Total Synthesis and Pharmacological Investigation of Cordyheptapeptide A
Abstract
:1. Introduction
2. Results
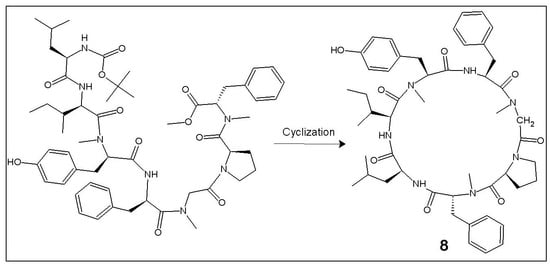
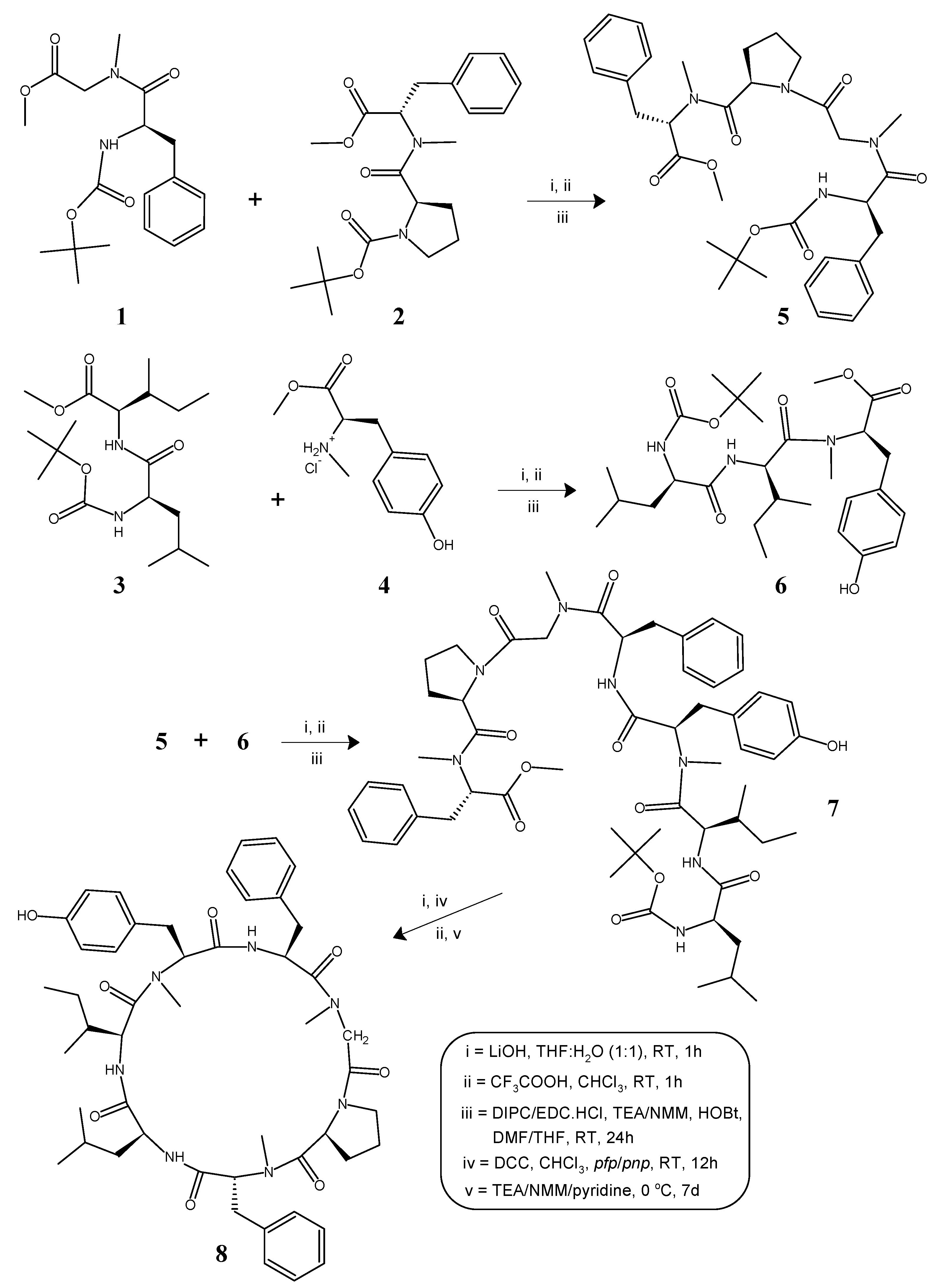
2.1. Chemistry
2.2. Pharmacological Activity Studies
3. Discussion
4. Materials and Methods
4.1. General Method for the Synthesis of Linear N-methylated Tri/Tetrapeptide Units (5, 6)
4.1.1. Tert-Butyloxycarbonyl-l-phenylalanyl-N(Me)glycyl-l-prolyl-d-N(Me)phenylalanine Methyl Ester (5)
4.1.2. Tert-Butyloxycarbonyl-l-leucyl-l-isoleucyl-l-N(Me)tyrosine Methyl Ester (6)
4.2. Deprotection of the Tetrapeptide Unit (5) at the Amino End and Tripeptide Unit (6) at the Carboxyl End
4.2.1. l-Phenylalanyl-N(Me)glycyl-l-prolyl-d-N(Me)phenylalanine Methyl Ester (5a)
4.2.2. Tert-Butyloxycarbonyl-l-leucyl-l-isoleucyl-l-N(Me)tyrosine (6a)
4.3. Procedure for the Preparation of the Linear Heptapeptide Unit and Its Cyclized Form (7, 8)
4.3.1. Tert-Butyloxycarbonyl-l-leucyl-l-isoleucyl-l-N(Me)tyrosyl-l-phenylalanyl-N(Me)glycyl-l-prolyl-d-N(Me)phenylalanine Methyl Ester (7)
4.3.2. Cyclo(l-leucyl-l-isoleucyl-l-N(Me)tyrosyl-l-phenylalanyl-N(Me)glycyl-l-prolyl-d-N(Me)phenylalanyl) (8)
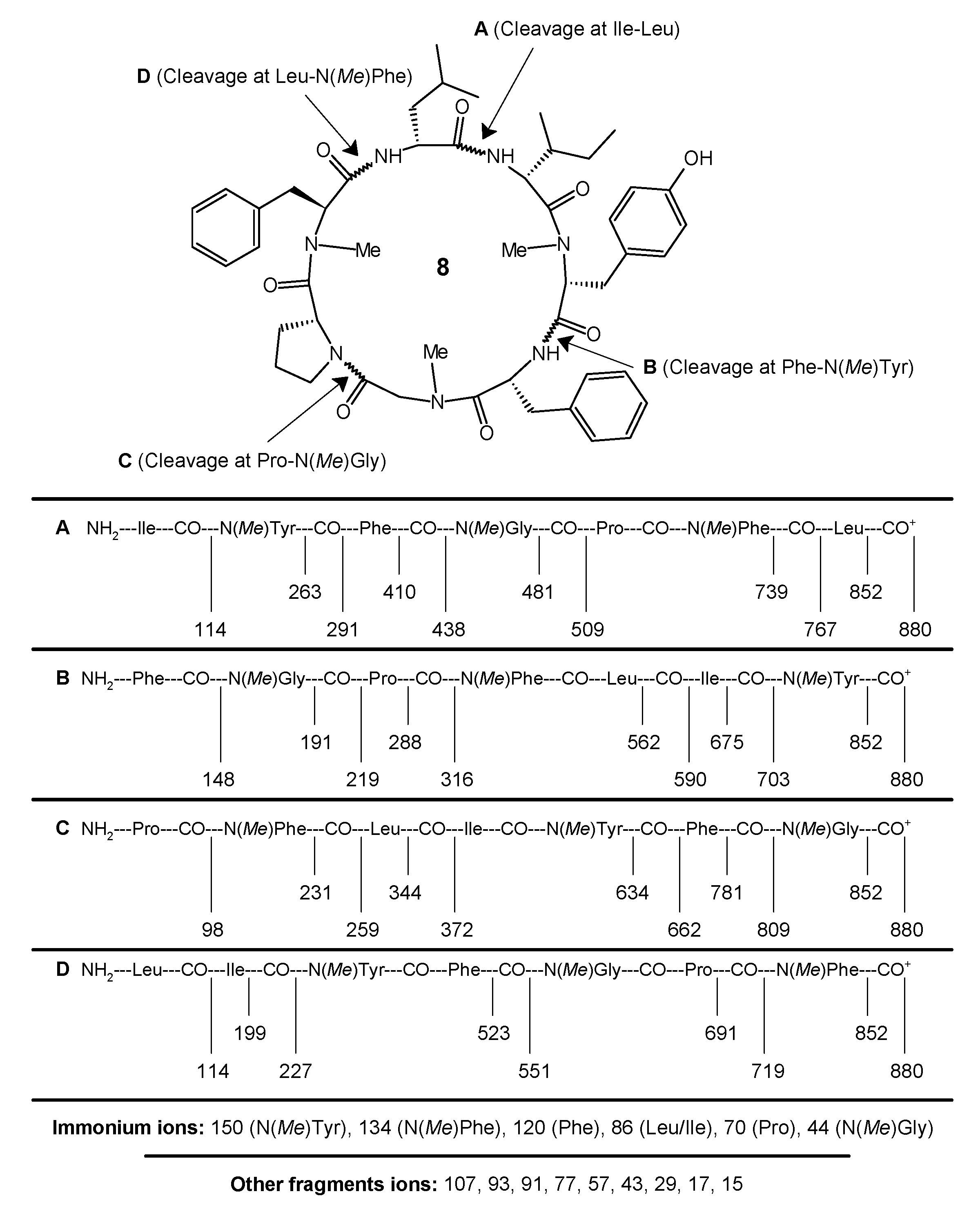
4.4. Preparation and Analysis of the Marfey Derivatives
4.5. Biological Evaluation Procedures
4.5.1. Cytotoxic Screening
4.5.2. Antimicrobial Screening
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Amedei, A.; D’Elios, M.M. New therapeutic approaches by using microorganism-derived compounds. Curr. Med. Chem. 2012, 19, 3822–3840. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, H.B. Natural products from microorganisms. Science 1980, 208, 1225–1229. [Google Scholar] [CrossRef] [PubMed]
- Leslie Gunatilaka, A.A. Natural products from plant-associated microorganisms: Distribution, structural diversity, bioactivity, and implications of their occurrence. J. Nat. Prod. 2006, 69, 509–526. [Google Scholar] [CrossRef] [PubMed]
- Dalsgaard, P.W.; Blunt, J.W.; Munro, M.H.; Larsen, T.O.; Christophersen, C. Psychrophilin B and C: Cyclic nitropeptides from the psychrotolerant fungus Penicillium rivulum. J. Nat. Prod. 2004, 67, 1950–1952. