Institute of Cytology, Russian Academy of Sciences, 194064 St. Petersburg, Russia
Int. J. Mol. Sci. 2022, 23(17), 9691; https://doi.org/10.3390/ijms23179691 - 26 Aug 2022
Cited by 21 | Viewed by 5252
Abstract
Polyploid cells demonstrate biological plasticity and stress adaptation in evolution; development; and pathologies, including cardiovascular diseases, neurodegeneration, and cancer. The nature of ploidy-related advantages is still not completely understood. Here, we summarize the literature on molecular mechanisms underlying ploidy-related adaptive features. Polyploidy can
[...] Read more.
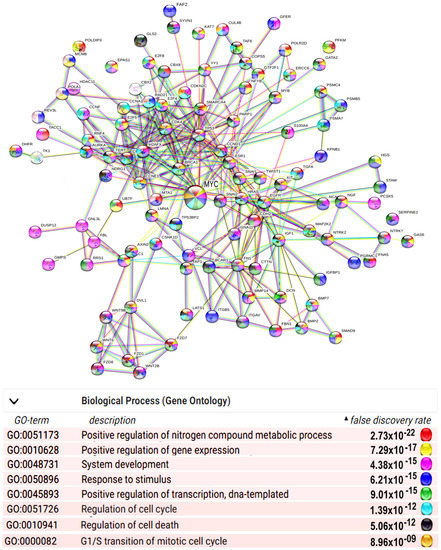
Polyploid cells demonstrate biological plasticity and stress adaptation in evolution; development; and pathologies, including cardiovascular diseases, neurodegeneration, and cancer. The nature of ploidy-related advantages is still not completely understood. Here, we summarize the literature on molecular mechanisms underlying ploidy-related adaptive features. Polyploidy can regulate gene expression via chromatin opening, reawakening ancient evolutionary programs of embryonality. Chromatin opening switches on genes with bivalent chromatin domains that promote adaptation via rapid induction in response to signals of stress or morphogenesis. Therefore, stress-associated polyploidy can activate Myc proto-oncogenes, which further promote chromatin opening. Moreover, Myc proto-oncogenes can trigger polyploidization de novo and accelerate genome accumulation in already polyploid cells. As a result of these cooperative effects, polyploidy can increase the ability of cells to search for adaptive states of cellular programs through gene regulatory network rewiring. This ability is manifested in epigenetic plasticity associated with traits of stemness, unicellularity, flexible energy metabolism, and a complex system of DNA damage protection, combining primitive error-prone unicellular repair pathways, advanced error-free multicellular repair pathways, and DNA damage-buffering ability. These three features can be considered important components of the increased adaptability of polyploid cells. The evidence presented here contribute to the understanding of the nature of stress resistance associated with ploidy and may be useful in the development of new methods for the prevention and treatment of cardiovascular and oncological diseases.
Full article
(This article belongs to the Special Issue Advances in Genome Regulation in Cancer)
▼
Show Figures