Reconstructing Phylogeny by Aligning Multiple Metabolic Pathways Using Functional Module Mapping
Abstract
:1. Introduction
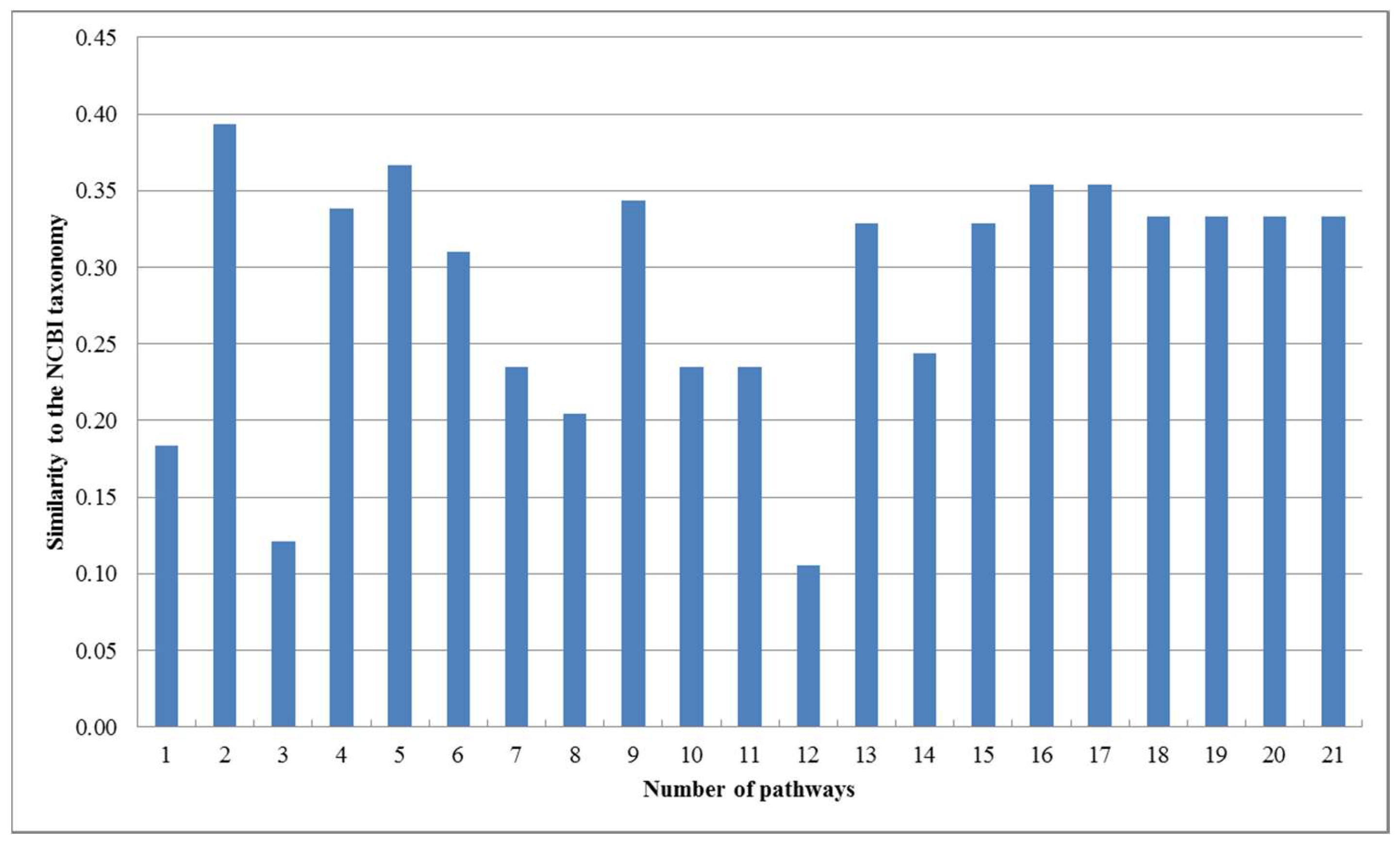
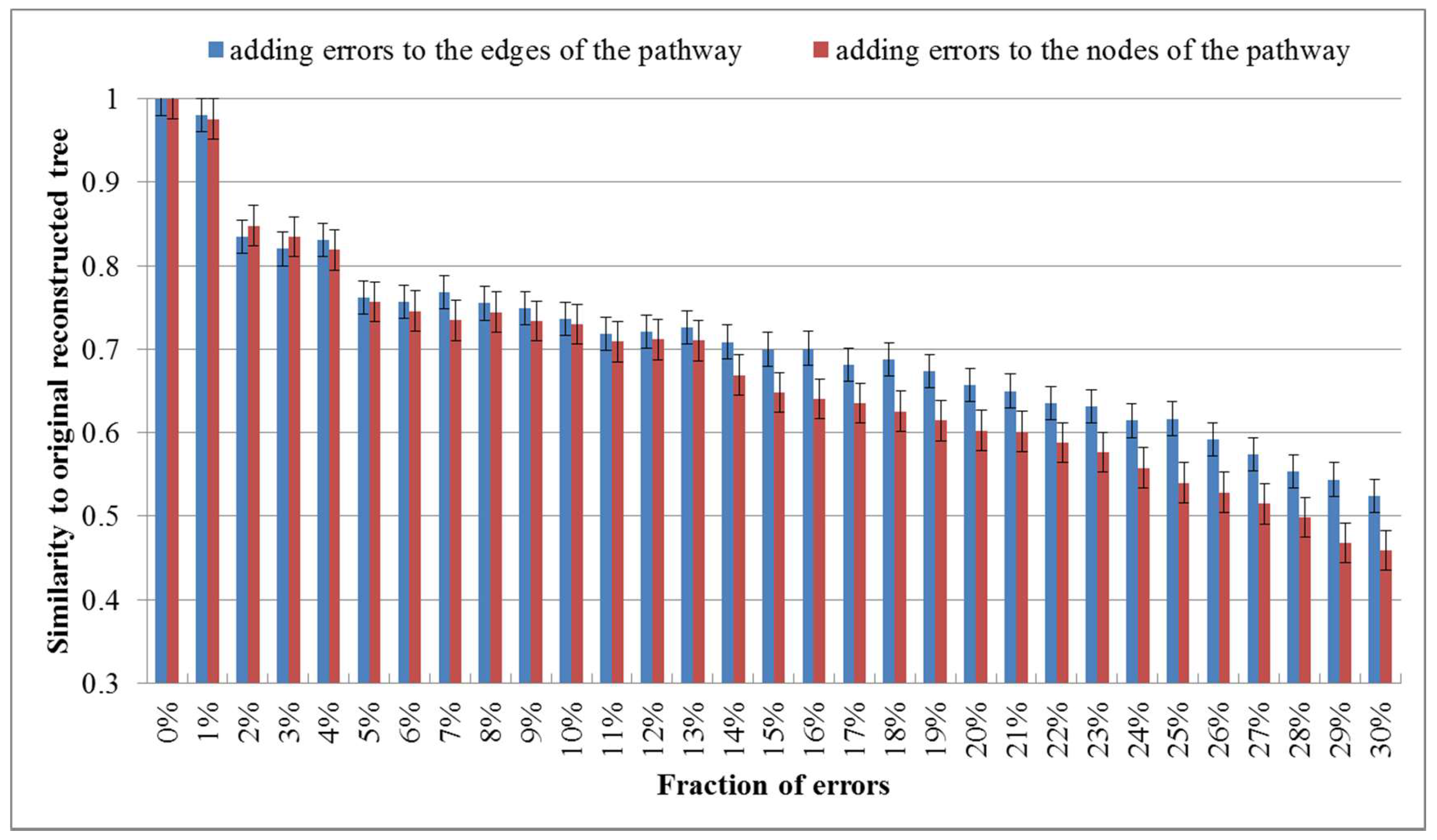
2. Results
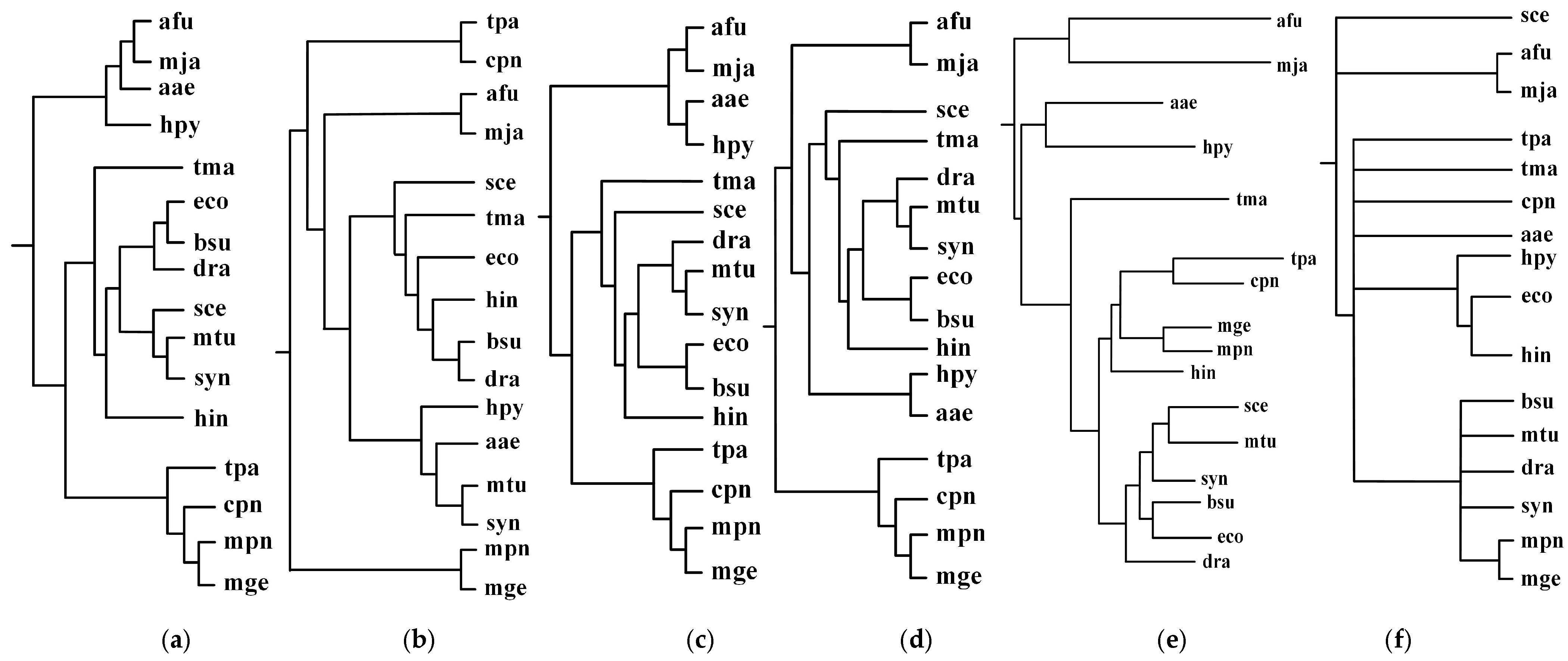
2.1. Comparison with the Classification Based on Substrate-Product Relationships and the Classification Based on EC Hierarchy
2.2. Comparison with the Classification Based on Comparing Single Pair of Metabolic Pathways
2.3. Comparison with the Classification Based on Global Alignment of Multiple Metabolic Pathways
2.3.1. Green Sulfur and Green Non-Sulfur Bacteria
2.3.2. Prochlorococcus and Synechococcus
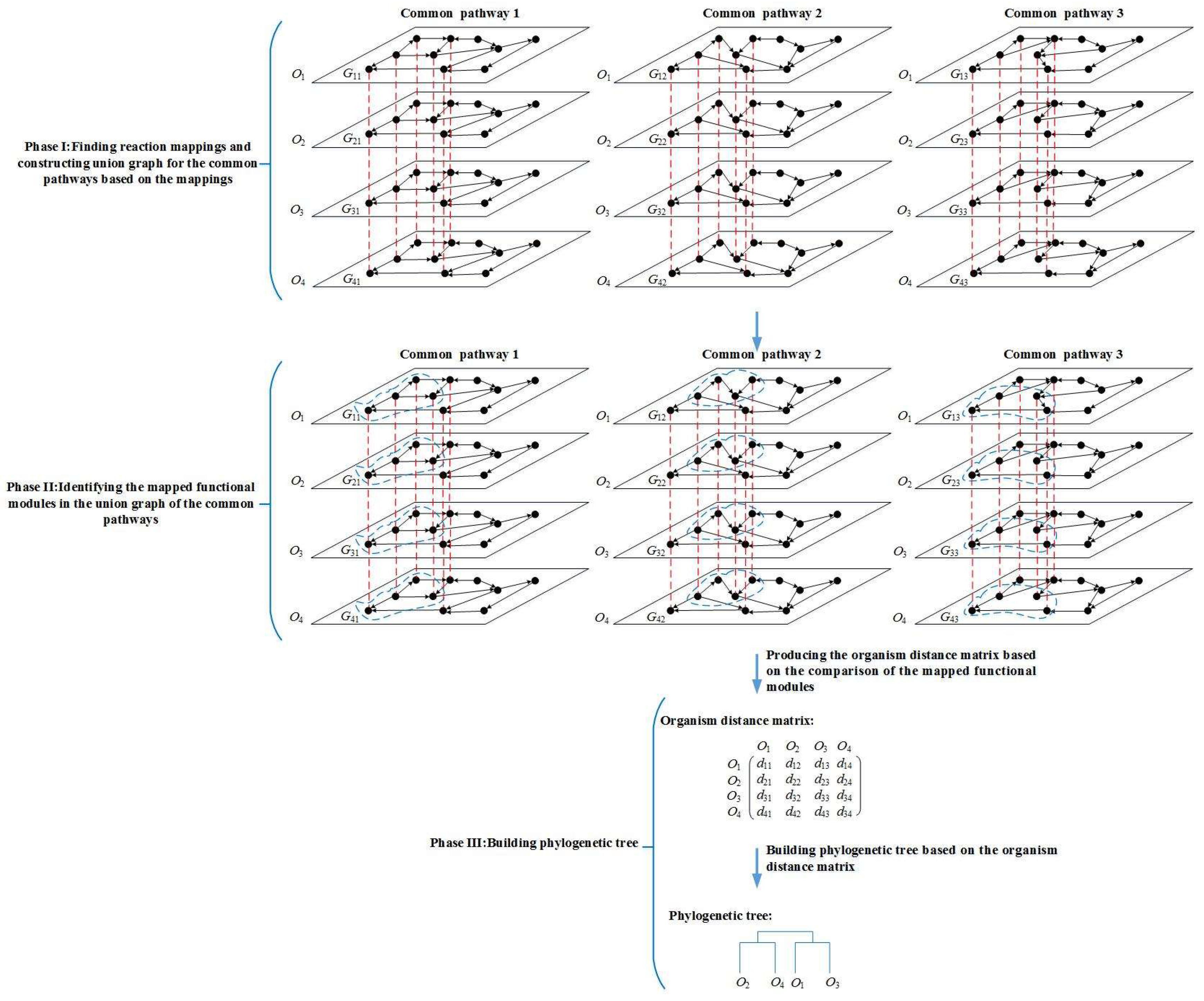
3. Methods
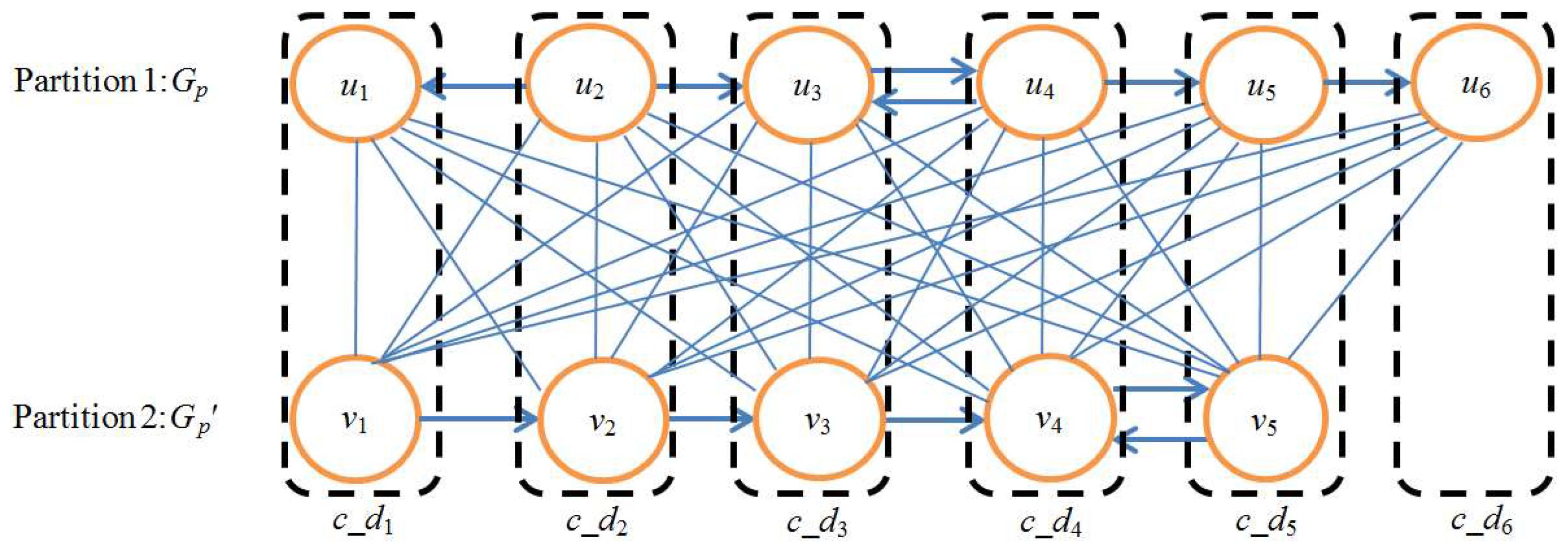
3.1. Phase I—Constructing Union Graph