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, G.; Lang, G.; Steffens, S.; Schaumann, K. Petrosifungins A and B, novel cyclodepsipeptides from a sponge-derived strain of Penicillium brevicompactum. J. Nat. Prod. 2004, 67, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Dalsgaard, P.W.; Larsen, T.O.; Christophersen, C. Bioactive cyclic peptides from the psychrotolerant fungus Penicillium algidum. J. Antibiot. 2005, 58, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Vongvanich, N.; Kittakoop, P.; Isaka, M.; Trakulnaleamsai, S.; Vimuttipong, S.; Tanticharoen, M.; Thebtaranonth, Y. Hirsutellide A, a new antimycobacterial cyclohexadepsipeptide from the entomopathogenic fungus Hirsutella kobayasii. J. Nat. Prod. 2002, 65, 1346–1348. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Chen, A.J.; Lan, J.; Zhang, H.; Chen, X.; Liu, X.; Zou, Z. Sesquiterpenes and cyclopeptides from the endophytic fungus Trichoderma asperellum Samuels, Lieckf. & Nirenberg. Chem. Biodivers. 2012, 9, 1205–1212. [Google Scholar] [PubMed]
- Zou, X.; Niu, S.; Ren, J.; Li, E.; Liu, X.; Che, Y. Verrucamides A-D, antibacterial cyclopeptides from Myrothecium verrucaria. J. Nat. Prod. 2011, 74, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, S.; Liu, H.; Liu, X.; Che, Y. Cycloaspeptides F and G, cyclic pentapeptides from a Cordyceps-colonizing isolate of Isaria farinosa. J. Nat. Prod. 2009, 72, 1364–1367. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Bao, J.; Zhang, X.Y.; Tu, Z.C.; Shi, Y.M.; Qi, S.H. Asperterrestide A, a cytotoxic cyclic tetrapeptide from the marine-derived fungus Aspergillus terreus SCSGAF0162. J. Nat. Prod. 2013, 76, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, R.; Pathak, D. Synthesis, characterization and biological evaluation of halolitoralin B—A natural cyclic peptide. Asian J. Chem. 2007, 19, 1499–1505. [Google Scholar]
- Rukachaisirikul, V.; Chantaruk, S.; Tansakul, C.; Saithong, S.; Chaicharernwimonkoon, L.; Pakawatchai, C.; Isaka, M.; Intereya, K. A cyclopeptide from the insect pathogenic fungus Cordyceps sp. BCC 1788. J. Nat. Prod. 2006, 69, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Isaka, I.; Srisanoh, U.; Lartpornmatulee, N.; Boonruangprapa, T. ES-242 derivatives and cycloheptapeptides from Cordyceps sp. strains BCC 16173 and BCC 16176. J. Nat. Prod. 2007, 70, 1601–1604. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, R.; Gautam, H. Solution phase synthesis and bioevaluation of cordyheptapeptide B. Bull. Pharm. Res. 2011, 1, 1–10. [Google Scholar]
