Chemical Synthesis and Functional Analysis of VarvA Cyclotide
Abstract
:1. Introduction
2. Results
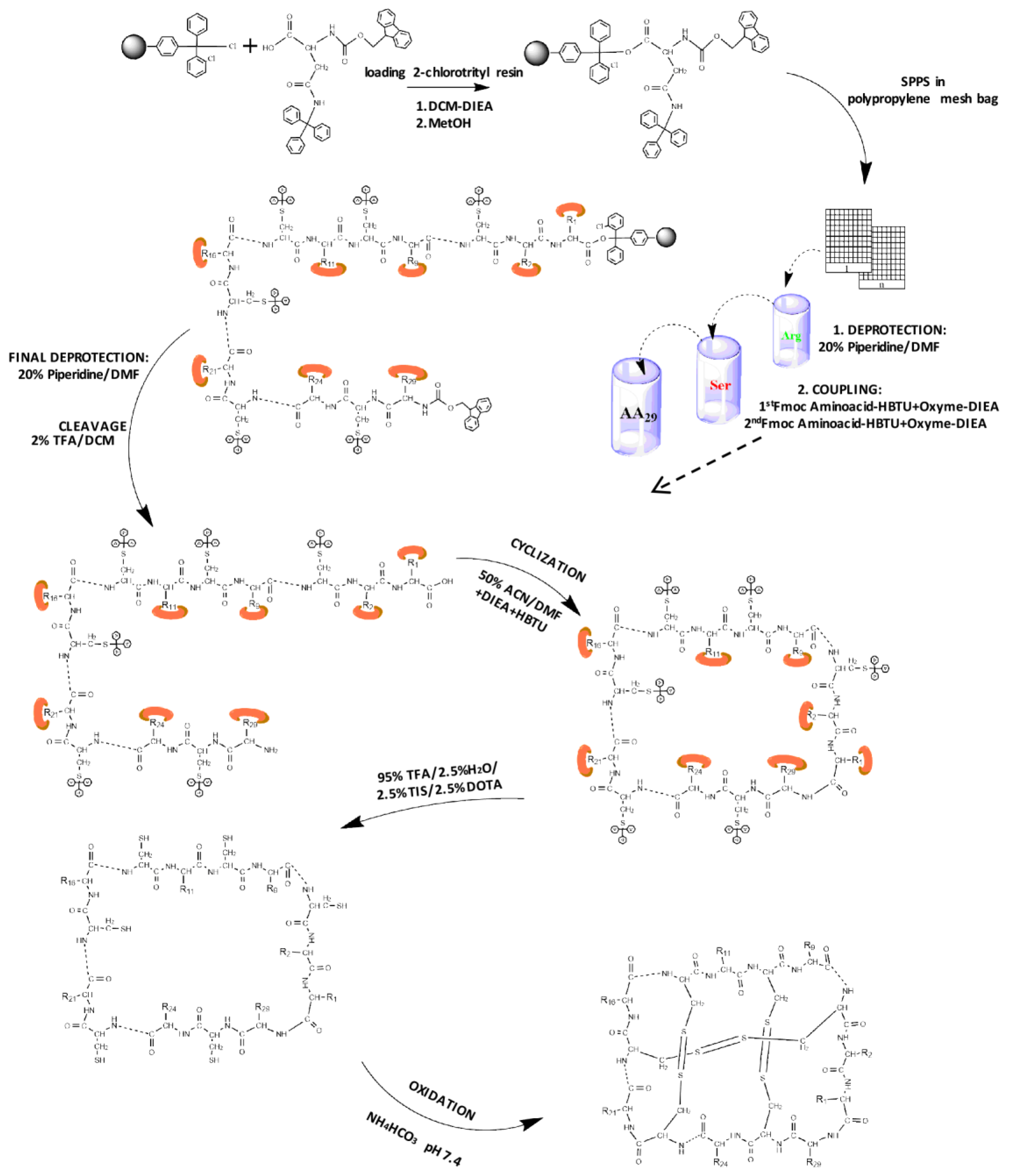
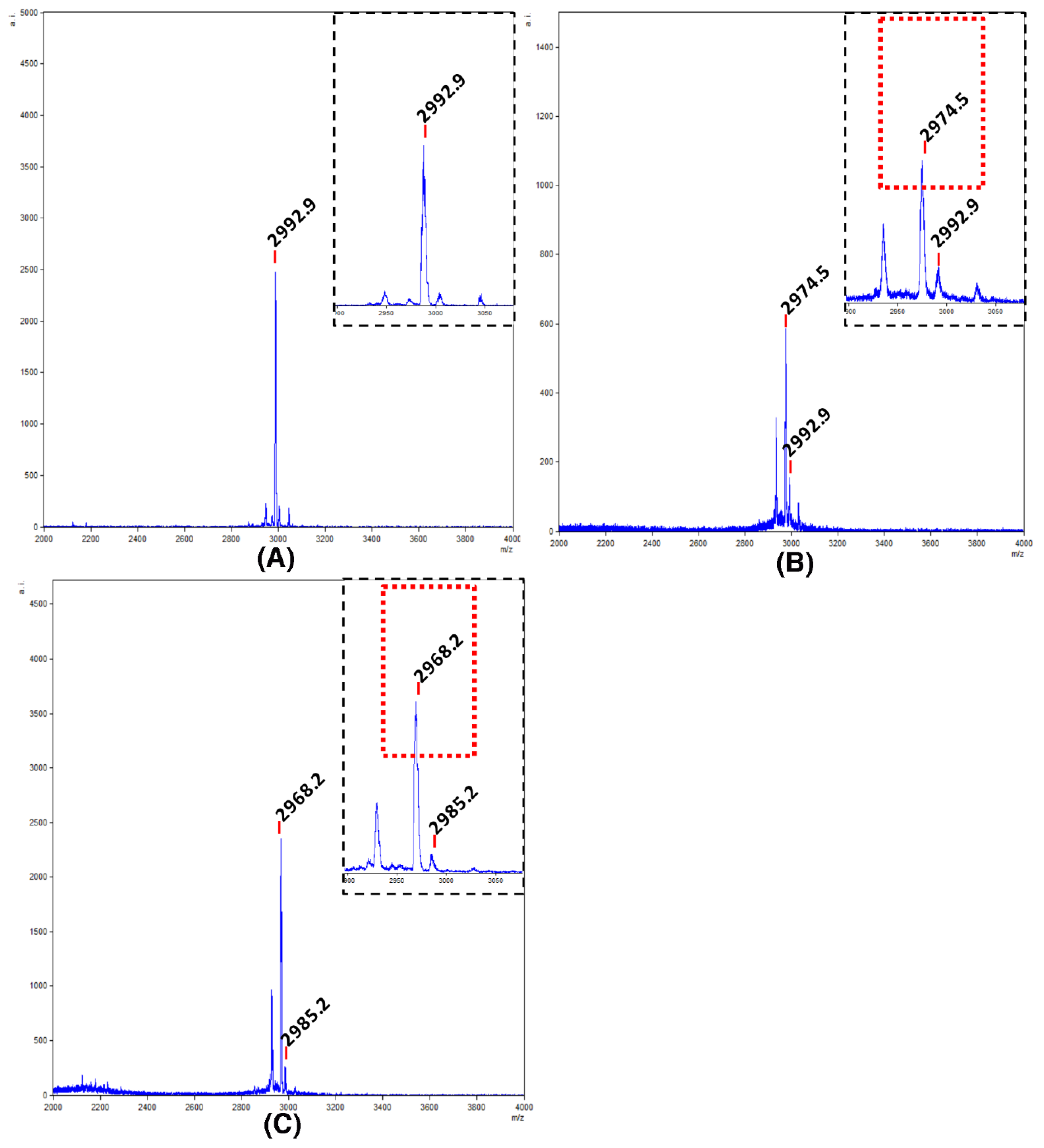
2.1. Chemical Synthesis of VarvA Cyclotide
2.2. Antimicrobial Activity of VarvA Synthetic Cyclotide against Fish Bacterial Pathogen
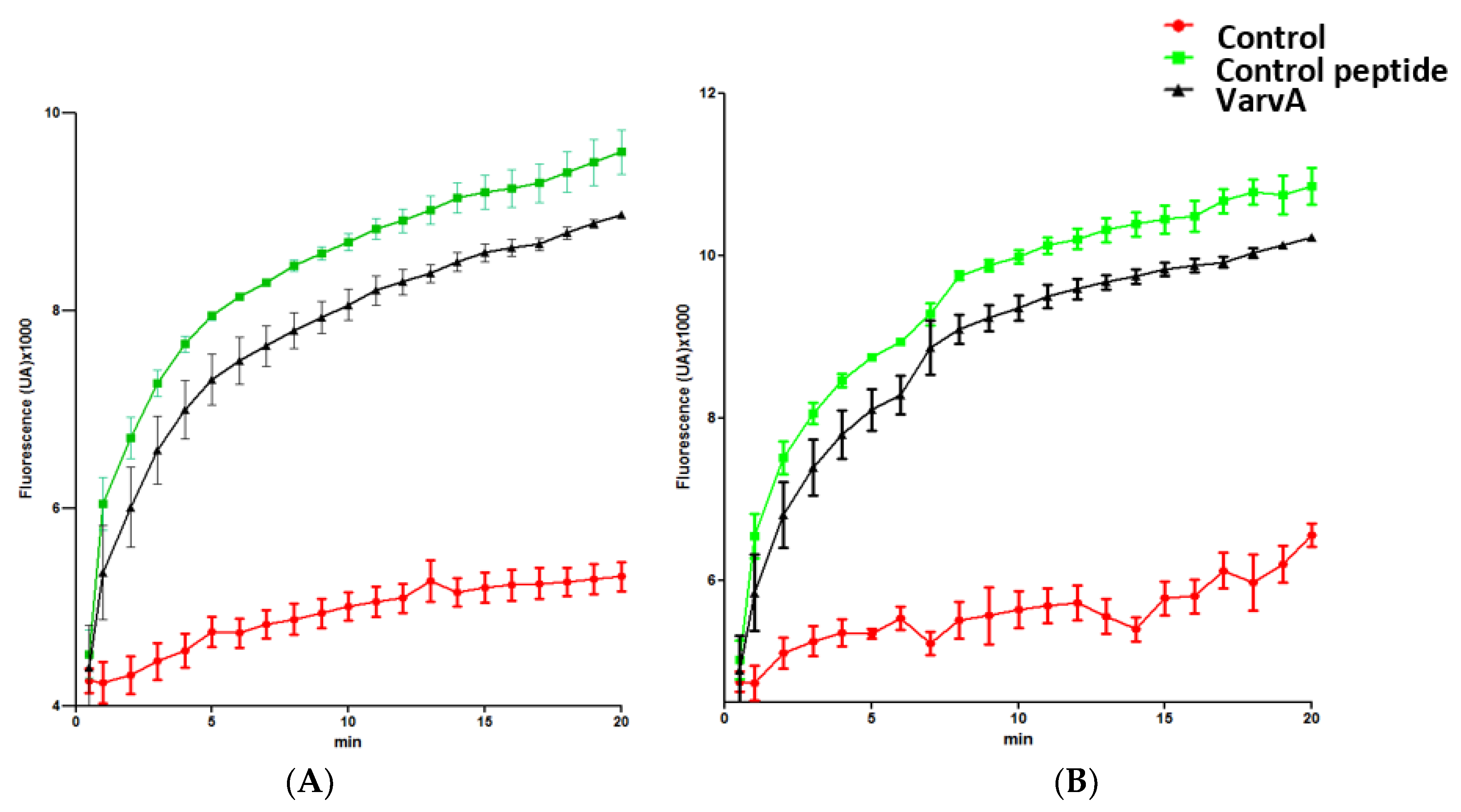
2.3. Bacterial Membrane Damage Induced by VarvA Synthetic Cyclotide
3. Discussion
4. Materials and Methods
4.1. Peptide Synthesis
4.2. Cyclotide Cyclization and Oxidative Folding
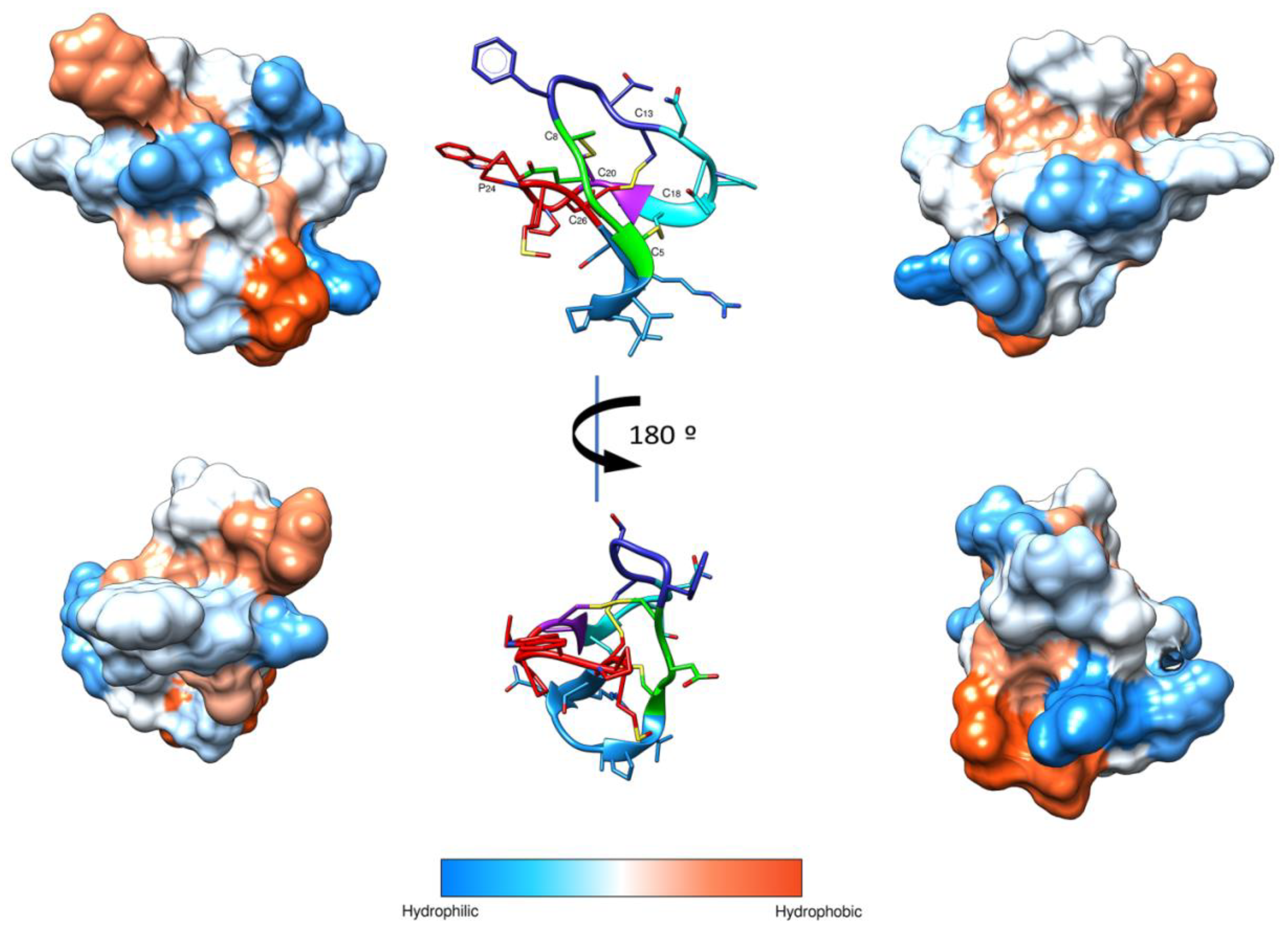
4.3. Prediction of Peptide Structure
4.4. Antibacterial Assay
4.5. SYTOX Green Bacteria Permeabilization Assay
4.6. Scanning Electron Microscopy (SEM)
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jad, Y.E.; El-Faham, A.; de la Torre, B.G.; Albericio, F. Solid-phase peptide synthesis, the state of the art: Challenges and opportunities. In Peptide-Based Drug Discovery: Challenges and New Therapeutics; Srivastava, V., Ed.; Royal Society of Chemistry, United Kingdom: London, UK, 2017; ISBN 978-1-78262-732-6. [Google Scholar]
- Gokhale, A.S.; Satyanarayanajois, S. Peptides and peptidomimetics as immunomodulators. Immunotherapy 2014, 6, 755–774. [Google Scholar] [CrossRef] [PubMed]
- Hamman, J.H.; Enslin, G.M.; Kotzé, A.F. Oral delivery of peptide drugs: Barriers and developments. BioDrugs 2005, 19, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Bogdanowich-Knipp, S.J.; Chakrabarti, S.; Williams, T.D.; Dillman, R.K.; Siahaan, T.J. Solution stability of linear vs. cyclic RGD peptides. J. Pept. Res. 1999, 53, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.H. Cyclic peptides as therapeutic agents and biochemical tools. Biomol. Ther. 2012, 20, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.K.; Craik, D.J. Cyclic peptide oral bioavailability: Lessons from the past. Biopolymers 2016, 106, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Camarero, J.A. Cyclotides, a versatile ultrastable micro-protein scaffold for biotechnological applications. Bioorg. Med. Chem. Lett. 2017, 27, 5089–5099. [Google Scholar] [CrossRef] [PubMed]