3.2. Phase II—Identifying Functionally Conserved Modules
3.3. Phase III—Building Phylogenetic Tree
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ma, C.-Y.; Lin, S.-H.; Lee, C.-C.; Tang, C.Y.; Berger, B.; Liao, C.-S. Reconstruction of phyletic trees by global alignment of multiple metabolic networks. BMC Bioinf. 2013, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
- Forst, C.V.; Schulten, K. Phylogenetic analysis of metabolic pathways. J. Mol. Evol. 2001, 52, 471–489. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.; Satou, K.; Valiente, G. Reconstruction of phylogenetic relationships from metabolic pathways based on the enzyme hierarchy and the gene ontology. Genome Inf. 2005, 16, 45–55. [Google Scholar]
- Oh, S.J.; Joung, J.-G.; Chang, J.-H.; Zhang, B.-T. Construction of phylogenetic trees by kernel-based comparative analysis of metabolic networks. BMC Bioinf. 2006, 7, 284. [Google Scholar] [CrossRef] [PubMed]
- Mano, A.; Tuller, T.; Béjà, O.; Pinter, R.Y. Comparative classification of species and the study of pathway evolution based on the alignment of metabolic pathways. BMC Bioinf. 2010, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Pinter, R.Y.; Rokhlenko, O.; Yeger-Lotem, E.; Ziv-Ukelson, M. Alignment of metabolic pathways. Bioinformatics 2005, 21, 3401–3408. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.-S.; Lu, K.; Baym, M.; Singh, R.; Berger, B. IsoRankN: spectral methods for global alignment of multiple protein networks. Bioinformatics 2009, 25, i253–i258. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Satou, K.; Valiente, G. Phylogenetic reconstruction from non-genomic data. Bioinformatics 2007, 23, e110–e115. [Google Scholar] [CrossRef] [PubMed]