- Chen, Z.; Song, Y.; Chen, Y.; Huang, H.; Zhang, W.; Ju, J. Cyclic heptapeptides, cordyheptapeptides C-E, from the marine-derived fungus Acremonium persicinum SCSIO 115 and their cytotoxic activities. J. Nat. Prod. 2012, 75, 1215–1219. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.Y.; Dahiya, R.; Qin, H.L.; Mourya, R.; Maharaj, S. Natural proline-rich cyclopolypeptides from marine organisms: Chemistry, synthetic methodologies and biological status. Mar. Drugs 2016, 14, 194. [Google Scholar] [CrossRef] [PubMed]
- Pathak, D.; Dahiya, R. Cyclic peptides as novel antineoplastic agents: A review. J. Sci. Pharm. 2003, 4, 125–131. [Google Scholar]
- Dahiya, R. Cyclopolypeptides with antifungal interest. Coll. Pharm. Commun. 2013, 1, 1–15. [Google Scholar]
- Dahiya, R.; Pathak, D. Synthetic studies on a natural cyclic tetrapeptide-halolitoralin C. J. Pharm. Res. 2006, 5, 69–73. [Google Scholar]
- Dahiya, R.; Pathak, D.; Himaja, M.; Bhatt, S. First total synthesis and biological screening of hymenamide E. Acta Pharm. 2006, 56, 399–415. [Google Scholar] [PubMed]
- Dahiya, R.; Pathak, D. First total synthesis and biological evaluation of halolitoralin A. J. Serbian Chem. Soc. 2007, 72, 101–107. [Google Scholar] [CrossRef]
- Dahiya, R. Synthesis, characterization and biological evaluation of a glycine-rich peptide—Cherimolacyclopeptide E. J. Chil. Chem. Soc. 2007, 52, 1224–1229. [Google Scholar] [CrossRef]
- Dahiya, R.; Kaur, K. Synthetic and biological studies on natural cyclic heptapeptide: Segetalin E. Arch. Pharm. Res. 2007, 30, 1380–1386. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, R. Synthesis of a phenylalanine-rich peptide as potential anthelmintic and cytotoxic agent. Acta Pol. Pharm. 2007, 64, 509–516. [Google Scholar] [PubMed]
- Dahiya, R. Synthetic and pharmacological studies on longicalycinin A. Pak. J. Pharm. Sci. 2007, 20, 317–323. [Google Scholar] [PubMed]
- Dahiya, R.; Kumar, A. Synthesis and biological activity of a potent analog of natural cyclopeptide. Int. J. Nat. Appl. Sci. 2007, 3, 433–440. [Google Scholar] [CrossRef]
- Dahiya, R. Synthesis, spectroscopic and biological investigation of cyclic octapeptide: Cherimolacyclopeptide G. Turk. J. Chem. 2008, 32, 205–215. [Google Scholar]
- Dahiya, R. Total synthesis and biological potential of psammosilenin A. Arch. Pharm. Chem. Life Sci. 2008, 341, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, R. Synthetic studies on a cyclic hexapeptide from Dianthus superbus. Chem. Pap. 2008, 62, 527–535. [Google Scholar] [CrossRef]