- De Veer, S.J.; Weidmann, J.; Craik, D.J. Cyclotides as tools in chemical biology. Acc. Chem. Res. 2017, 50, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Ireland, D.C.; Colgrave, M.L.; Nguyencong, P.; Daly, N.L.; Craik, D.J. Discovery and characterization of a linear cyclotide from Viola odorata: Implications for the processing of circular proteins. J. Mol. Biol. 2006, 357, 1522–1535. [Google Scholar] [CrossRef] [PubMed]
- Poth, A.G.; Colgrave, M.L.; Philip, R.; Kerenga, B.; Daly, N.L.; Anderson, M.A.; Craik, D.J. Discovery of Cyclotides in the Fabaceae plant family provides new insights into the cyclization, evolution, and distribution of circular proteins. ACS Chem. Biol. 2011, 6, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Ravipati, A.S.; Poth, A.G.; Troeira Henriques, S.; Bhandari, M.; Huang, Y.-H.; Nino, J.; Colgrave, M.L.; Craik, D.J. Understanding the diversity and distribution of cyclotides from plants of varied genetic origin. J. Nat. Prod. 2017, 80, 1522–1530. [Google Scholar] [CrossRef] [PubMed]
- Burman, R.; Yeshak, M.Y.; Larsson, S.; Craik, D.J.; Rosengren, K.J.; Göransson, U. Distribution of circular proteins in plants: Large-scale mapping of cyclotides in the Violaceae. Front. Plant Sci. 2015, 6, 855. [Google Scholar] [CrossRef] [PubMed]
- Craik, D.J.; Clark, R.J.; Daly, N.L. Potential therapeutic applications of the cyclotides and related cystine knot mini-proteins. Expert Opin. Investig. Drugs 2007, 16, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Burman, R.; Gunasekera, S.; Strömstedt, A.A.; Göransson, U. Chemistry and biology of cyclotides: Circular plant peptides outside the box. J. Nat. Prod. 2014, 77, 724–736. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.E.; Muir, T.W.; Clark-Lewis, I.; Kent, S.B. Synthesis of proteins by native chemical ligation. Science 1994, 266, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Lu, Y.A.; Yu, Q. Thia Zip reaction for synthesis of large cyclic peptides: mechanisms and applications. J. Am. Chem. Soc. 1999, 121, 4316–4324. [Google Scholar] [CrossRef]
- Reinwarth, M.; Nasu, D.; Kolmar, H.; Avrutina, O. Chemical synthesis, backbone cyclization and oxidative folding of cystine-knot peptides—promising scaffolds for applications in drug design. Molecules 2012, 17, 12533–12552. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Wong, C.T.T. Chemical synthesis of circular proteins. J. Biol. Chem. 2012, 287, 27020–27025. [Google Scholar] [CrossRef] [PubMed]
