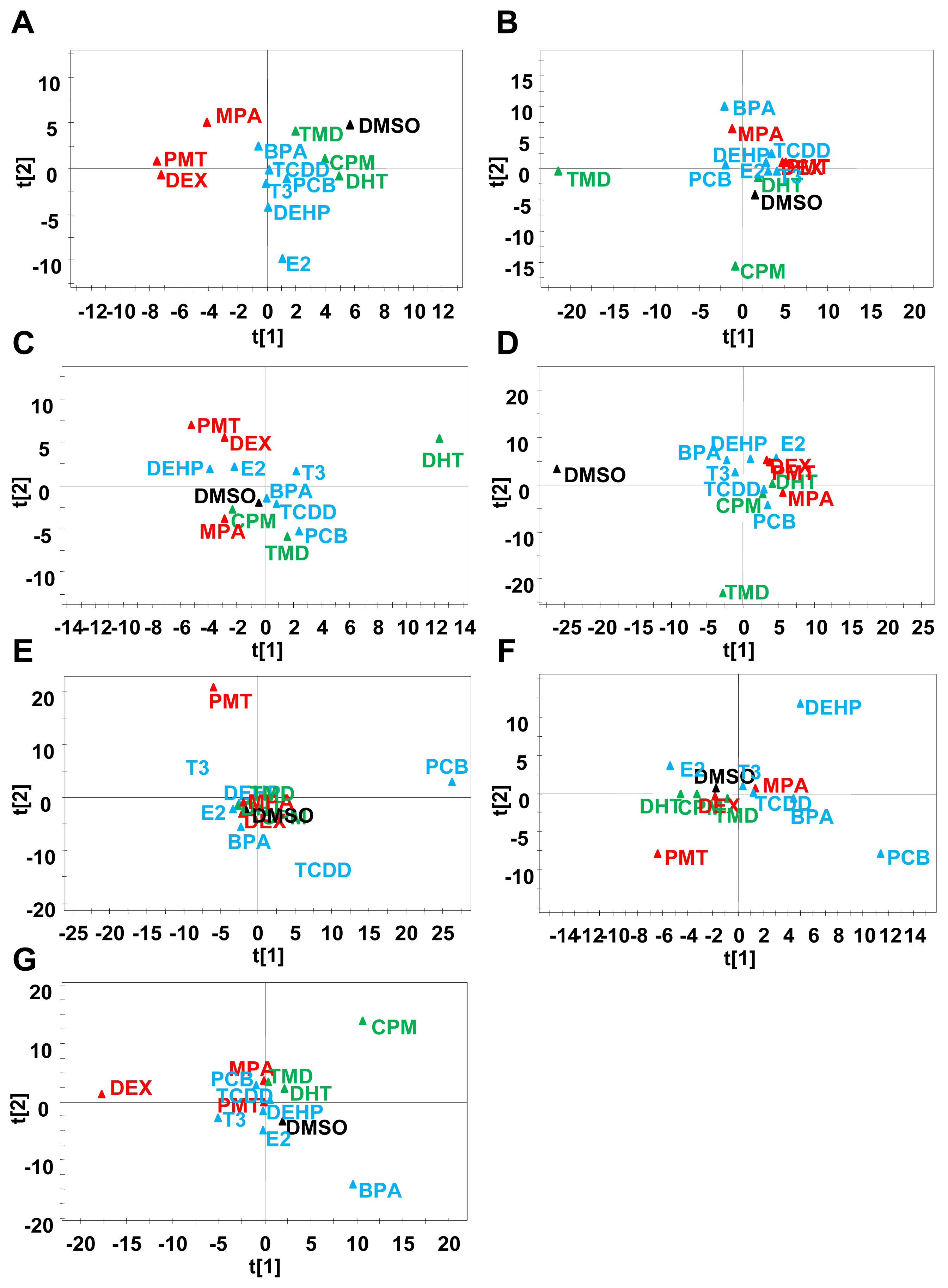
2.3. Classification of Chemicals Based on the Values of the Parameters of the Comprehensive Networks
2.4. Discussion for Future Work
3. Experimental Section
3.1. Selection of Test Chemicals
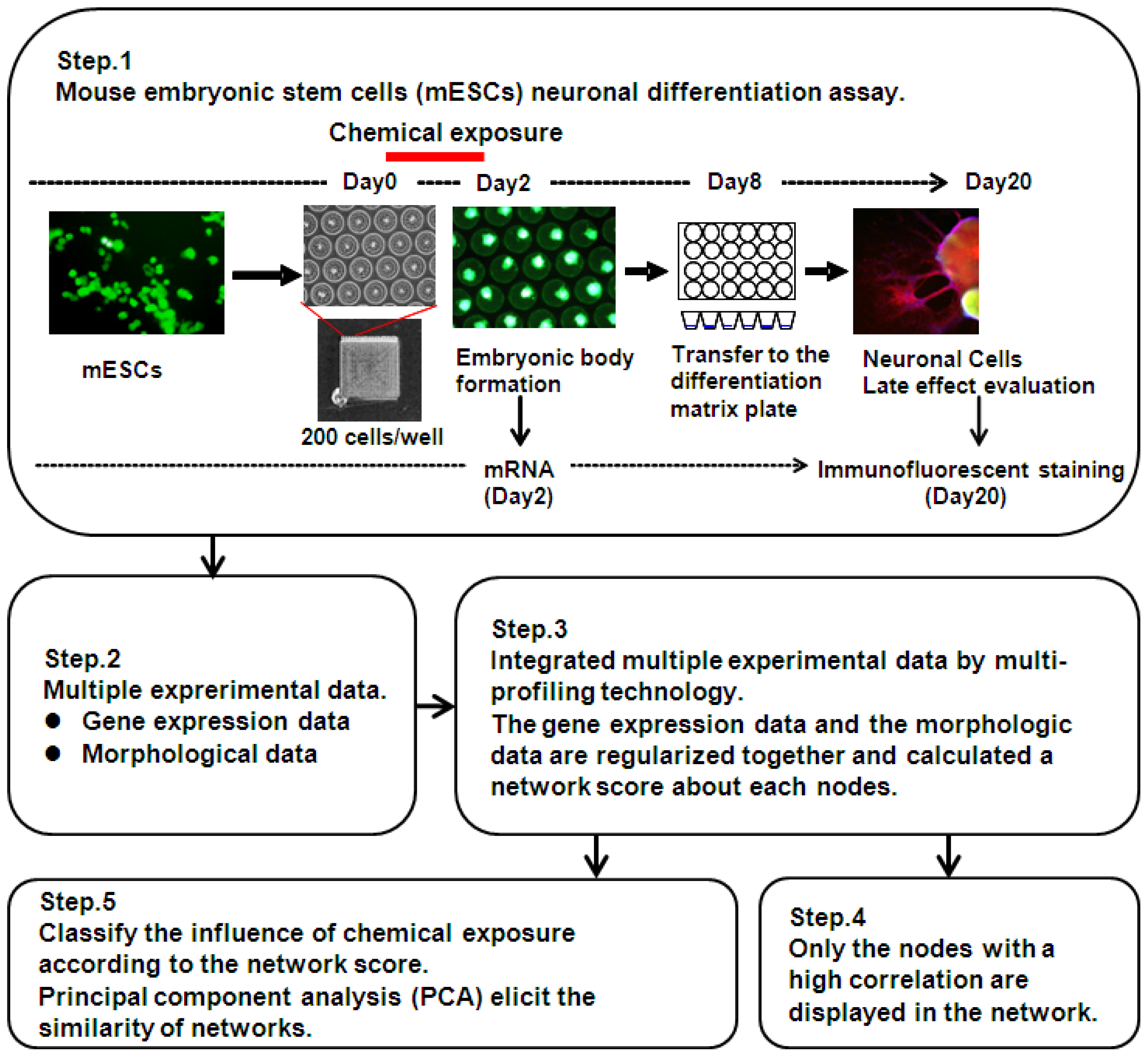
3.2. Design of Multi-Parametric Profiling Networks Analysis for Detecting Developmental Neuronal Toxicity of Chemicals That Effects Fetal Programming
3.3. mESC Culture and Maintenance
3.4. EB Formation from mESC and Chemical Treatment
3.5. Immunofluorescence
3.6. Morphological Analysis of mESC, EB and Neuronal Cell Lineages
3.7. Gene Expression Analysis and Creation of Candidate Gene Sets
3.8. Gene and Morphology Interaction Network Analysis
3.9. Statistical Analysis
4. Conclusions
Acknowledgments
References
- Guilloteau, P.; Zabielski, R.; Hammon, H.M.; Metges, C.C. Adverse effects of nutritional programming during prenatal and early postnatal life, some aspects of regulation and potential prevention and treatments. J. Physiol. Pharmacol 2009, 60, 17–35. [Google Scholar]
- Hales, C.N.; Barker, D.J. The thrifty phenotype hypothesis. Br. Med. Bull 2001, 60, 5–20. [Google Scholar]
- Heindel, J.J. Role of exposure to environmental chemicals in the developmental basis of reproductive disease and dysfunction. Semin. Reprod. Med 2006, 24, 168–177. [Google Scholar]
- McMillen, I.C.; Robinson, J.S. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol. Rev 2005, 85, 571–633. [Google Scholar]
- Crofton, K.M.; Mundy, W.R.; Lein, P.J.; Bal-Price, A.; Coecke, S.; Seiler, A.E.; Knaut, H.; Buzanska, L.; Goldberg, A. Developmental neurotoxicity testing: Recommendations for developing alternative methods for the screening and prioritization of chemicals. ALTEX 2011, 28, 9–15. [Google Scholar]
