Optimization of Stress-Based Microfluidic Testing for Methicillin Resistance in Staphylococcus aureus Strains
Abstract
:1. Introduction
2. Materials and Methods
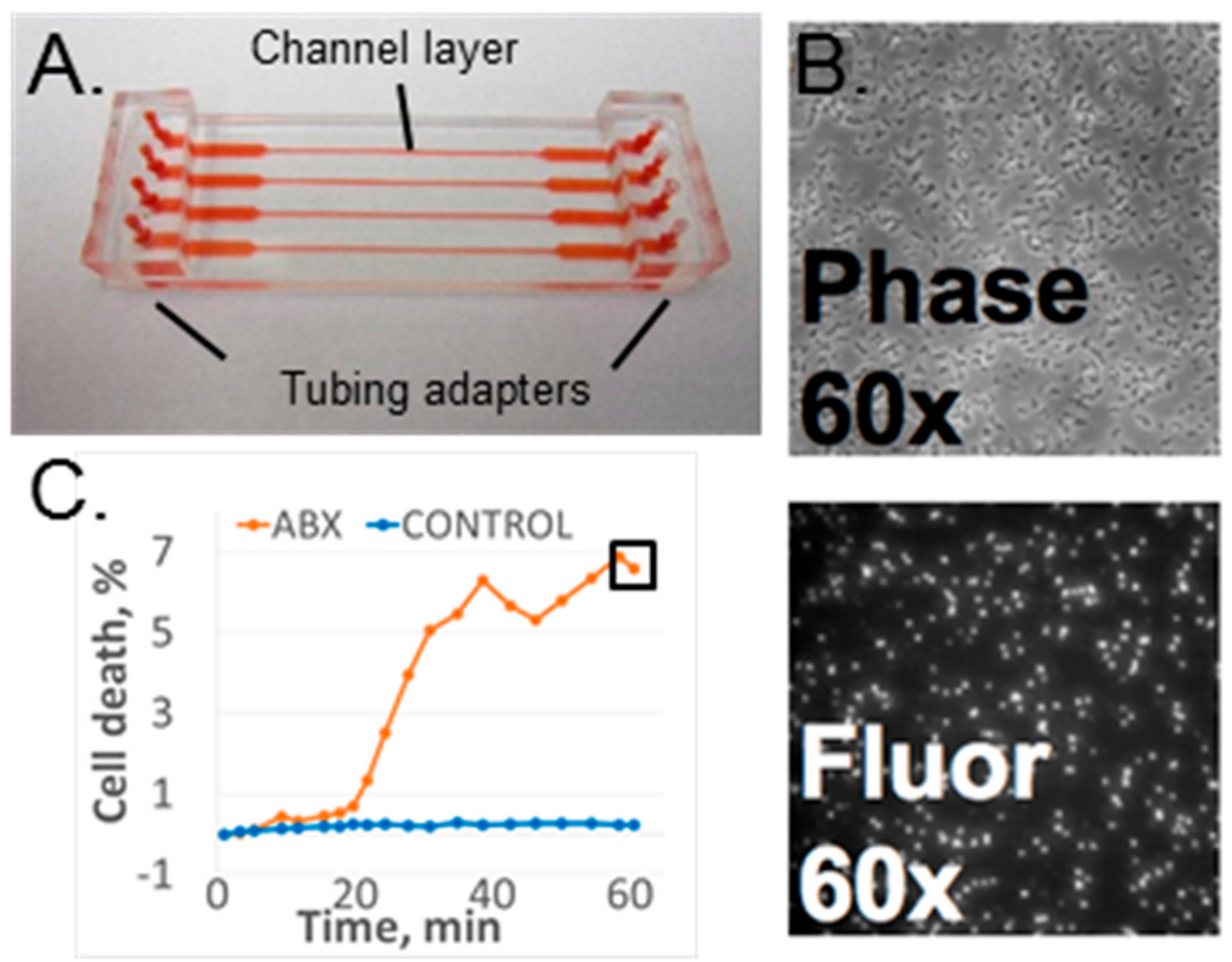
2.1. Microfluidic Flow Cell
2.2. Experimental Setup
2.2.1. Bacteria and Media
2.2.2. Imaging Hardware
2.2.3. Data Acquisition and Analysis
