Molecular Cloning of cpcU and Heterodimeric Bilin Lyase Activity Analysis of CpcU and CpcS for Attachment of Phycocyanobilin to Cys-82 on the β-Subunit of Phycocyanin in Arthrospira platensis FACHB314
Abstract
:1. Introduction
2. Results
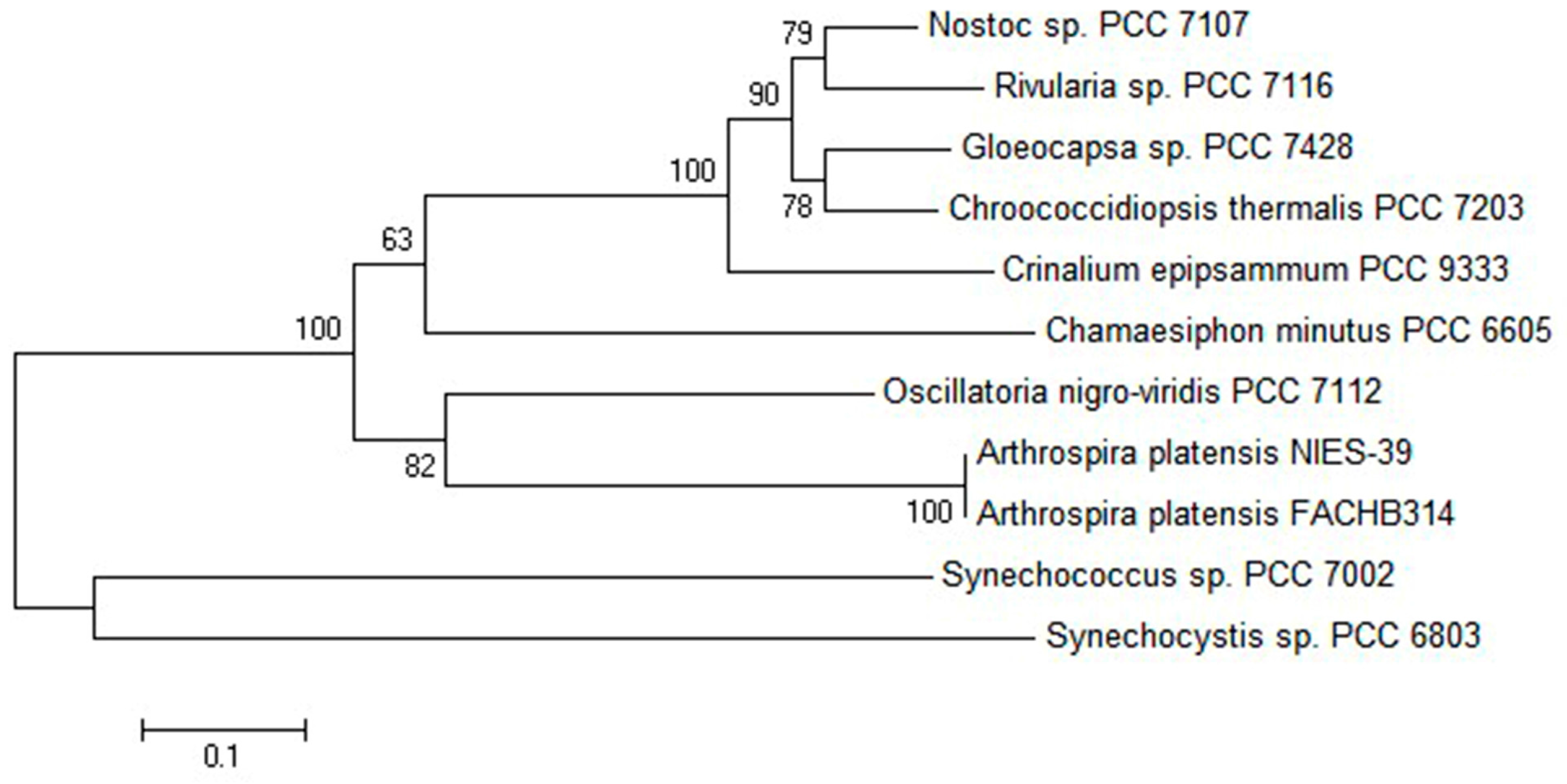
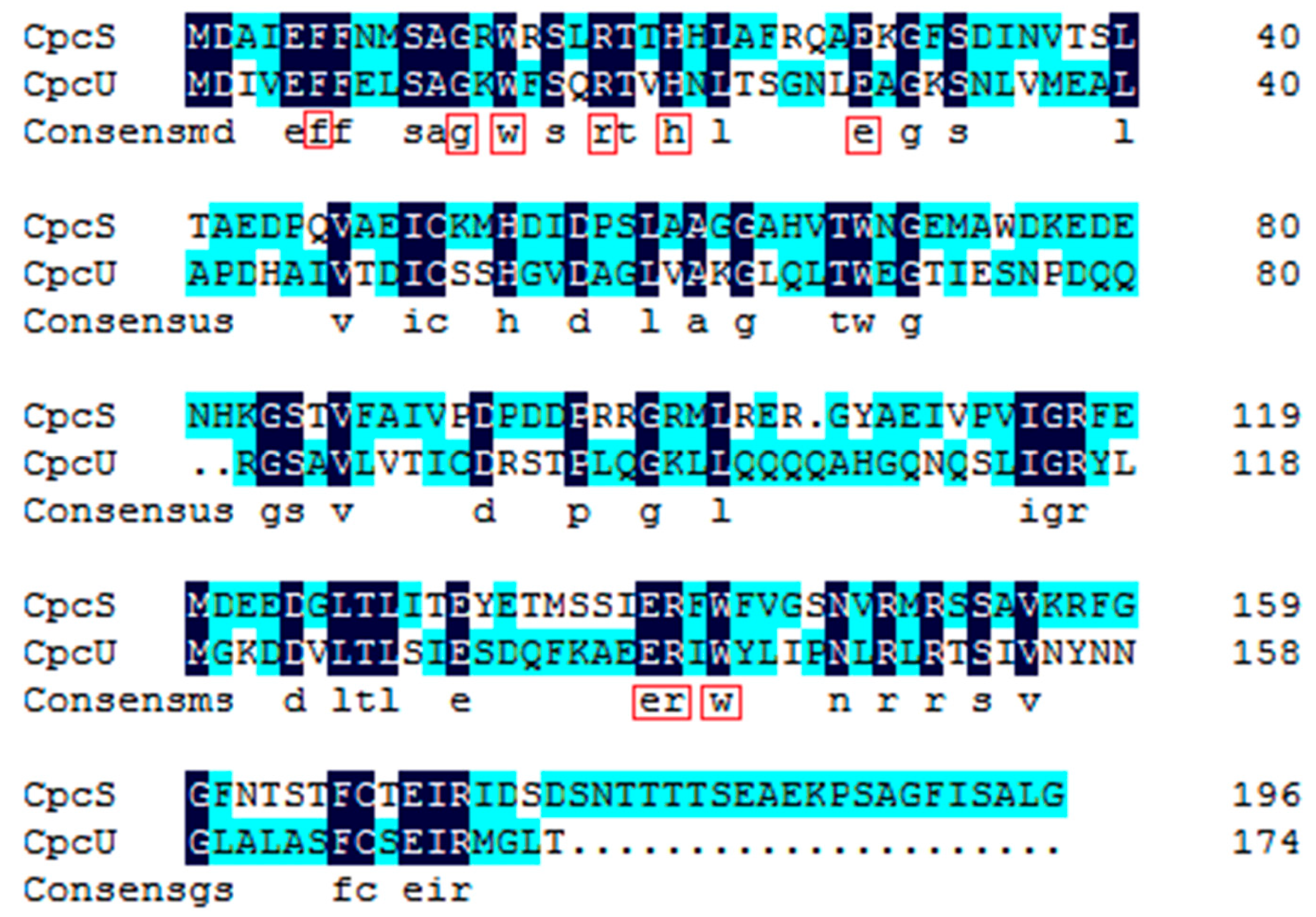
2.1. Comparative Bioinformatics Analysis of cpcU
2.2. The Construction of the Recombinant Expression Strains
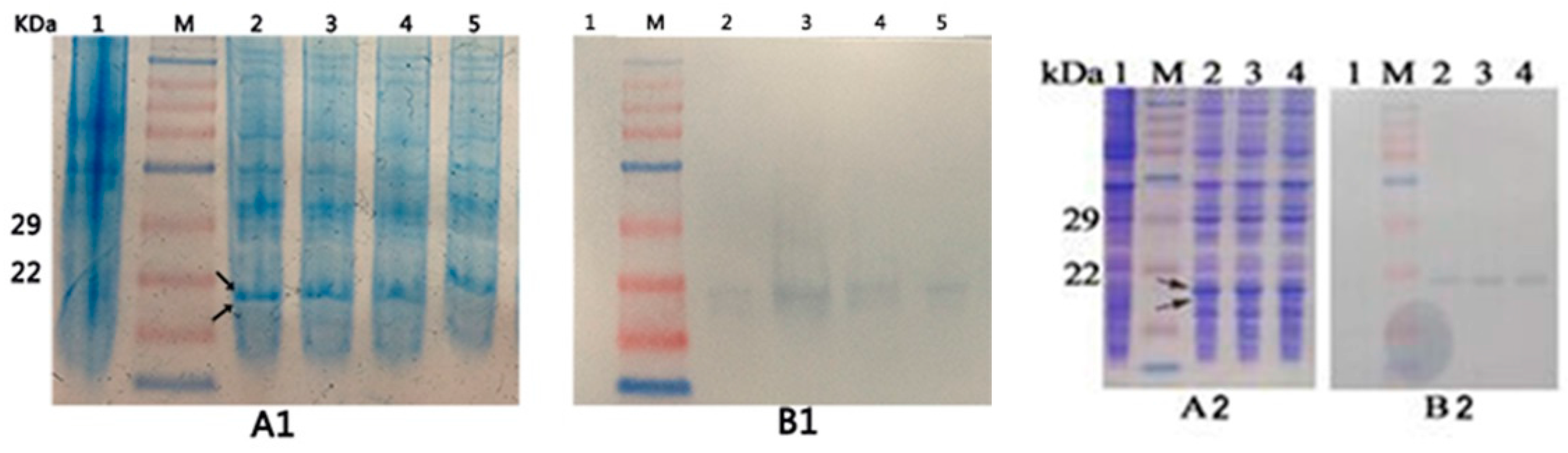
2.3. Expression of the Recombinant Proteins
2.4. Fluorescence Emission Spectra
3. Discussion
4. Materials and Methods
4.1. Strains and Plasmids
4.2. Genomic DNA Isolation and Gene Cloning of cpcU
4.3. Construction of the Recombinant Expression Vectors
4.4. Plasmid Transformation and Protein Expression
4.5. Recombinant Protein Analysis
4.6. Fluorescence Emission Spectra
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Glazer, A.N. Light harvesting by phycobilisomes. Ann. Rev. Biophys. Biophys. Chem. 1985, 14, 47–77. [Google Scholar] [CrossRef] [PubMed]
- Glazer, A.N. Light guides. Directional energy transfer in a photosynthetic antenna. J. Biol. Chem. 1989, 264, 1–4. [Google Scholar] [PubMed]
- Sidler, W.A. The Molecular Biology of Cyanobacteria; Springer Netherlands: Dordrecht, The Netherlands, 1994; pp. 139–216. [Google Scholar]
- Schirmer, T.; Bode, W.; Huber, R. Refined three-dimensional structures of two cyanobacterial C-phycocyanins at 2.1 and 2.5 A resolution. A common principle of phycobilin-protein interaction. J. Mol. Biol. 1987, 196, 677–695. [Google Scholar] [CrossRef]