- Witherup, K.M.; Bogusky, M.J.; Anderson, P.S.; Ramjit, H.; Ransom, R.W.; Wood, T.; Sardana, M. Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J. Nat. Prod. 1994, 57, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Lindholm, P.; Göransson, U.; Johansson, S.; Claeson, P.; Gullbo, J.; Larsson, R.; Bohlin, L.; Backlund, A. Cyclotides: A novel type of cytotoxic agents. Mol. Cancer Ther. 2002, 1, 365–369. [Google Scholar] [PubMed]
- Jennings, C.V.; Rosengren, K.J.; Daly, N.L.; Plan, M.; Stevens, J.; Scanlon, M.J.; Waine, C.; Norman, D.G.; Anderson, M.A.; Craik, D.J. Isolation, Solution structure, and insecticidal activity of Kalata B2, a circular protein with a twist: Do Möbius strips exist in nature? Biochemistry 2005, 44, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Colgrave, M.L.; Huang, Y.H.; Craik, D.J.; Kotze, A.C. Cyclotide interactions with the nematode external surface. Antimicrob. Agents Chemother. 2010, 54, 2160–2166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, N.L.; Clark, R.J.; Plan, M.R.; Craik, D.J. Kalata B8, a novel antiviral circular protein, exhibits conformational flexibility in the cystine knot motif. Biochem. J. 2006, 393, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Strömstedt, A.A.; Park, S.; Burman, R.; Göransson, U. Bactericidal activity of cyclotides where phosphatidylethanolamine-lipid selectivity determines antimicrobial spectra. Biochim. Biophys. Acta 2017, 1859, 1986–2000. [Google Scholar] [CrossRef] [PubMed]
- Pranting, M.; Loov, C.; Burman, R.; Goransson, U.; Andersson, D.I. The cyclotide cycloviolacin O2 from Viola odorata has potent bactericidal activity against Gram-negative bacteria. J. Antimicrob. Chemother. 2010, 65, 1964–1971. [Google Scholar] [CrossRef] [PubMed]
- Petersen, A.; Andersen, J.S.; Kaewmak, T.; Somsiri, T.; Dalsgaard, A. Impact of integrated fish farming on antimicrobial resistance in a pond environment. Appl. Environ. Microbiol. 2002, 68, 6036–6042. [Google Scholar] [CrossRef] [PubMed]
- Smith, P. Antimicrobial resistance in aquaculture. Rev. Sci. Tech. 2008, 27, 243–264. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.E.M.; Schreier, H.J.; Lanska, L.; Hale, M.S. The rising tide of antimicrobial resistance in aquaculture: Sources, sinks and solutions. Mar. Drugs 2017, 15. [Google Scholar] [CrossRef] [PubMed]