- Chapin, R.E.; Stedman, D.B. Endless possibilities: Stem cells and the vision for toxicology testing in the 21st century. Toxicol. Sci 2009, 112, 17–22. [Google Scholar]
- Seiler, A.E.; Buesen, R.; Visan, A.; Spielmann, H. Use of murine embryonic stem cells in embryotoxicity assays: the embryonic stem cell test. Methods Mol. Biol 2006, 329, 371–395. [Google Scholar]
- van Dartel, D.A.; Piersma, A.H. The embryonic stem cell test combined with toxicogenomics as an alternative testing model for the assessment of developmental toxicity. Reprod. Toxicol 2011, 32, 235–244. [Google Scholar]
- Theunissen, P.T.; Schulpen, S.H.W.; van Dartel, D.A.M.; Hermsen, S.A.B.; van Schooten, F.J.; Piersma, A.H. An abbreviated protocol for multilineage neural differentiation of murine embryonic stem cells and its perturbation by methyl mercury. Reprod. Toxicol 2010, 29, 383–392. [Google Scholar]
- Zimmer, B.; Kuegler, P.B.; Baudis, B.; Genewsky, A.; Tanavde, V.; Koh, W.; Tan, B.; Waldmann, T.; Kadereit, S.; Leist, M. Coordinated waves of gene expression during neuronal differentiation of embryonic stem cells as basis for novel approaches to developmental neurotoxicity testing. Cell Death Differ 2011, 18, 383–395. [Google Scholar]
- Coecke, S.; Goldberg, A.M.; Allen, S.; Buzanska, L.; Calamandrei, G.; Crofton, K.; Hareng, L.; Hartung, T.; Knaut, H.; Honegger, P.; et al. Workgroup report: incorporating in vitro alternative methods for developmental neurotoxicity into international hazard and risk assessment strategies. Environ. Health Perspect 2007, 115, 924–931. [Google Scholar]
- Friedman, N.; Linial, M.; Nachman, I.; Pe’er, D. Using Bayesian networks to analyze expression data. J. Comput. Biol 2000, 7, 601–620. [Google Scholar]