- Tooley, A.J.; Glazer, A.N. Biosynthesis of the cyanobacterial light-harvesting polypeptide phycoerythrocyanin holo-α-subunit in a heterologous host. J. Bacteriol. 2002, 184, 4666–4671. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.H.; Gasparich, G.E.; Stirewalt, V.L.; Bryant, D.A. The cpcE and cpcF genes of Synechococcus sp. PCC 7002. J. Biol. Chem. 1992, 267, 16138–16145. [Google Scholar] [PubMed]
- Yi, J.J.; Zang, X.N.; Zhang, X.C.; Yuan, D.Y.; Zhao, B.R.; Tang, L. Recombinant expression of a fluorescent phycocyanin holo-α-subunit from Arthrospira platensis in Escherichia coli. Period. Ocean Univ. China 2011, 41, 59–65. (In Chinese) [Google Scholar]
- Shen, G.; Saunee, N.A.; Gallo, E.F.; Begovic, Z.; Schluchter, W.M.; Bryant, D.A. Photosynthesis 2004 Light-harvesting Systems Workshop; Niederman, R.A., Frank, H., Robert, B., van Grondelle, R., Eds.; Saint Adele, QC, Canada, 2004; pp. 14–15. [Google Scholar]
- Zhao, K.H.; Su, P.; Tu, J.M.; Wang, X.; Liu, H.; Plöscher, M.; Eichacker, L.; Yang, B.; Zhou, M.; Scheer, H. Phycobilin: Cystein-84 biliprotein lyase, a near-universal lyase for cysteine-84-binding sites in cyanobacterial phycobiliproteins. Proc. Natl. Acad. Sci. USA 2007, 104, 14300–14305. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.Z.; Saunée, N.A.; Williams, S.R.; Gallo, E.F.; Schluchter, W.M. Identification and characterization of a new class of bilin lyase responsible for attachment of phycocyanobilin to Cys 153 on the β-subunit of phycocyanin in Synechococcus sp. PCC 7002. J. Biol. Chem. 2006, 27, 16644–16722. [Google Scholar]
- Zhao, K.H.; Wu, D.; Zhang, L.; Zhou, M.; Böhm, S.; Bubenzer, C.; Scheer, H. Chromophore attachment in phycocyanin. Functional amino acids of phycocyanobilin-alphaphycocyanin lyase and evidence for chromophore binding. FEBS J. 2006, 273, 1262–1274. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.Z.; Schluchter, W.M.; Bryant, D.A. Biogenesis of phycobiliproteins I. CpcS-I and CpcU mutants of the cyanobacterium Synechococcus sp. PCC 7002 define a heterodimeric phycocyanobilin lyase specific for β-phycocyanin and allophycocyanin subunits. J. Biol. Chem. 2008, 283, 7503–7512. [Google Scholar] [CrossRef] [PubMed]
- Ong, L.J.; Glazer, A.N. Structural studies of phycobiliproteins in unicellular marine cyanobacteria. In Light-Energy Transduction in Photosynthesis: Higher Plant and Bacterial Models; Stevens, S.E., Bryant, D.A., Jr., Eds.; American Society of Plant Physiologists: Rockville, MD, USA, 1988; pp. 102–121. [Google Scholar]
- Zhao, K.H.; Zhang, J.; Tu, J.M.; Böhm, S.; Plöscher, M.; Eichacker, L.; Bubenzer, C.; Scheer, H.; Wang, X.; Zhou, M. Lyase activities of CpcS- and CpcT-like proteins from Nostoc PCC 7120 and sequential reconstitution of binding sites of phycoerythrocyanin and phycocyanin β-subunits. J. Biol. Chem. 2007, 282, 34093–34103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Feng, X.T.; Wu, F.; Ding, Y.; Zang, X.N.; Zhang, X.C.; Yuan, D.Y.; Zhao, B.R. Molecular cloning and expression analysis of a new bilin lyase: The cpcT gene encoding a bilin lyase responsible for attachment of phycocyanobilin to Cys-153 on the β-subunit of phycocyanin in Arthrospira platensis FACHB314. Gene 2014, 544, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Fairchild, C.D. Oligomeric structure, enzyme kinetics, and substrate specificity of the phycocyanin a subunit phycocyanobilin lyase. J. Biol. Chem. 1994, 269, 8686–8694. [Google Scholar] [PubMed]