- Mazurie, A.; Bonchev, D.; Schwikowski, B.; Buck, G.A. Phylogenetic distances are encoded in networks of interacting pathways. Bioinformatics 2008, 24, 2579–2585. [Google Scholar] [CrossRef] [PubMed]
- Borenstein, E.; Kupiec, M.; Feldman, M.W.; Ruppin, E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc. Natl. Acad. Sci. USA 2008, 105, 14482–14487. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-W.; Lyu, P.-C.; Arita, M. Reconstructing phylogeny from metabolic substrate-product relationships. BMC Bioinf. 2011, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhong, C.; Lin, H.X.; Wang, J. A Method for Finding Metabolic Pathways Using Atomic Group Tracking. PLoS ONE 2017, 12, e0168725. [Google Scholar] [CrossRef] [PubMed]
- Plotree, D.; Plotgram, D. PHYLIP-phylogeny inference package (version 3.2). Cladistics 1989, 5, 163–166. [Google Scholar]
- Page, R.D. Visualizing phylogenetic trees using TreeView. Curr. Protoc. Bioinf. 2002, 6, 1–15. [Google Scholar]
- Shasha, D.; Wang, J.T.; Zhang, S. Unordered tree mining with applications to phylogeny. In Proceedings of the 20th International Conference on Data Engineering (ICDE), Boston, MA, USA, 2 April 2004; pp. 708–719. [Google Scholar]
- Heymans, M.; Singh, A.K. Deriving phylogenetic trees from the similarity analysis of metabolic pathways. Bioinformatics 2003, 19, i138–i146. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, S.; Skogerbø, G.; Zhang, Z.; Zhu, X.; Zhang, Z.; Sun, S.; Lu, H.; Shi, B.; Chen, R. Phylophenetic properties of metabolic pathway topologies as revealed by global analysis. BMC Bioinf. 2006, 7, 1. [Google Scholar]
