Life 2015, 5(1), 403-417; https://doi.org/10.3390/life5010403 - 4 Feb 2015
Cited by 38 | Viewed by 7543
Abstract
Marine Synechococcus and Prochlorococcus are picocyanobacteria predominating in subtropical, oligotrophic marine environments, a niche predicted to expand with climate change. When grown under common low light conditions Synechococcus WH 8102 and Prochlorococcus MED 4 show similar Cytochrome b6f and Photosystem I
[...] Read more.
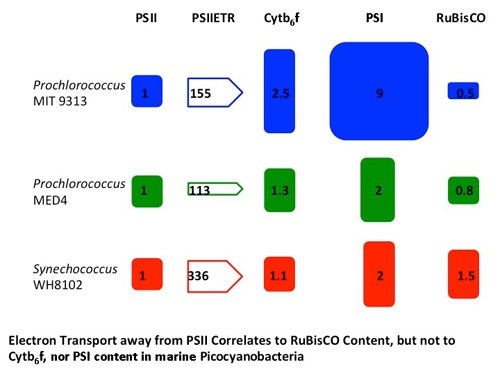
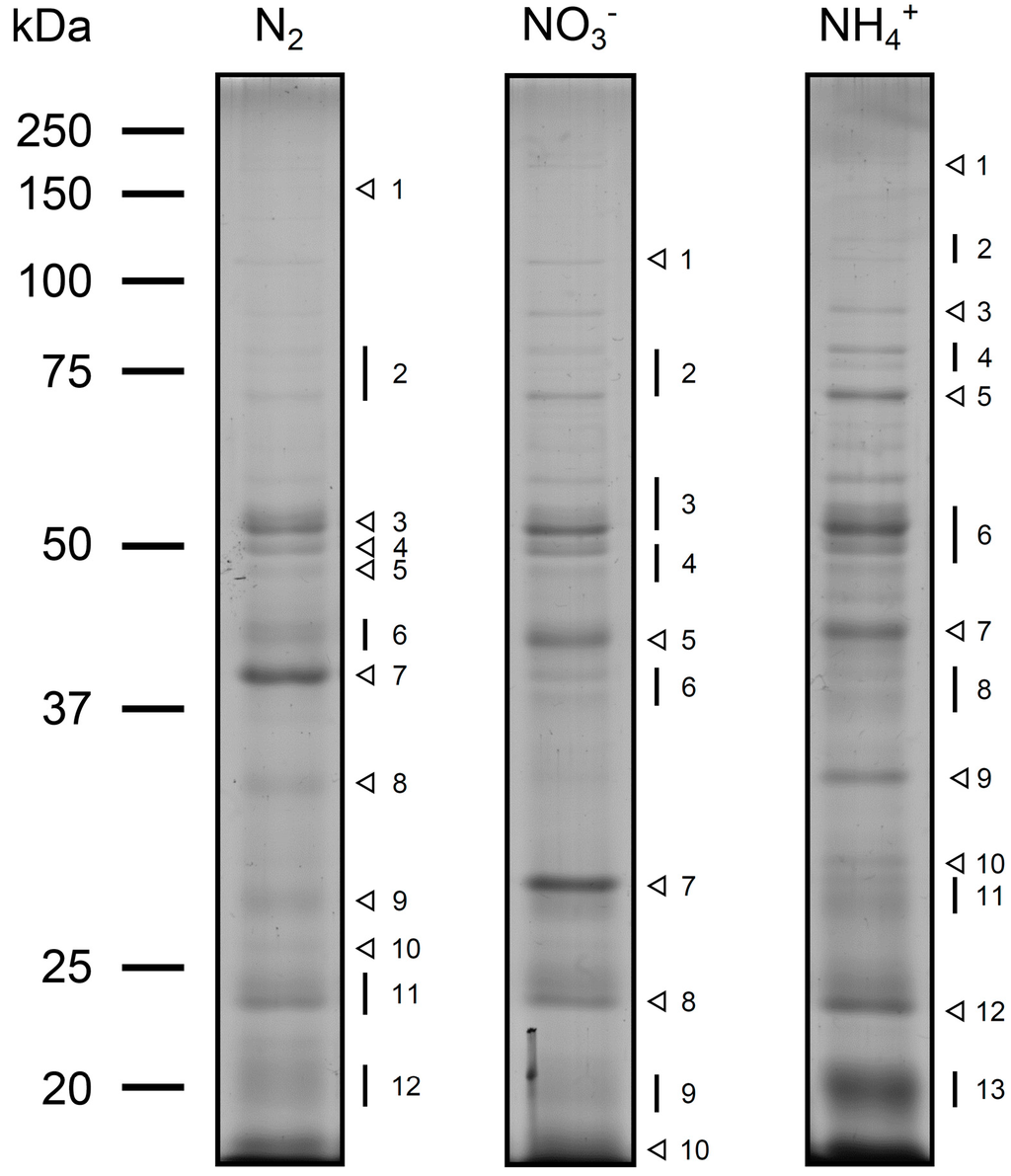
Marine Synechococcus and Prochlorococcus are picocyanobacteria predominating in subtropical, oligotrophic marine environments, a niche predicted to expand with climate change. When grown under common low light conditions Synechococcus WH 8102 and Prochlorococcus MED 4 show similar Cytochrome b6f and Photosystem I contents normalized to Photosystem II content, while Prochlorococcus MIT 9313 has twice the Cytochrome b6f content and four times the Photosystem I content of the other strains. Interestingly, the Prochlorococcus strains contain only one third to one half of the RUBISCO catalytic subunits compared to the marine Synechococcus strain. The maximum Photosystem II electron transport rates were similar for the two Prochlorococcus strains but higher for the marine Synechococcus strain. Photosystem II electron transport capacity is highly correlated to the molar ratio of RUBISCO active sites to Photosystem II but not to the ratio of cytochrome b6f to Photosystem II, nor to the ratio of Photosystem I: Photosystem II. Thus, the catalytic capacity for the rate-limiting step of carbon fixation, the ultimate electron sink, appears to limit electron transport rates. The high abundance of Cytochrome b6f and Photosystem I in MIT 9313, combined with the slower flow of electrons away from Photosystem II and the relatively low level of RUBISCO, are consistent with cyclic electron flow around Photosystem I in this strain.
Full article
(This article belongs to the Special Issue Cyanobacteria: Ecology, Physiology and Genetics)
►
Show Figures