2.3. Optimization Experiments
2.3.1. Bacterial Strains
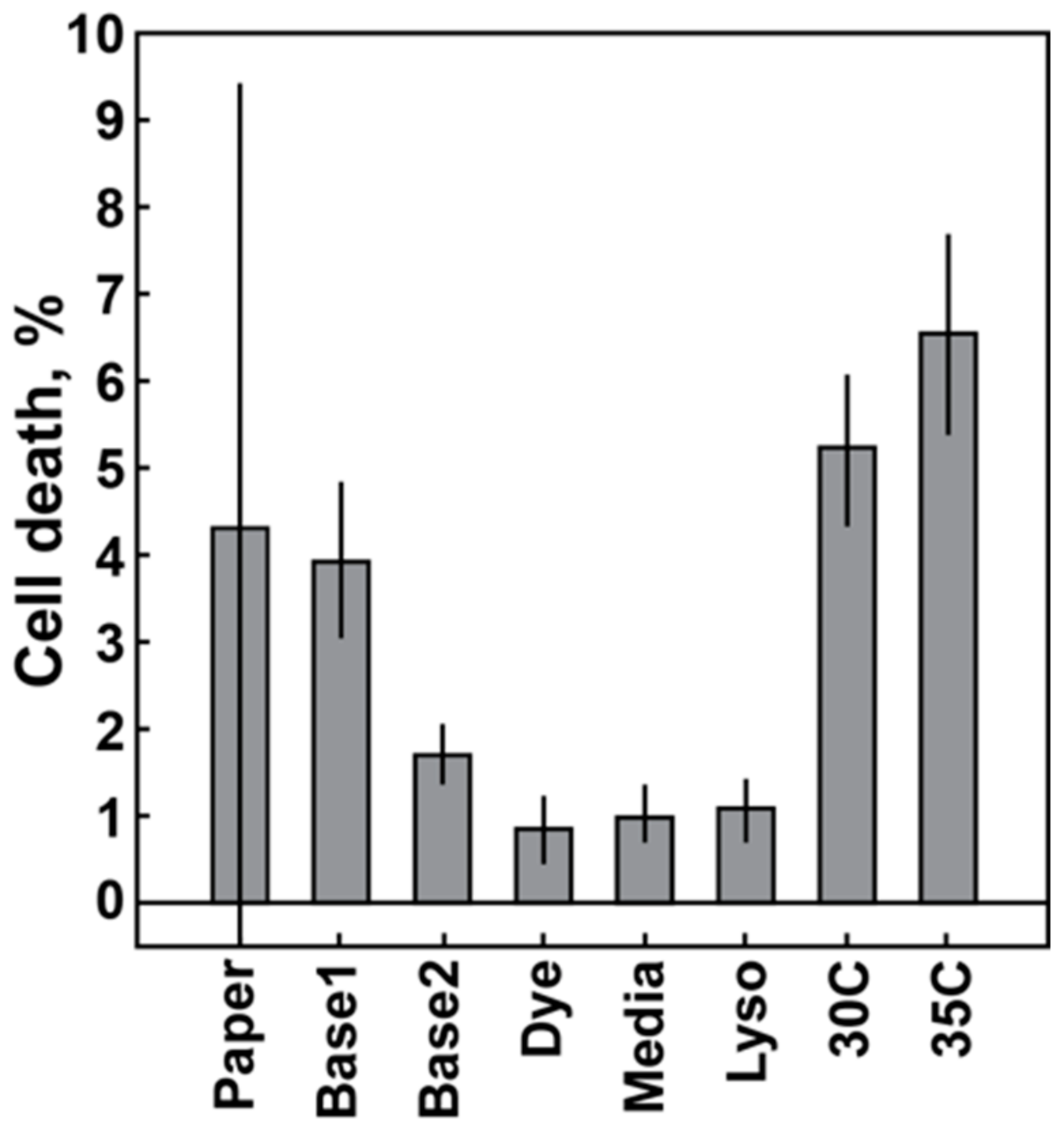
2.3.2. Baseline Variation: Temperature Control, Component Quality
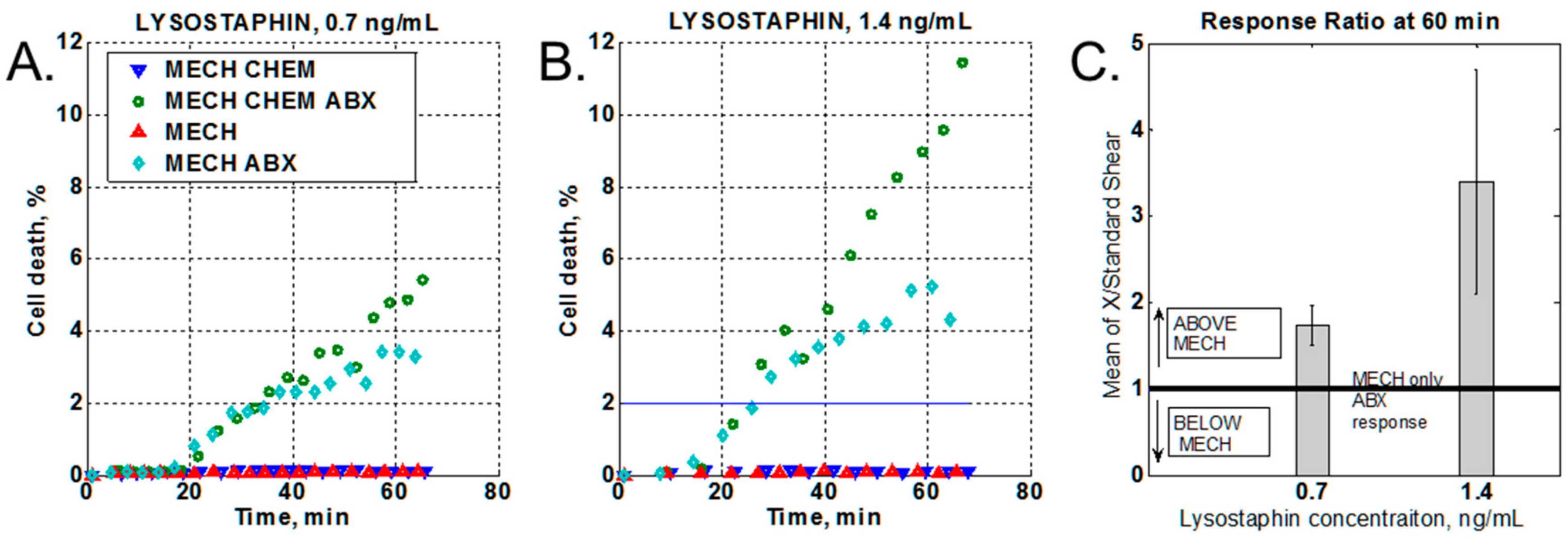
2.3.3. Effect of a Chemical Stressor
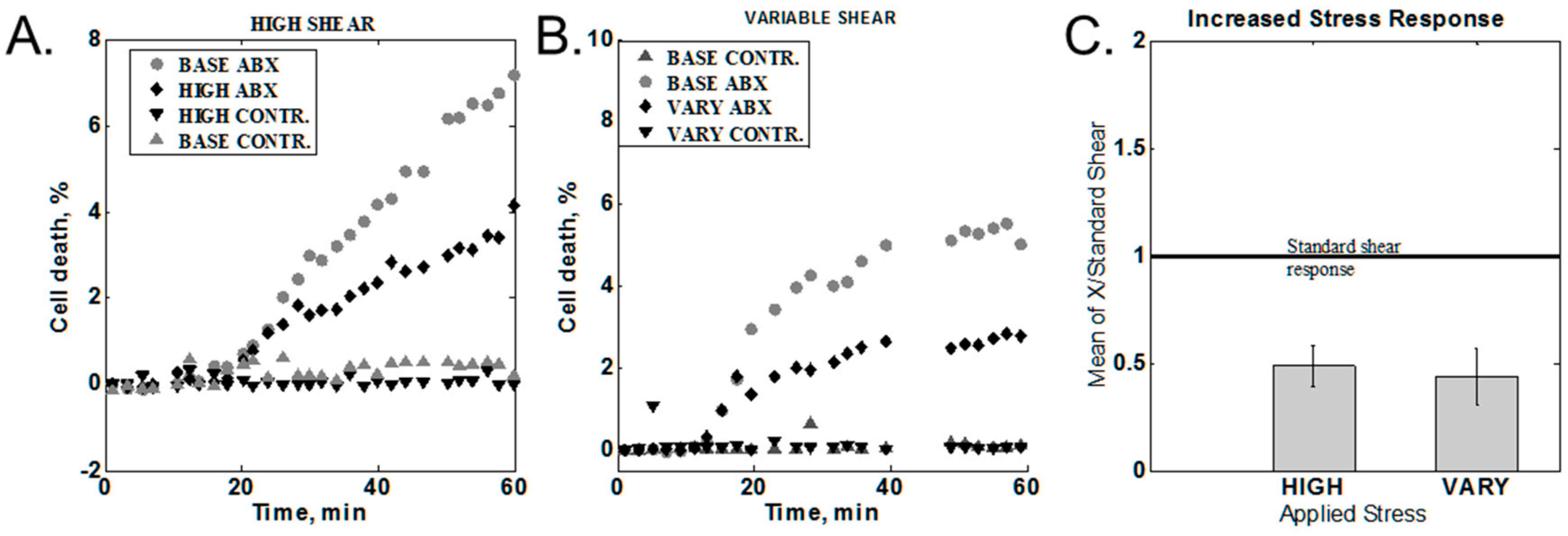
2.3.4. Increased Shear Stress
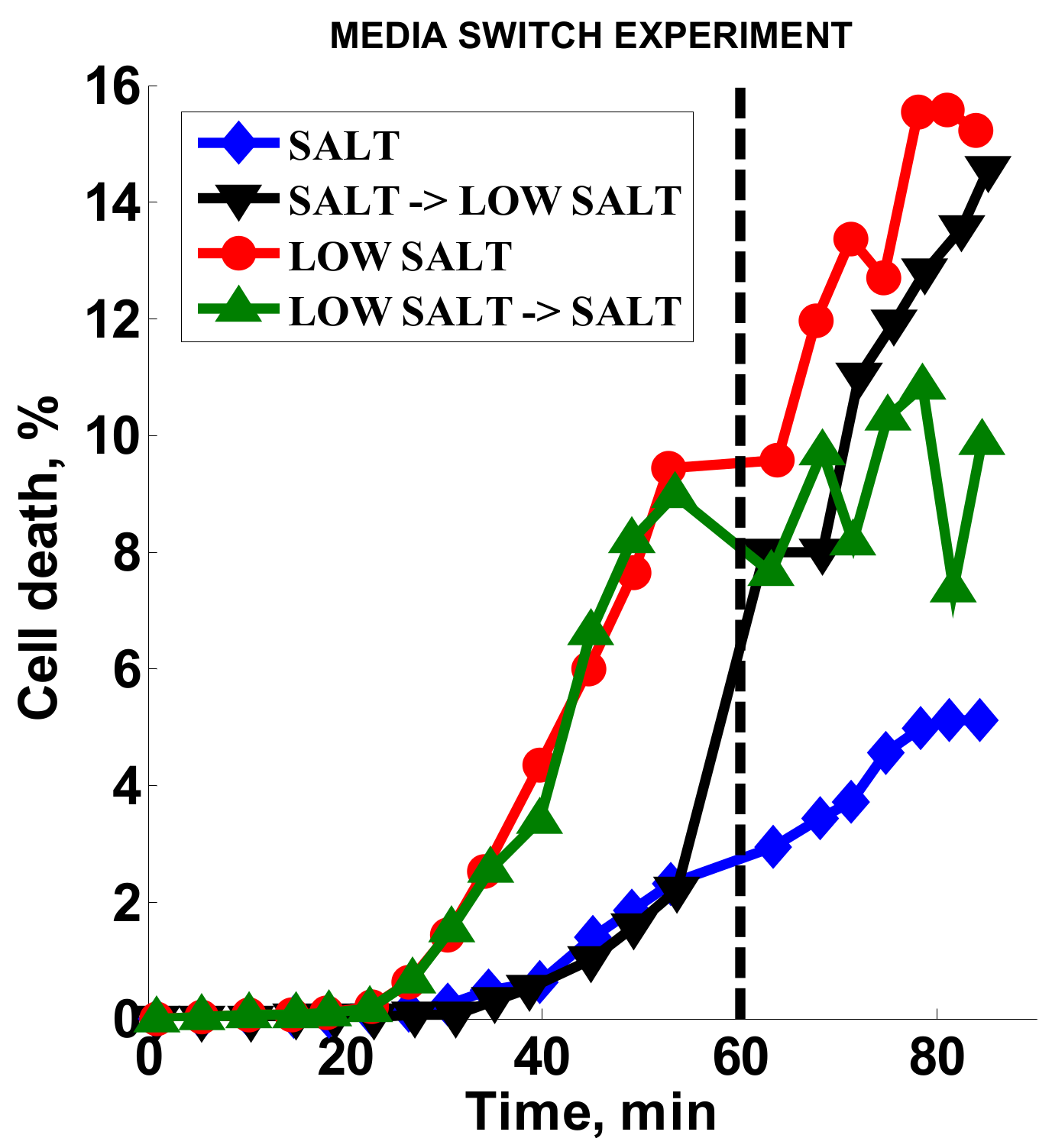