- Fairchild, C.D.; Zhao, J.D.; Zhou, J.H.; Colson, S.E.; Bryant, D.A.; Glazer, A.N. Phycocyanin α-subunit phycocyanobilin lyase. Proc. Natl. Acad. Sci. USA. 1992, 89, 7017–7021. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, F.T.; Forreiter, C.; Hurtado Pico, A.; Lamparter, T.; Hughes, J. Recombinant holophytochrome in Escherichia coli. FEBS Lett. 2001, 508, 459–462. [Google Scholar] [CrossRef]
- Shen, G.Z.; Saunee, N.A.; Williams, S.R.; Gallo, E.F.; Schluchter, W.M.; Bryant, D.A. Identification and characterization of a new class of bilin lyase: The cpcT gene encodes a bilin lyase responsible for attachment of phycocyanobilin to Cys-153 on the β-subunit of phycocyanin in Synechococcus sp. PCC 7002. J. Biol. Chem. 2006, 281, 17768–17778. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, D.L.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; Federhen, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2006, 34, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Tooley, A.J.; Cai, Y.A.; Glazer, A.N. Biosynthesis of a fluorescent cyanobacterial C-phycocyanin holo-α subunit in a heterologous host. Proc. Natl. Acad. Sci. USA 2001, 98, 10560–10565. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.Z.; Bryant, D.A. Characterization of a Synechococcus sp. strain PCC 7002 mutant lacking Photosystem I. Protein assembly and energy distribution in the absence of the Photosystem I reaction center core complex. Photosynth. Res. 1995, 41, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Patel, A.; Zhao, Y.; Reuter, W. Structural basis for the photochemistry of α-Phycoerythrocyanin. Biochemistry 2007, 46, 416–423. [Google Scholar] [CrossRef] [PubMed]
- MacColl, R.; Csatorday, K.; Berns, D.S.; Traeger, E. Chromophore interactions in allophycocyanin. Biochemistry 1980, 19, 2817–2820. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds are available from the authors.