- Muziasari, W.I.; Pärnänen, K.; Johnson, T.A.; Lyra, C.; Karkman, A.; Stedtfeld, R.D.; Tamminen, M.; Tiedje, J.M.; Virta, M. Aquaculture changes the profile of antibiotic resistance and mobile genetic element associated genes in Baltic Sea sediments. FEMS Microbiol. Ecol. 2016, 92, fiw052. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.Q.A.; Cabello, F.C.; L’Abée-Lund, T.M.; Tomova, A.; Godfrey, H.P.; Buschmann, A.H.; Sørum, H. Antimicrobial resistance and antimicrobial resistance genes in marine bacteria from salmon aquaculture and non-aquaculture sites. Environ. Microbiol. 2014, 16, 1310–1320. [Google Scholar] [CrossRef] [PubMed]
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef] [PubMed]
- Kathleen, M.M.; Samuel, L.; Felecia, C.; Reagan, E.L.; Kasing, A.; Lesley, M.; Toh, S.C. Antibiotic resistance of diverse bacteria from aquaculture in Borneo. Int. J. Microbiol. 2016, 2016, 2164761. [Google Scholar] [CrossRef] [PubMed]
- Göransson, U.; Luijendijk, T.; Johansson, S.; Bohlin, L.; Claeson, P. Seven novel macrocyclic polypeptides from Viola a rvensis. J. Nat. Prod. 1999, 62, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Flegel, M.; Sheppard, R.C. A sensitive, general method for quantitative monitoring of continuous flow solid phase peptide synthesis. J. Chem. Soc. Chem. Commun. 1990, 536–538. [Google Scholar] [CrossRef]
- Cheneval, O.; Schroeder, C.I.; Durek, T.; Walsh, P.; Huang, Y.H.; Liras, S.; Price, D.A.; Craik, D.J. Fmoc-Based synthesis of disulfide-rich cyclic peptides. J. Org. Chem. 2014, 79, 5538–5544. [Google Scholar] [CrossRef] [PubMed]
- Calce, E.; Vitale, R.M.; Scaloni, A.; Amodeo, P.; De Luca, S. Air oxidation method employed for the disulfide bond formation of natural and synthetic peptides. Amino Acids 2015, 47, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Bourbon, C.; Bry, C.; Roggemans, C.; Soulard, C.; Thizon, C.; Garbay, B. Use of a real-time polymerase chain reaction thermocycler to study bacterial cell permeabilization by antimicrobial peptides. Anal. Biochem. 2008, 381, 279–281. [Google Scholar] [CrossRef] [PubMed]
- Murillo, L.A.; Lan, C.; Agabian, N.M.; Larios, S.; Lomonte, B. Fungicidal activity of a phospholipase-A2-derived synthetic peptide variant against Candida albicans. Rev. Esp. Quimioter. 2007, 20, 330–333. [Google Scholar] [PubMed]