2.3.5. Effect of Media Alteration
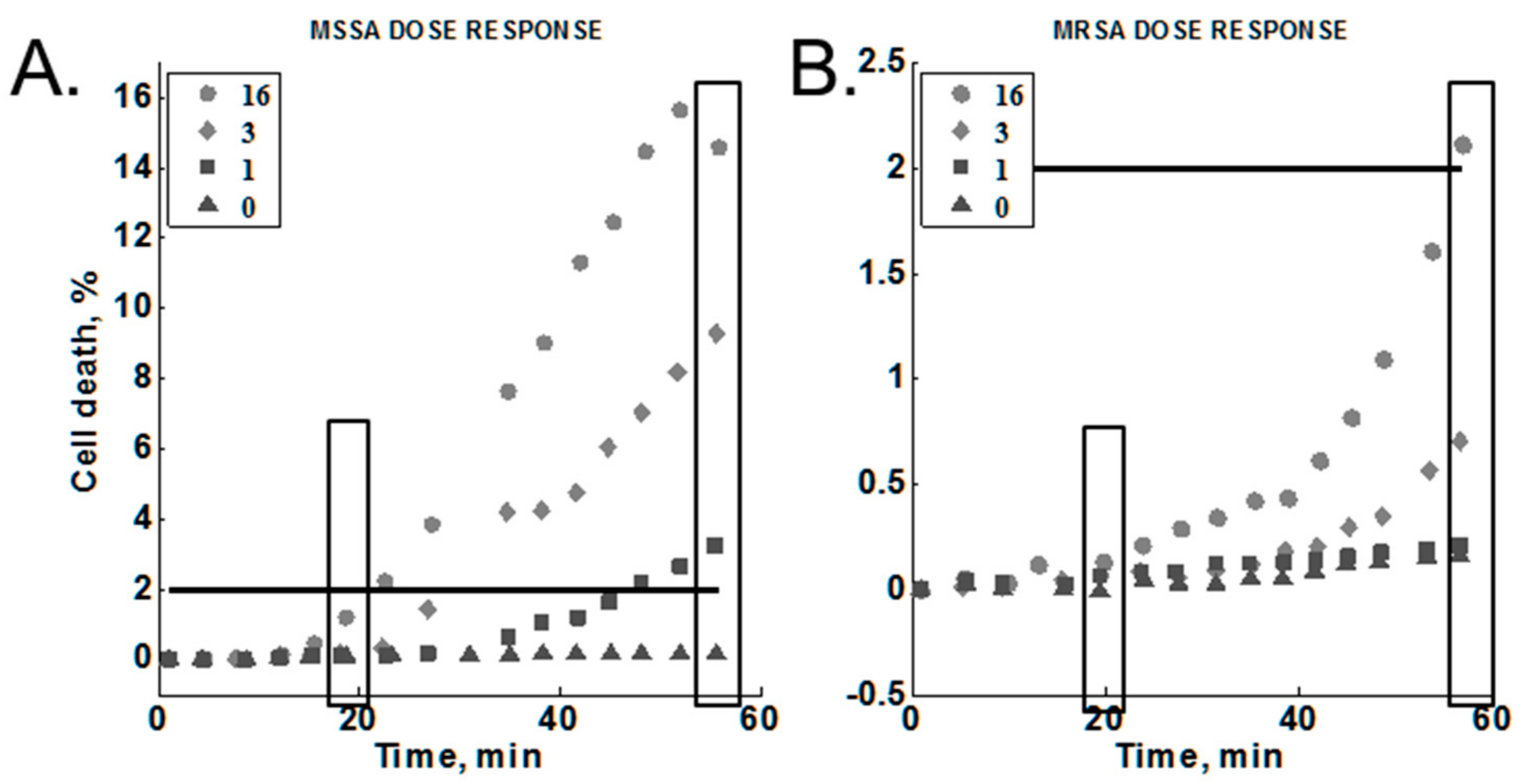
2.3.6. Cumulative Effect on Susceptibility Testing
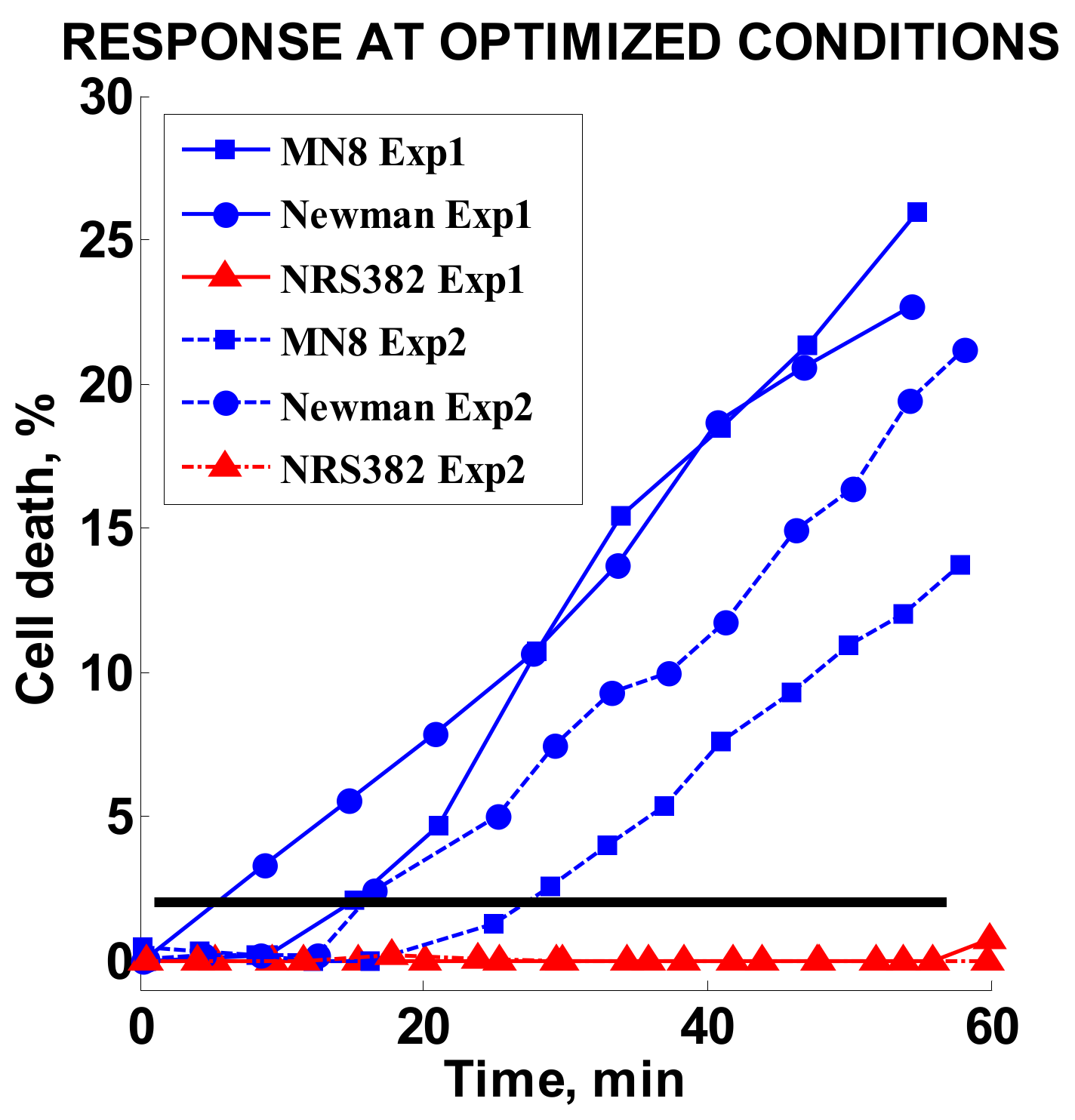
2.3.7. Optimized Conditions Test
2.4. Lysostaphin and Salt Influence on Oxacillin MIC Determination
3. Results
3.1. Baseline Variation
3.2. Effect of Chemical Stressor