- Dahiya, R. Synthesis and in vitro cytotoxic activity of a natural peptide of plant origin. J. Iran. Chem. Soc. 2008, 5, 445–452. [Google Scholar] [CrossRef]
- Dahiya, R.; Sharma, R.D. Synthesis and bioactivity of a novel cyclic hexapeptide from Stellaria delavayi. Eur. J. Sci. Res. 2008, 21, 277–287. [Google Scholar]
- Dahiya, R.; Kumar, A. Synthetic and biological studies on a cyclopolypeptide of plant origin. J. Zhejiang Univ. Sci. B 2008, 9, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, R.; Kaur, K. Synthetic and Pharmacological Investigation of Segetalin C as a Novel Antifungal and Cytotoxic Agent. Arzneim. Forsch. 2008, 58, 29–34. [Google Scholar]
- Dahiya, R.; Maheshwari, M.; Kumar, A. Toward the synthesis and biological evaluation of hirsutide. Monatsh. Chem. 2009, 140, 121–127. [Google Scholar] [CrossRef]
- Dahiya, R.; Kumar, A.; Gupta, R. Synthesis, cytotoxic and antimicrobial screening of a proline-rich cyclopolypeptide. Chem. Pharm. Bull. 2009, 57, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, R.; Maheshwari, M.; Yadav, R. Synthetic, cytotoxic and antimicrobial activity studies on annomuricatin B. Z. Naturforsch. 2009, 64b, 237–244. [Google Scholar] [CrossRef]
- Dahiya, R.; Gautam, H. Total synthesis and antimicrobial activity of a natural cycloheptapeptide of marine-origin. Mar. Drugs 2010, 8, 2384–2394. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, R.; Gautam, H. Synthetic and pharmacological studies on a natural cyclopeptide from Gypsophila arabica. J. Med. Plants Res. 2010, 4, 1960–1966. [Google Scholar]
- Dahiya, R.; Gautam, H. Toward the first total synthesis of gypsin D: A natural cyclopolypeptide from Gypsophila arabica. Am. J. Sci. Res. 2010, 11, 150–158. [Google Scholar]
- Dahiya, R.; Gautam, H. Synthesis, characterization and biological evaluation of cyclomontanin D. Afr. J. Pharm. Pharmacol. 2011, 5, 447–453. [Google Scholar] [CrossRef]
- Dahiya, R.; Gautam, H. Toward the synthesis and biological screening of a cyclotetrapeptide from marine bacteria. Mar. Drugs 2011, 9, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, R.; Singh, S. First total synthesis and biological potential of a heptacyclopeptide of plant origin. Chin. J. Chem. 2016, 34, 1158–1164. [Google Scholar] [CrossRef]