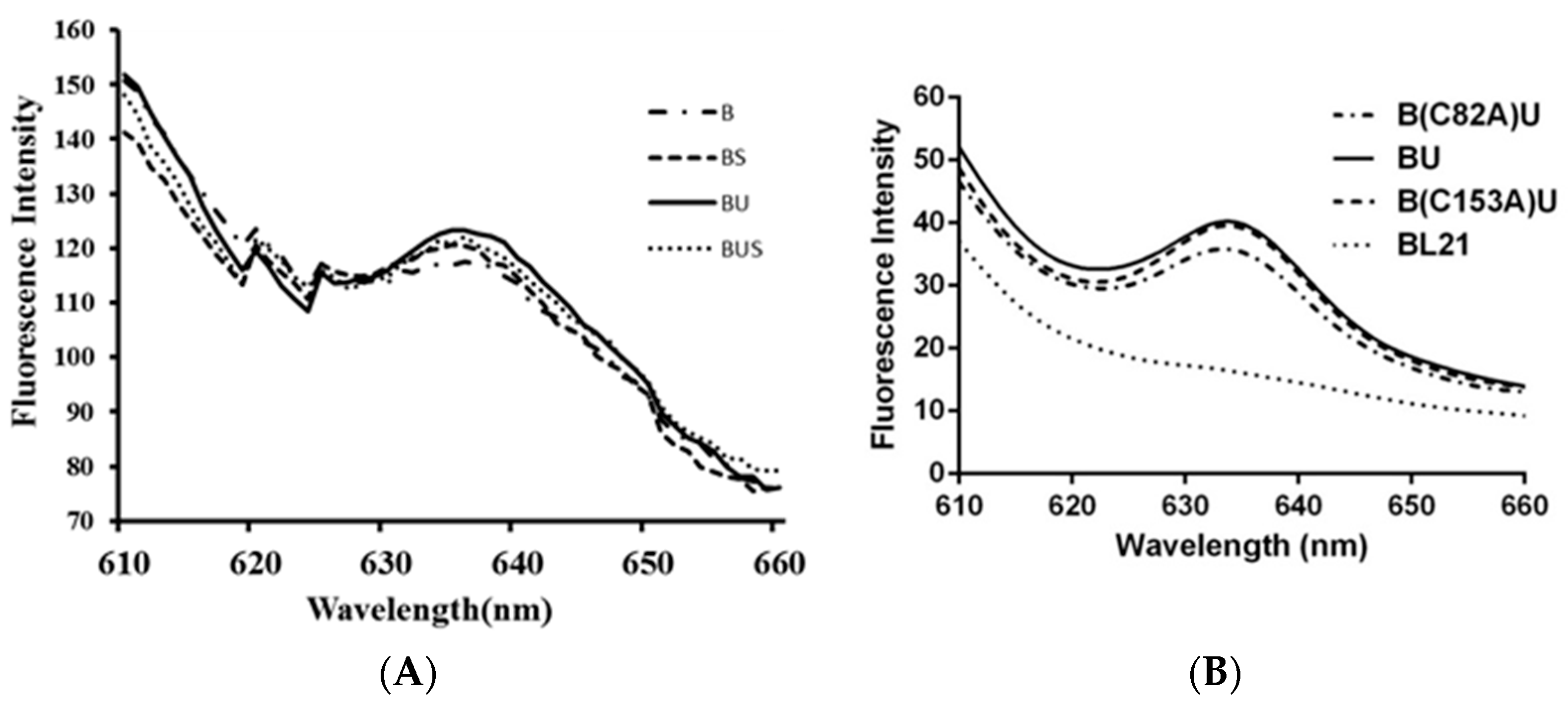


| Recombinant Strain | Total Protein Concentration (mg∙mL−1) | Phycocyanin (%) | Phycocyanin Concentration (mg∙mL−1) | Phycocyanin Fluorescence Intensity per Unit Mass |
|---|---|---|---|---|
| E. coli/B | 42.07 | 1.98 | 0.83 | 117.55 |
| E. coli/BU | 37.96 | 2.63 | 1.11 | 123.39 |
| E. coli/BS | 43.28 | 2.13 | 0.92 | 120.68 |
| E. coli/BUS | 41.44 | 2.27 | 0.94 | 122.24 |
| Recombinant Strain | Total Protein Concentration (mg∙mL−1) | Phycocyanin (%) | Phycocyanin Concentration (mg∙mL−1) | Phycocyanin Fluorescence Intensity per Unit Mass |
|---|---|---|---|---|
| E. coli/BU | 50.45 | 2.42 | 1.22 | 39.98 |
| E. coli/B(C82)U | 49.17 | 2.10 | 1.03 | 35.82 |
| E. coli/B(C153)U | 50.02 | 2.38 | 1.19 | 40.22 |
| Strain Source Application | ||
|---|---|---|
| pMD18-T | TaKaRa (Dalian, China) | Cloning vector |
| pACYCDuet-1 | Novagen (Germany) | Expression vector |
| E. coli DH5α | TaKaRa (Dalian, China) | Cloning strain |
| E. coli BL21 | TaKaRa (Dalian, China) | Expression strain |
| pET-hoxI-pcyA | Our laboratory | Expression the chromophore synthase |
| pACYCDuet-cpcB | Our laboratory | Expression the β-PC |
| pACYCDuet-cpcB-cpcS | Our laboratory | Expression the β-PC and the chromophore lyase CpcS |
| Names of the Transformed | Expression Vectors | |
|---|---|---|
| E. coli Strains | ||
| B | pACYCDuet-cpcB | pET-hoxI-pcyA |