- Husmeier, D. Sensitivity and specificity of inferring genetic regulatory interactions from microarray experiments with dynamic Bayesian networks. Bioinformatics 2003, 19, 2271–2282. [Google Scholar]
- Rogers, S.; Girolami, M. A Bayesian regression approach to the inference of regulatory networks from gene expression data. Bioinformatics 2005, 21, 3131–3137. [Google Scholar]
- Toyoshiba, H.; Yamanaka, T.; Sone, H.; Parham, F.M.; Walker, N.J.; Martinez, J.; Portier, C.J. Gene interaction network suggests dioxin induces a significant linkage between aryl hydrocarbon receptor and retinoic acid receptor beta. Environ. Health Perspect 2004, 112, 1217–1224. [Google Scholar]
- Yamanaka, T.; Toyoshiba, H.; Sone, H.; Parham, F.M.; Portier, C.J. The TAO-Gen algorithm for identifying gene interaction networks with application to SOS repair in E. coli. Environ. Health Perspect 2004, 112, 1614–1621. [Google Scholar]
- Jayawardhana, B.; Kell, D.B.; Rattray, M. Bayesian inference of the sites of perturbations in metabolic pathways via Markov chain Monte Carlo. Bioinformatics 2008, 24, 1191–1197. [Google Scholar]
- Tang, W.; Wu, X.; Jiang, R.; Li, Y. Epistatic module detection for case-control studies: A Bayesian model with a Gibbs sampling strategy. PLoS Genet 2009, 5. [Google Scholar] [CrossRef]
- Anchang, B.; Sadeh, M.J.; Jacob, J.; Tresch, A.; Vlad, M.O.; Oefner, P.J.; Spang, R. Modeling the temporal interplay of molecular signaling and gene expression by using dynamic nested effects models. Proc. Natl. Acad. Sci. USA 2009, 106, 6447–6452. [Google Scholar]
- Harikrishnan, K.N.; Chow, M.Z.; Baker, E.K.; Pal, S.; Bassal, S.; Brasacchio, D.; Wang, L.; Craig, J.M.; Jones, P.L.; Sif, S.; et al. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet 2005, 37, 254–264. [Google Scholar]