- Gould, A.; Ji, Y.; Aboye, T.L.; Camarero, J.A. Cyclotides, a novel ultrastable polypeptide scaffold for drug discovery. Curr. Pharm. Des. 2011, 17, 4294–4307. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Chaousis, S.; Cheneval, O.; Craik, D.J.; Henriques, S.T. Optimization of the cyclotide framework to improve cell penetration properties. Front. Pharmacol. 2015, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Chatzi, K.B.O.; Gatos, D.; Stavropoulos, G. 2-Chlorotrityl chloride resin. Studies on anchoring of Fmoc-amino acids and peptide cleavage Int. J. Pept. Protein Res. 1991, 6, 513–520. [Google Scholar]
- Malesevic, M.; Strijowski, U.; Bächle, D.; Sewald, N. An improved method for the solution cyclization of peptides under pseudo-high dilution conditions. J. Biotechnol. 2004, 112, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Yudin, A.K. Macrocycles: Lessons from the distant past, recent developments, and future directions. Chem. Sci. 2015, 6, 30–49. [Google Scholar] [CrossRef] [PubMed]
- Otaka, A.; Koide, T.; Shide, A.; Fujii, N. Application of dimethylsulphoxide(dms0) / trifluoroacetic acid(TFA) oxidation to the synthesis of cystine-containing peptide. Temhedron Lett. 1991, 32, 1223–1226. [Google Scholar] [CrossRef]
- Kudryavtseva, E.V.; Sidorova, M.V.; Ovchinnikov, M.V.; Bespalova, Z.D.; Bushuev, V.N. Comparative evaluation of different methods for disulfide bond formation in synthesis of the HIV-2 antigenic determinant. J. Pept. Res. 1997, 49, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Annis, I.; Barany, G. Disulfide bond formation in peptides. Curr. Protoc. Protein Sci. 2001, 18, 198–221. [Google Scholar] [CrossRef]
- Harris, K.M.; Flemer, S.; Hondal, R.J.; Hondal, R.J. Studies on deprotection of cysteine and selenocysteine side-chain protecting groups. J. Pept. Sci. 2007, 13, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.T.T.; Taichi, M.; Nishio, H.; Nishiuchi, Y.; Tam, J.P. Optimal oxidative folding of the novel antimicrobial cyclotide from Hedyotis biflora requires high alcohol concentrations. Biochemistry 2011, 50, 7275–7283. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, S.M.; Sando, L.; Ireland, D.C.; Colgrave, M.L.; Bharathi, R.; Göransson, U.; Craik, D.J. A continent of plant defense peptide diversity: Cyclotides in Australian Hybanthus (Violaceae). Plant Cell 2005, 17, 3176–3189. [Google Scholar] [CrossRef] [PubMed]