- Dahiya, R.; Singh, S. Synthesis, characterization and biological screening of diandrine A. Acta Pol. Pharm. 2017, 3. accepted. [Google Scholar]
- Dahiya, R.; Singh, S. Synthesis, characterization, and biological activity studies on fanlizhicyclopeptide A. Iran. J. Pharm. Res. 2017. accepted. [Google Scholar]
- Das, P.; Himaja, M. Design and synthesis of 4-[2′-(5′-nitro)imidazolyl]benzoyl (N-methyl) amino acids and peptides. Int. J. Drug Dev. Res. 2010, 2, 364–370. [Google Scholar]
- Bodanzsky, M.; Bodanzsky, A. The Practice of Peptide Synthesis; Springer: New York, NY, USA, 1984; pp. 78–143. [Google Scholar]
- Kuttan, R.; Bhanumathy, P.; Nirmala, K.; George, M.C. Potential anticancer activity of turmeric (Curcuma longa). Cancer Lett. 1985, 29, 197–202. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1996, 45, 493–496. [Google Scholar]
- Joo, S.N. Cyclic peptides as therapeutic agents and biochemical tools. Biomol. Ther. (Seoul) 2012, 20, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, R.; Singh, S. Toward the synthesis and pharmacological screening of a natural cycloheptapeptide of plant origin. Nat. Prod. Commun. 2017, 12, 379–383. [Google Scholar]
- Dahiya, R.; Pathak, D.; Bhatt, S. Synthesis and biological evaluation of a novel series of 2-(4-chloro-3-methylphenoxy) acetyl amino acids and peptides. J. Saudi Chem. Soc. 2006, 10, 165–176. [Google Scholar]
- Dahiya, R.; Pathak, D. Synthetic studies on novel benzimidazolopeptides with antimicrobial, cytotoxic and anthelmintic potential. Eur. J. Med. Chem. 2007, 42, 772–798. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, R.; Singh, S.; Sharma, A.; Chennupati, S.V.; Maharaj, S. First total synthesis and biological screening of a proline-rich cyclopeptide from a Caribbean marine sponge. Mar. Drugs 2016, 14, 228. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, R.; Kumar, A.; Yadav, R. Synthesis and biological activity of peptide derivatives of iodoquinazolinones/nitroimidazoles. Molecules 2008, 13, 958–976. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available. |
| Compd. | Conc. (μg/mL) | Dalton’s Lymphoma Ascites (DLA) Cells | Ehrlich’s Ascites Carcinoma (EAC) Cells | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Live Cells Counted | No. of Dead Cells | % GI a | CTC50 (μM) b | Live Cells Counted | No. of Dead Cells | % GI | CTC50 (μM) | ||