- Zhang, A.; Shen, C.H.; Ma, S.Y.; Ke, Y.; El Idrissi, A. Altered expression of Autism-associated genes in the brain of Fragile X mouse model. Biochem. Biophys. Res. Commun 2009, 379, 920–923. [Google Scholar]
- Chao, H.T.; Chen, H.; Samaco, R.C.; Xue, M.; Chahrour, M.; Yoo, J.; Neul, J.L.; Gong, S.; Lu, H.C.; Heintz, N.; et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature 2010, 468, 263–269. [Google Scholar]
- Goncharova, E.A.; Goncharov, D.A.; Li, H.; Pimtong, W.; Lu, S.; Khavin, I.; Krymskay, V.P. mTORC2 is required for proliferation and survival of TSC2-null cells. Mol. Cell. Biol 2011, 31, 2484–2498. [Google Scholar]
- Nasuti, C.; Gabbianelli, R.; Falcioni, M.L.; di Stefano, A.; Sozio, P.; Cantalamessa, F. Dopaminergic system modulation, behavioral changes, and oxidative stress after neonatal administration of pyrethroids. Toxicology 2007, 229, 194–205. [Google Scholar]
- Elwan, M.A.; Richardson, J.R.; Guillot, T.S.; Caudle, W.M.; Miller, G.W. Pyrethroid pesticide-induced alterations in dopamine transporter function. Toxicol. Appl. Pharmacol 2006, 211, 188–197. [Google Scholar]
- Kyoto Encyclopedia of Genes and Genomes. Available online: http://www.genome.jp/kegg/pathway/hsa/hsa05012.html (accessed on 9 January 2011).
- Profiles of Chemical Effects on Cells system. Available online: http://project.nies.go.jp/eCA/cgi-bin/index.cgi (accessed on 28 February 2007).
- Sone, H.; Okura, M.; Zaha, H.; Fujibuchi, W.; Taniguchi, T.; Akanuma, H.; Nagano, R.; Ohsako, S.; Yonemoto, J. Profiles of Chemical Effects on Cells (pCEC): A toxicogenomics database with a toxicoinformatics system for risk evaluation and toxicity prediction of environmental chemicals. J. Toxicol. Sci 2010, 35, 115–123. [Google Scholar]
- Gene Expression Omnibus. Available online: http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE18503 (accessed on 10 October 2009).