3.3. Variation of the Shear Stress
3.4. Effect of Media Composition
3.5. Cumulative Effect on Susceptibility Testing of Optimized Conditions
3.6. Optimized Conditions Test
3.7. MIC Studies Results
4. Discussion and Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Marston, H.D.; Dixon, D.M.; Knisely, J.M.; Palmore, T.N.; Fauci, A.S. Antimicrobial Resistance. JAMA 2016, 316, 1193–1204. [Google Scholar] [CrossRef] [PubMed]
- National Strategy. Available online: https://www.cdc.gov/drugresistance/federal-engagement-in-ar/national-strategy/index.html (accessed on 24 January 2017).
- Wood, K.; Angus, D. Pharmacoeconomic implications of new therapies in sepsis. Pharmacoeconomics 2004, 22, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Chun, K.; Syndergaard, C.; Damas, C.; Trubey, R.; Mukindaraj, A.; Qian, S.; Jin, X.; Breslow, S.; Niemz, A. Sepsis Pathogen Identification. J. Lab. Autom. 2015, 20, 539–561. [Google Scholar] [CrossRef] [PubMed]
- Sievert, D.M.; Ricks, P.; Edwards, J.R.; Schneider, A.; Patel, J.; Srinivasan, A.; Kallen, A.; Limbago, B.; Fridkin, S. Antimicrobial-Resistant Pathogens Associated with Healthcare-Associated Infections Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect. Control Hosp. Epidemiol. 2013, 34, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Biggest Threats/Antibiotic/Antimicrobial Resistance; CDC: Atlanta, GA, USA, 2016.