- Garcia, A.E.; Camarero, J.A. Biological activities of natural and engineered cyclotides, a novel molecular scaffold for peptide-based therapeutics. Curr. Mol. Pharmacol. 2010, 3, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.K.T.; Lian, Y.; Pang, E.W.H.; Nguyen, P.Q.T.; Tran, T.D.; Tam, J.P. Discovery of linear cyclotides in monocot plant Panicum laxum of Poaceae family provides new insights into evolution and distribution of cyclotides in plants. J. Biol. Chem. 2013, 288, 3370–3380. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, K.; McKee, T.; Bokesch, H. Anti-HIV Cyclotides. Curr. Protein Pept. Sci. 2004, 5, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Jennings, C.; West, J.; Waine, C.; Craik, D.; Anderson, M. Biosynthesis and insecticidal properties of plant cyclotides: The cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. USA 2001, 98, 10614–10619. [Google Scholar] [CrossRef] [PubMed]
- Sudheesh, P.S.; Al-Ghabshi, A.; Al-Mazrooei, N.; Al-Habsi, S. Comparative pathogenomics of bacteria causing infectious diseases in fish. Int. J. Evol. Biol. 2012, 2012, 457264. [Google Scholar] [CrossRef] [PubMed]
- Gran, L.; Sletten, K.; Skjeldal, L. Cyclic peptides from Oldenlandia affinis DC. Molecular and biological properties. Chem. Biodivers. 2008, 5, 2014–2022. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Lu, Y.A.; Yang, J.L.; Chiu, K.W. An unusual structural motif of antimicrobial peptides containing end-to-end macrocycle and cystine-knot disulfides. Proc. Natl. Acad. Sci. USA 1999, 96, 8913–8918. [Google Scholar] [CrossRef] [PubMed]
- Burman, R.; Strömstedt, A.A.; Malmsten, M.; Göransson, U. Cyclotide-membrane interactions: Defining factors of membrane binding, depletion and disruption. Biochim. Biophys. Acta. 2011, 1808, 2665–2673. [Google Scholar] [CrossRef] [PubMed]
- Henriques, S.T.; Huang, Y.H.; Castanho, M.A.R.B.; Bagatolli, L.A.; Sonza, S.; Tachedjian, G.; Daly, N.L.; Craik, D.J. Phosphatidylethanolamine binding is a conserved feature of cyclotide-membrane interactions. J. Biol. Chem. 2012, 287, 33629–33643. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Berditsch, M.; Hawecker, J.; Ardakani, M.F.; Gerthsen, D.; Ulrich, A.S. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob. Agents Chemother. 2010, 54, 3132–3142. [Google Scholar] [CrossRef] [PubMed]