- Green, K.N.; Billings, L.M.; Roozendaal, B.; McGaugh, J.L.; LaFerla, F.M. Glucocorticoids increase amyloid-beta and tau pathology in a mouse model of Alzheimer’s disease. J. Neurosci 2006, 26, 9047–9056. [Google Scholar]
- Arguelles, S.; Herrera, A.J.; Carreno-Muller, E.; de Pablos, R.M.; Villaran, R.F.; Espinosa-Oliva, A.M.; Machado, A.; Cano, J. Degeneration of dopaminergic neurons induced by thrombin injection in the substantia nigra of the rat is enhanced by dexamethasone: Role of monoamine oxidase enzyme. Neurotoxicology 2010, 31, 55–66. [Google Scholar]
- Cummings, J.L.; Vinters, H.V.; Cole, G.M.; Khachaturian, Z.S. Alzheimer’s disease: Etiologies, pathophysiology, cognitive reserve, and treatment opportunities. Neurology 1998, 51, S2–17, discussion S65–67. [Google Scholar]
- van den Eeden, S.K.; Tanner, C.M.; Bernstein, A.L.; Fross, R.D.; Leimpeter, A.; Bloch, D.A.; Nelson, L.M. Incidence of Parkinson’s disease: Variation by age, gender, and race/ethnicity. Am. J. Epidemiol 2003, 157, 1015–1022. [Google Scholar]
- van Dartel, D.A.; Pennings, J.L.; de la Fonteyne, L.J.; van Herwijnen, M.H.; van Delft, J.H.; van Schooten, F.J.; Piersma, A.H. Monitoring developmental toxicity in the embryonic stem cell test using differential gene expression of differentiation-related genes. Toxicol. Sci 2010, 116, 130–139. [Google Scholar]
- Chandler, K.J.; Barrier, M.; Jeffay, S.; Nichols, H.P.; Kleinstreuer, N.C.; Singh, A.V.; Reif, D.M.; Sipes, N.S.; Judson, R.S.; Dix, D.J.; et al. Evaluation of 309 environmental chemicals using a mouse embryonic stem cell adherent cell differentiation and cytotoxicity assay. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Gohlke, J.M.; Armant, O.; Parham, F.M.; Smith, M.V.; Zimmer, C.; Castro, D.S.; Nguyen, L.; Parker, J.S.; Gradwohl, G.; Portier, C.J.; et al. Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol 2008, 6. [Google Scholar] [CrossRef]
- Zhao, L.; Morgan, M.A.; Parsels, L.A.; Maybaum, J.; Lawrence, T.S.; Normolle, D. Bayesian hierarchical changepoint methods in modeling the tumor growth profiles in xenograft experiments. Clin. Cancer Res 2011, 17, 1057–1064. [Google Scholar]
- Landers, J.P.; Bunce, N.J. The Ah receptor and the mechanism of dioxin toxicity. Biochem. J 1991, 276, 273–287. [Google Scholar]
- Gould, A.; Missailidis, S. Targeting the hedgehog pathway: The development of cyclopamine and the development of anti-cancer drugs targeting the hedgehog pathway. Mini. Rev. Med. Chem 2011, 11, 200–213. [Google Scholar]
- Okada, Y.; Shimazaki, T.; Sobue, G.; Okano, H. Retinoic-acid-concentration-dependent acquisition of neural cell identity during in vitro differentiation of mouse embryonic stem cells. Dev. Biol 2004, 275, 124–142. [Google Scholar]
- Vargesson, N. Thalidomide-induced limb defects: Resolving a 50-year-old puzzle. Bioessays 2009, 31, 1327–1336. [Google Scholar]
- Rodier, P.M.; Ingram, J.L.; Tisdale, B.; Nelson, S.; Romano, J. Embryological origin for autism: Developmental anomalies of the cranial nerve motor nuclei. J. Comp. Neurol 1996, 370, 247–261. [Google Scholar]
- Stromland, K.; Nordin, V.; Miller, M.; Akerstrom, B.; Gillberg, C. Autism in thalidomide embryopathy: A population study. Dev. Med. Child Neurol 1994, 36, 351–356. [Google Scholar]