| BU | pACYCDuet-cpcB-cpcU | pET-hoxI-pcyA |
| B(C82A)U | pACYCDuet-cpcB(C82A)-cpcU | pET-hoxI-pcyA |
| B(C153A)U | pACYCDuet-cpcB(C153A)-cpcU | pET-hoxI-pcyA |
| BS | pACYCDuet-cpcB-cpcS | pET-hoxI-pcyA |
| BUS | pACYCDuet-cpcB-cpcU-cpcS | pET-hoxI-pcyA |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, F.; Zang, X.; Zhang, X.; Zhang, R.; Huang, X.; Hou, L.; Jiang, M.; Liu, C.; Pang, C. Molecular Cloning of cpcU and Heterodimeric Bilin Lyase Activity Analysis of CpcU and CpcS for Attachment of Phycocyanobilin to Cys-82 on the β-Subunit of Phycocyanin in Arthrospira platensis FACHB314. Molecules 2016, 21, 357. https://doi.org/10.3390/molecules21030357
Wu F, Zang X, Zhang X, Zhang R, Huang X, Hou L, Jiang M, Liu C, Pang C. Molecular Cloning of cpcU and Heterodimeric Bilin Lyase Activity Analysis of CpcU and CpcS for Attachment of Phycocyanobilin to Cys-82 on the β-Subunit of Phycocyanin in Arthrospira platensis FACHB314. Molecules. 2016; 21(3):357. https://doi.org/10.3390/molecules21030357
Chicago/Turabian StyleWu, Fei, Xiaonan Zang, Xuecheng Zhang, Ran Zhang, Xiaoyun Huang, Lulu Hou, Minjie Jiang, Chang Liu, and Chunhong Pang. 2016. "Molecular Cloning of cpcU and Heterodimeric Bilin Lyase Activity Analysis of CpcU and CpcS for Attachment of Phycocyanobilin to Cys-82 on the β-Subunit of Phycocyanin in Arthrospira platensis FACHB314" Molecules 21, no. 3: 357. https://doi.org/10.3390/molecules21030357