| 7 | 62.50 | 0 | 38 | 100.0 | 0 | 28 | 100.0 | ||
| 31.25 | 11 | 27 | 71.05 | 8 | 20 | 71.43 | |||
| 15.63 | 21 | 17 | 44.74 | 19.9 | 17 | 11 | 39.29 | 22.6 | |
| 7.81 | 25 | 12 | 34.21 | 21 | 7 | 25.00 | |||
| 3.91 | 34 | 4 | 10.53 | 24 | 4 | 14.29 | |||
| 8 | 62.50 | 0 | 38 | 100.0 | 0 | 28 | 100.0 | ||
| 31.25 | 7 | 31 | 81.58 | 4 | 24 | 85.71 | |||
| 15.63 | 15 | 23 | 60.53 | 10.6 | 11 | 17 | 60.71 | 14.6 | |
| 7.81 | 22 | 16 | 42.11 | 18 | 10 | 35.71 | |||
| 3.91 | 27 | 10 | 28.95 | 22 | 6 | 21.43 | |||
| Control | 62.50 | 38 | 0 | – | 28 | 0 | – | ||
| 31.25 | 38 | 0 | – | 28 | 0 | – | |||
| 15.63 | 38 | 0 | – | – | 28 | 0 | – | – | |
| 7.81 | 38 | 0 | – | 28 | 0 | – | |||
| 3.91 | 38 | 0 | – | 28 | 0 | – | |||
| Std. | 62.50 | 0 | 38 | 100.0 | 0 | 28 | 100.0 | ||
| (5-FU) | 31.25 | 0 | 38 | 100.0 | 0 | 28 | 100.0 | ||
| 15.63 | 10 | 28 | 73.68 | 37.4 | 11 | 17 | 60.71 | 90.6 | |
| 7.81 | 13 | 25 | 65.79 | 19 | 9 | 32.14 | |||
| 3.91 | 22 | 16 | 42.11 | 23 | 5 | 17.86 | |||
| Compound | Diameter of the Zone of Inhibition (mm) | |||||||
|---|---|---|---|---|---|---|---|---|
| Bacterial Strains | Fungal Strains | |||||||
| B. sub. | S. aur. | P. aeru. | K. pneu. | C. alb. | M. audo. | A. niger | T. menta. | |
| 7 | 10(50) | – | 21(6) | 23(6) | 12(25) | 18(6) | – | 16(6) |
| 8 | 12(50) | 10(25) | 25(6) | 26(6) | 15(25) | 22(6) | 9(25) | 20(6) |
| DMF † | – | – | – | – | ||||
| DMSO ‡ | – | – | – | – | ||||
| Gatifloxacin | 18(12.5) * | 27(6) | 23(6) | 25(6) | – | – | – | – |
| Griseofulvin | – | – | – | – | 20(6) | 18(6) | 20(12.5) | 19(6) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, S.; Dahiya, R.; Khokra, S.L.; Mourya, R.; Chennupati, S.V.; Maharaj, S. Total Synthesis and Pharmacological Investigation of Cordyheptapeptide A. Molecules 2017, 22, 682. https://doi.org/10.3390/molecules22060682
Kumar S, Dahiya R, Khokra SL, Mourya R, Chennupati SV, Maharaj S. Total Synthesis and Pharmacological Investigation of Cordyheptapeptide A. Molecules. 2017; 22(6):682. https://doi.org/10.3390/molecules22060682
Chicago/Turabian StyleKumar, Suresh, Rajiv Dahiya, Sukhbir Lal Khokra, Rita Mourya, Suresh V. Chennupati, and Sandeep Maharaj. 2017. "Total Synthesis and Pharmacological Investigation of Cordyheptapeptide A" Molecules 22, no. 6: 682. https://doi.org/10.3390/molecules22060682