- Meincken, M.; Holroyd, D.L.; Rautenbach, M. Atomic force microscopy study of the effect of antimicrobial peptides on the cell envelope of Escherichia coli. Antimicrob. Agents Chemother. 2005, 49, 4085–4092. [Google Scholar] [CrossRef] [PubMed]
- Hale, J.D.; Hancock, R.E. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev. Anti. Infect. Ther. 2007, 5, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Biopolymers 2002, 66, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Newcomb, W.S.; Deegan, T.L.; Miller, W.; Porco, J.A., Jr. Analysis of 9-fluorenylmethoxycarbonyl (FMOC) loading of solid-phase synthesis resins by gas chromatography. Biotechnol. Bioeng. 1998, 61, 55–60. [Google Scholar]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Mitta, G.; Vandenbulcke, F.; Hubert, F.; Roch, P. Mussel defensins are synthesised and processed in granulocytes then released into the plasma after bacterial challenge. J. Cell Sci. 1999, 112, 4233–4242. [Google Scholar] [PubMed]
- Schmitt, P.; Mercado, L.; Díaz, M.; Guzmán, F.; Arenas, G.; Marshall, S.H. Characterization and functional recovery of a novel antimicrobial peptide (CECdir-CECret) from inclusion bodies after expression in Escherichia coli. Peptides 2008, 29, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Jofré, C.; Guzmán, F.; Cárdenas, C.; Albericio, F.; Marshall, S.H. A natural peptide and its variants derived from the processing of infectious pancreatic necrosis virus (IPNV) displaying enhanced antimicrobial activity: A novel alternative for the control of bacterial diseases. Peptides 2011, 32, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, C.; Acosta, F.; Montero, D.; Guzmán, F.; Torres, E.; Vega, B.; Mercado, L. Synthetic hepcidin from fish: Uptake and protection against Vibrio anguillarum in sea bass (Dicentrarchus labrax). Fish Shellfish Immunol. 2016, 55, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.A.; Walker, R.D.; Baya, A.; Clemens, K.; Coles, M.; Hawke, J.P.; Henricson, B.E.; Hsu, H.M.; Mathers, J.J.; Oaks, J.L.; Papapetropoulou, M.; Reimschuessel, R. Antimicrobial susceptibility testing of aquatic bacteria: Quality control disk diffusion ranges for Escherichia coli ATCC 25922 and Aeromonas salmonicida subsp. salmonicida ATCC 33658 at 22 and 28 degrees C. J. Clin. Microbiol. 2003, 41, 4318–4323. [Google Scholar] [CrossRef] [PubMed]