- Narita, M.; Oyabu, A.; Imura, Y.; Kamada, N.; Yokoyama, T.; Tano, K.; Uchida, A.; Narita, N. Nonexploratory movement and behavioral alterations in a thalidomide or valproic acid-induced autism model rat. Neurosci. Res 2010, 66, 2–6. [Google Scholar]
- Meerts, I.A.; Lilienthal, H.; Hoving, S.; van den Berg, J.H.; Weijers, B.M.; Bergman, A.; Koeman, J.H.; Brouwer, A. Developmental exposure to 4-hydroxy-2,3,3′,4′,5-pentachlorobiphenyl (4-OH-CB107): Long-term effects on brain development, behavior, and brain stem auditory evoked potentials in rats. Toxicol. Sci 2004, 82, 207–218. [Google Scholar]
- Stump, D.G.; Beck, M.J.; Radovsky, A.; Garman, R.H.; Freshwater, L.L.; Sheets, L.P.; Marty, M.S.; Waechter, J.M., Jr; Dimond, S.S.; van Miller, J.P.; et al. Developmental neurotoxicity study of dietary bisphenol A in Sprague-Dawley rats. Toxicol. Sci 2010, 115, 167–182. [Google Scholar]
- Dalgaard, M.; Ostergaard, G.; Lam, H.R.; Hansen, E.V.; Ladefoged, O. Toxicity study of di(2-ethylhexyl) phthalate (DEHP) in combination with acetone in rats. Pharmacol. Toxicol 2000, 86, 92–100. [Google Scholar]
- Kakko, I.; Toimela, T.; Tahti, H. The synaptosomal membrane bound ATPase as a target for the neurotoxic effects of pyrethroids, permethrin and cypermethrin. Chemosphere 2003, 51, 475–480. [Google Scholar]
- Numis, A.L.; Major, P.; Montenegro, M.A.; Muzykewicz, D.A.; Pulsifer, M.B.; Thiele, E.A. Identification of risk factors for autism spectrum disorders in tuberous sclerosis complex. Neurology 2011, 76, 981–987. [Google Scholar]
- O’Roak, B.J.; State, M.W. Autism genetics: Strategies, challenges, and opportunities. Autism Res 2008, 1, 4–17. [Google Scholar]
- Muhle, R.; Trentacoste, S.V.; Rapin, I. The genetics of autism. Pediatrics 2004, 113, e472–e486. [Google Scholar]
- Wang, J.; Rao, S.; Chu, J.; Shen, X.; Levasseur, D.N.; Theunissen, T.W.; Orkin, S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature 2006, 444, 364–368. [Google Scholar]
- Muller, F.J.; Laurent, L.C.; Kostka, D.; Ulitsky, I.; Williams, R.; Lu, C.; Park, I.H.; Rao, M.S.; Shamir, R.; Schwartz, P.H.; et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature 2008, 455, 401–405. [Google Scholar]
- GeneSpring, version GX10.02; Agilent Technologies: Palo Alto, CA, USA, 2010.
| Chemical Name | Ellipsis | Intended Use | Physiological Effect and Toxicity | Mode of Action | Target Protein |
|---|---|---|---|---|---|
| Triiodotyronine | T3 | Endogenenous hormne | Pseudo thyroid hormone | transcriptional regulation | Thyroid hormone receptor (TR)α, TRβ |
| Dexamethazone | DEX | Medicinal drug | Pseudo corticosteroid hormone | transcriptional regulation | Glucocorticoid receptor (GR) |
| 17b-Estradiol | E2 | Endogenenous hormne | transcriptional regulation | Estrogen receptor (ER)α, ERβ | |
| 5a-Dihydrotestosterone | DHT | Endogenenous hormne | transcriptional regulation | Androgen receptor (AR) | |
| 2,3,7,8-tetrachlorodibenzo-p-dioxin | TCDD | Unintentional chemical | Multi-toxicity | transcriptional regulation | Aryl hydrocarbon receptor (AhR) |
| Methoprene acid | MPA | Pesticides | Teretogenecity | transcriptional regulation | Retinoid X receptor (RXR)α, RXRβ, RXRγ |
| Cyclopamine | CPM | Medicinal drug | Teretogenecity | Signal inhibition | Hadgehog signaling pathway |
| Thalidmide | TMD | Medicinal drug | Teretogenecity and Autism | Unknown | Oxidative stress |