- Alberich, R.; Llabrés, M.; Sánchez, D.; Simeoni, M.; Tuduri, M. MP-Align: alignment of metabolic pathways. BMC Syst. Biol. 2014, 8, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Zhong, C.; Lin, H.X.; Huang, J. Aligning Metabolic Pathways Exploiting Binary Relation of Reactions. PloS ONE 2016, 11, e0168044. [Google Scholar] [CrossRef] [PubMed]
- Ay, F.; Kellis, M.; Kahveci, T. SubMAP: Aligning metabolic pathways with subnetwork mappings. J. Comput. Biol. 2011, 18, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Hattori, M.; Okuno, Y.; Goto, S.; Kanehisa, M. Development of a chemical structure comparison method for integrated analysis of chemical and genomic information in the metabolic pathways. J. Am. Chem. Soc. 2003, 125, 11853–11865. [Google Scholar] [CrossRef] [PubMed]
- Sankowski, P. Maximum weight bipartite matching in matrix multiplication time. Theor. Comput. Sci. 2009, 410, 4480–4488. [Google Scholar] [CrossRef]
- Frey, B.J.; Dueck, D. Clustering by passing messages between data points. Science 2007, 315, 972–976. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available. |

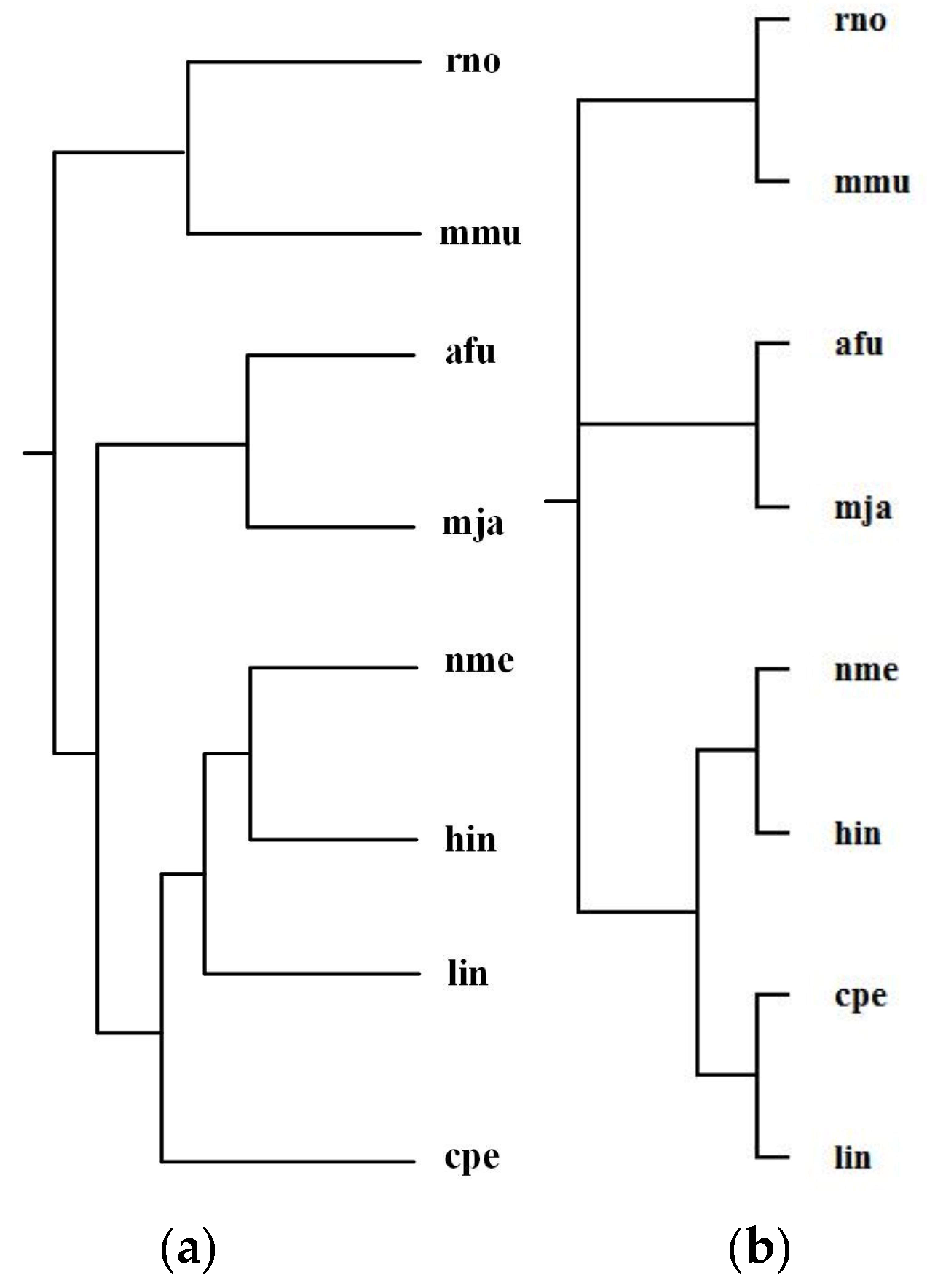
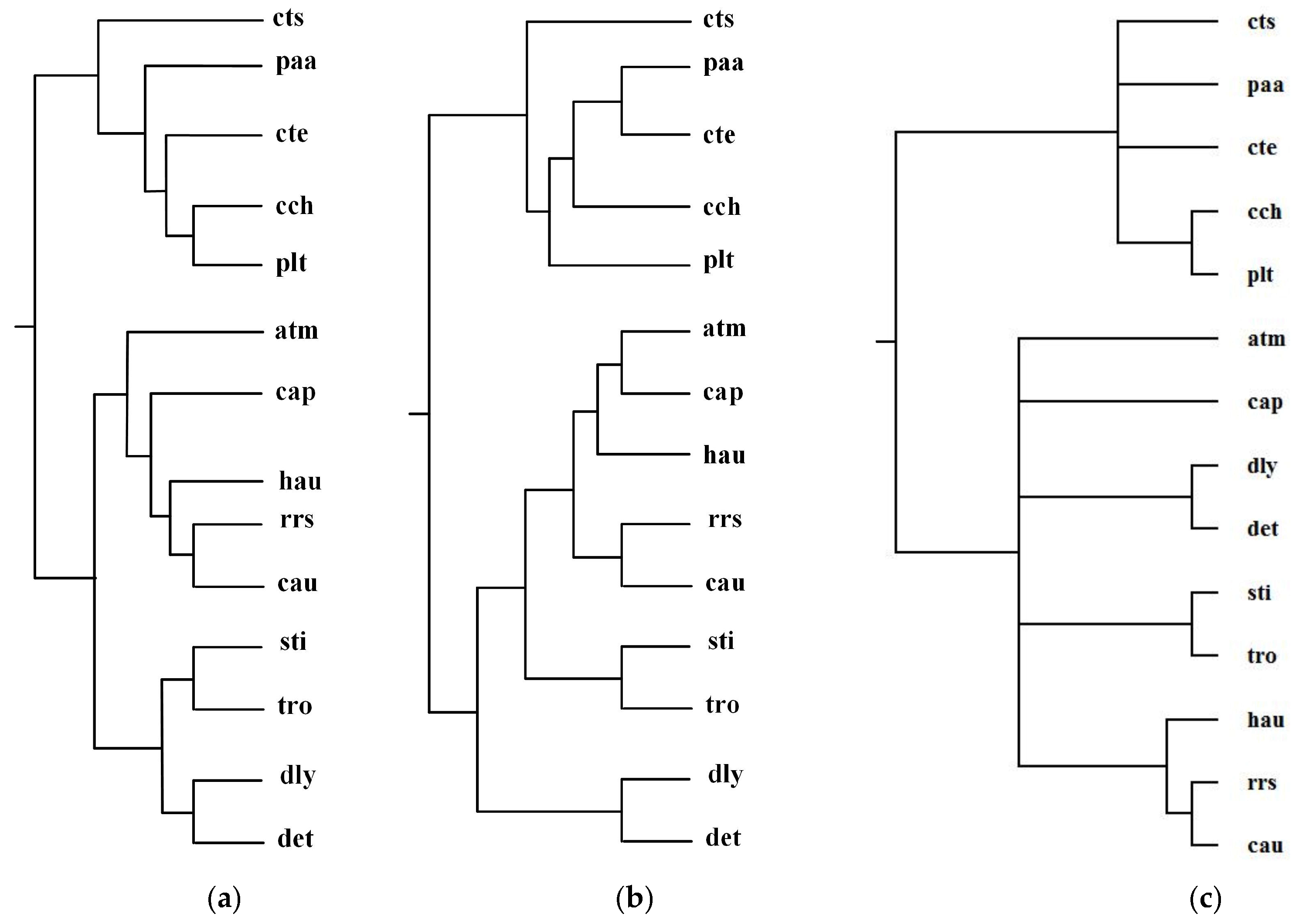
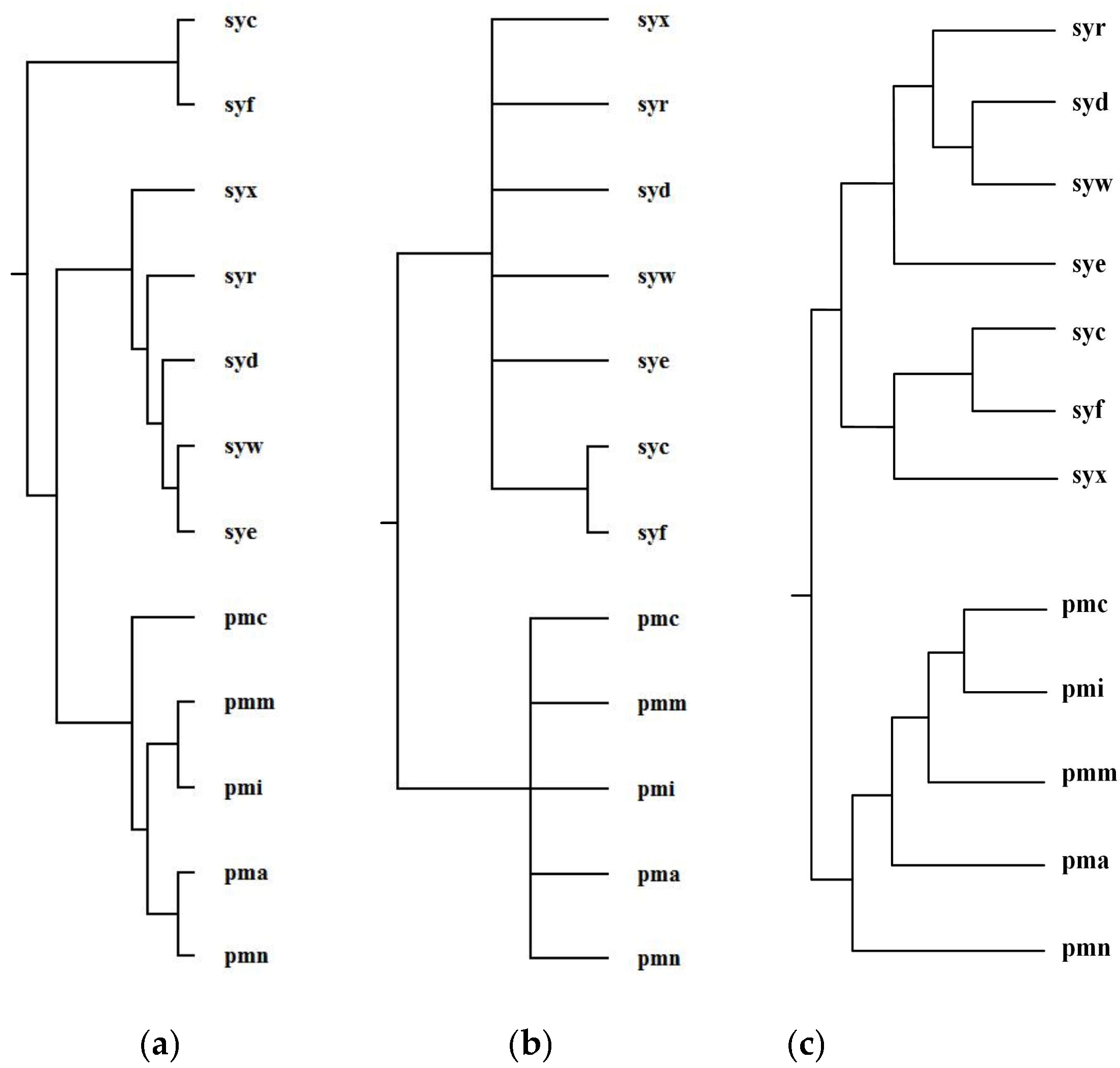
| Reconstructed Tree | Similarity |
|---|---|
| Our tree T1 | 0.38 |
| Chang et al.’s tree T2 | 0.19 |
| Clemente et al.’s tree T3 | 0.16 |
| Reconstructed Tree | Similarity |
|---|---|
| Our tree T1 | 0.20 |
| Ma et al.’s tree T2 | 0.12 |
| Reconstructed Tree | Similarity |
|---|---|
| Our tree T1 | 0.14 |
| Ma et al.’s tree T2 | 0.10 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Zhong, C.; Lin, H.X.; Wang, J.; Peng, Y. Reconstructing Phylogeny by Aligning Multiple Metabolic Pathways Using Functional Module Mapping. Molecules 2018, 23, 486. https://doi.org/10.3390/molecules23020486
Huang Y, Zhong C, Lin HX, Wang J, Peng Y. Reconstructing Phylogeny by Aligning Multiple Metabolic Pathways Using Functional Module Mapping. Molecules. 2018; 23(2):486. https://doi.org/10.3390/molecules23020486
Chicago/Turabian StyleHuang, Yiran, Cheng Zhong, Hai Xiang Lin, Jianyi Wang, and Yuzhong Peng. 2018. "Reconstructing Phylogeny by Aligning Multiple Metabolic Pathways Using Functional Module Mapping" Molecules 23, no. 2: 486. https://doi.org/10.3390/molecules23020486