- Dumetz, F.; Duchaud, E.; LaPatra, S.E.; Le Marrec, C.; Claverol, S.; Urdaci, M.-C.; Le Hénaff, M. A protective immune response is generated in rainbow trout by an OmpH-like surface antigen (P18) of Flavobacterium psychrophilum. Appl. Environ. Microbiol. 2006, 72, 4845–4852. [Google Scholar] [CrossRef] [PubMed]
- Giacometti, A.; Cirioni, O.; Barchiesi, F.; Del Prete, M.S.; Fortuna, M.; Caselli, F.; Scalise, G. In Vitro susceptibility tests for cationic peptides: Comparison of broth microdilution methods for bacteria that grow aerobically. Antimicrob Agents Chemother. 2000, 44, 1694–1696. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.A.; Guzmán, F.; Cárdenas, C.; Marshall, S.H.; Mercado, L. Antimicrobial activity of trout hepcidin. Fish Shellfish Immunol. 2014, 41, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Santana, P.A.; Salinas, N.; Álvarez, C.A.; Mercado, L.A.; Guzmán, F. Alpha-helical domain from IL-8 of salmonids: Mechanism of action and identification of a novel antimicrobial function. Biochem. Biophys. Res. Commun. 2018, 498, 803–809. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds reported in this paper are available from the authors. |

| Gram-Negative Bacterial Strain | MIC (μM) |
|---|---|
| A. salmonicida | 22.5 ± 0.7 |
| A. hydrophila | 22.5 ± 0.3 |
| V. anguillarum | 30 ± 1.2 |
| V. ordalii | 30 ± 0.8 |
| F. psychrophilum | 12.5 ± 0.3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez, C.A.; Santana, P.A.; Luna, O.; Cárdenas, C.; Albericio, F.; Romero, M.S.; Guzmán, F. Chemical Synthesis and Functional Analysis of VarvA Cyclotide. Molecules 2018, 23, 952. https://doi.org/10.3390/molecules23040952
Álvarez CA, Santana PA, Luna O, Cárdenas C, Albericio F, Romero MS, Guzmán F. Chemical Synthesis and Functional Analysis of VarvA Cyclotide. Molecules. 2018; 23(4):952. https://doi.org/10.3390/molecules23040952
Chicago/Turabian StyleÁlvarez, Claudio A., Paula A. Santana, Omar Luna, Constanza Cárdenas, Fernando Albericio, María S. Romero, and Fanny Guzmán. 2018. "Chemical Synthesis and Functional Analysis of VarvA Cyclotide" Molecules 23, no. 4: 952. https://doi.org/10.3390/molecules23040952