| 4(OH)-2′,3,3′,4′,5′-pentachlorobephenyl 107 | PCB | Metabolite of PBC | Multi-toxicity | Unknown | Unknown (ERα, oxidativestress) |
| Permethrin | PMT | Pesticides | Neuro-toxicity | Unknown | Oxidative stress |
| Bisphenol A | BPA | Plastic materials | Reproductive and Neuro-toxicity? | Unknown | Unknown (ERα, ERRγ) |
| Bis(2-ethylhexyl) phthalate | DEHP | Plastic materials | Reproductive and Neuro-toxicity? | Unknown | Unknown [Peroxisome proliferator-activated receptor (PPAR)α, antiTR] |
| Alzheimer | Autism | Parkinson | Axon Guidance | Pluripotent | Neural Development | Oxidative-Stress |
|---|---|---|---|---|---|---|
| AR | AR | AR | 1500003O03Rik | Arid3b | Atbf1 | Aass |
| ApoE | Cntnap2 | Casp3 | Abl1 | Esrrb | Cdyl | Als2 |
| App | En2 | Casp7 | Ablim1 | Fkbp3 | Fos | Apoe |
| Bace | Esr1 | Casp9 | Cfl1 | Hdac2 | Gbx2 | Ctsb |
| Casp3 | Esr2 | Esr1 | Cxcl12 | Klf4 | Gfap | Dnm2 |
| Casp7 | Fmr1 | Esr2 | Efna4 | Mybbp1a | Hras1 | Fancc |
| Esr1 | Foxp2 | Park2 | Epha2 | Nacc1 | Map2 | Gpx7 |
| Esr2 | Gabrb3 | Park7 | Ephb1 | Nanog | Mapk1 | Gpx8 |
| Ide | Mecp2 | RARa | Nfatc2 | Nfkbib | Mapk3 | Gusb |
| Il1r1 | Nlgn3 | RARb | Nfatc3 | Nr0b1 | Nestin | Hprt1 |
| Mme | RARa | RARg | Ntng1 | Nr5a2 | Pla2g6 | Kif9 |
| Psen | RARb | Slc6a3 | Sema3a | Pou5f1 | Raf1 | Noxo1 |
| RARa | RARg | Snca | Sema3b | Rex1 | Rhog | Nxn |
| RARb | Reln | Th | Sema3d | Sall4 | Rif1 | Park7 |
| RARg | Slc6a4 | Uchl1 | Sema3f | Smarcad1 | Rps6ka1 | Ppp1r15b |
| Tnfrsf1a | Tsc1 | Sema3g | Smarcc1 | Sall1 | Prdx2 | |
| Tsc2 | Sema6a | Sox2 | Shc1 | Prdx6-rs1 | ||
| Ube3a | Sema6b | Sp1 | Smarcad1 | Psmb5 | ||
| Sema6d | Spag1 | Sox2 | Recql4 | |||
| Srgap3 | Trim28 | Tuj1 | Scd1 | |||
| Unc5d | Zfp281 | Map2k1 | Slc41a3 | |||
| c-Myc | Sod1 | |||||
| Sod3 | ||||||
| Txnip | ||||||
| Txnrd1 | ||||||
| Xpa |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nagano, R.; Akanuma, H.; Qin, X.-Y.; Imanishi, S.; Toyoshiba, H.; Yoshinaga, J.; Ohsako, S.; Sone, H. Multi-Parametric Profiling Network Based on Gene Expression and Phenotype Data: A Novel Approach to Developmental Neurotoxicity Testing. Int. J. Mol. Sci. 2012, 13, 187-207. https://doi.org/10.3390/ijms13010187
Nagano R, Akanuma H, Qin X-Y, Imanishi S, Toyoshiba H, Yoshinaga J, Ohsako S, Sone H. Multi-Parametric Profiling Network Based on Gene Expression and Phenotype Data: A Novel Approach to Developmental Neurotoxicity Testing. International Journal of Molecular Sciences. 2012; 13(1):187-207. https://doi.org/10.3390/ijms13010187
Chicago/Turabian StyleNagano, Reiko, Hiromi Akanuma, Xian-Yang Qin, Satoshi Imanishi, Hiroyoshi Toyoshiba, Jun Yoshinaga, Seiichiroh Ohsako, and Hideko Sone. 2012. "Multi-Parametric Profiling Network Based on Gene Expression and Phenotype Data: A Novel Approach to Developmental Neurotoxicity Testing" International Journal of Molecular Sciences 13, no. 1: 187-207. https://doi.org/10.3390/ijms13010187
APA StyleNagano, R., Akanuma, H., Qin, X.-Y., Imanishi, S., Toyoshiba, H., Yoshinaga, J., Ohsako, S., & Sone, H. (2012). Multi-Parametric Profiling Network Based on Gene Expression and Phenotype Data: A Novel Approach to Developmental Neurotoxicity Testing. International Journal of Molecular Sciences, 13(1), 187-207. https://doi.org/10.3390/ijms13010187