- Patel, J.B. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement; Clinical & Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]
- Yagupsky, P.; Nolte, F.S. Quantitative aspects of septicemia. Clin. Microbiol. Rev. 1990, 3, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Sabui, T.; Tudehope, D.I.; Tilse, M. Clinical significance of quantitative blood cultures in newborn infants. J. Paediatr. Child Health 1999, 35, 578–581. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.H.; Huang, Y.C.; Lin, T.Y. Evaluation of the BD GeneOhm StaphSR assay for detection of Staphylococcus aureus in patients in intensive care units. J. Microbiol. Immunol. Infect. 2011, 44, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Katayama, Y.; Takeuchi, F.; Ito, T.; Ma, X.X.; Ui-Mizutani, Y.; Kobayashi, I.; Hiramatsu, K. Identification in Methicillin-Susceptible Staphylococcus hominis of an Active Primordial Mobile Genetic Element for the Staphylococcal Cassette Chromosome mec of Methicillin-Resistant Staphylococcus aureus. J. Bacteriol. 2003, 185, 2711–2722. [Google Scholar] [CrossRef] [PubMed]
- Köser, C.U.; Ellington, M.J.; Peacock, S.J. Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 2014, 30, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Horinouchi, T.; Furusawa, C. Prediction of antibiotic resistance by gene expression profiles. Nat. Commun. 2014, 5, 5792. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.; McBeth, C.; Kalashnikov, M.; Boardman, A.K.; Sharon, A.; Sauer-Budge, A.F. Microfluidic advances in phenotypic antibiotic susceptibility testing. Biomed. Microdevices 2016, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.G.; Lee, J.S.; Lee, S.; Joo, S.; Song, S.H.; Kim, E.C.; Lee, J.C. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6, 267ra174. [Google Scholar] [CrossRef] [PubMed]
- Kalashnikov, M.; Campbell, J.; Lee, J.C.; Sharon, A.; Sauer-Budge, A.F. Stress-induced Antibiotic Susceptibility Testing on a Chip. J. Vis. Exp. 2014, 8, e50828. [Google Scholar] [CrossRef] [PubMed]
- Kalashnikov, M.; Lee, J.C.; Campbell, J.; Sharon, A.; Sauer-Budge, A.F. A microfluidic platform for rapid, stress-induced antibiotic susceptibility testing of Staphylococcus aureus. Lab Chip 2012, 12, 4523–4532. [Google Scholar] [CrossRef] [PubMed]
- Kalashnikov, M.; Mueller, M.; McBeth, C.; Lee, J.C.; Campbell, J.; Sharon, A.; Sauer-Budge, A.F. Rapid phenotypic stress-based microfluidic antibiotic susceptibility testing of Gram-negative clinical isolates. Sci. Rep. 2017, 7, 8031. [Google Scholar] [CrossRef] [PubMed]
- Santiso, R.; Tamayo, M.; Gosálvez, J.; Bou, G.; del Carmen Fernández, M.; Fernández, J.L. A rapid in situ procedure for determination of bacterial susceptibility or resistance to antibiotics that inhibit peptidoglycan biosynthesis. BMC Microbiol. 2011, 11, 191. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-H.; Ning, X.; Wang, X.; Murthy, N.; Tzeng, Y.L.; Dickson, R.M. Rapid Cytometric Antibiotic Susceptibility Testing Utilizing Adaptive Multidimensional Statistical Metrics. Anal. Chem. 2015, 87, 1941–1949. [Google Scholar] [CrossRef] [PubMed]
- Nonejuie, P.; Burkart, M.; Pogliano, K.; Pogliano, J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. USA 2013, 110, 16169–16174. [Google Scholar] [CrossRef] [PubMed]
- Longo, G.; Alonso-Sarduy, L.; Rio, L.M.; Bizzini, A.; Trampuz, A.; Notz, J.; Dietler, G.; Kasas, S. Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotechnol. 2013, 8, 522–526. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.C.; Whitesides, G.M. Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc. Chem. Res. 2002, 35, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [PubMed]
- Eng, R.H.; Padberg, F.T.; Smith, S.M.; Tan, E.N.; Cherubin, C.E. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 1991, 35, 1824–1828. [Google Scholar] [CrossRef] [PubMed]
- Mascio, C.T.M.; Alder, J.D.; Silverman, J.A. Bactericidal Action of Daptomycin against Stationary-Phase and Nondividing Staphylococcus aureus Cells. Antimicrob. Agents Chemother. 2007, 51, 4255–4260. [Google Scholar] [CrossRef] [PubMed]
- Francius, G.; Domenech, O.; Mingeot-Leclercq, M.P.; Dufrene, Y.F. Direct Observation of Staphylococcus aureus Cell Wall Digestion by Lysostaphin. J. Bacteriol. 2008, 190, 7904–7909. [Google Scholar] [CrossRef] [PubMed]
- Armitage, B.A. DNA Binders and Related Subjects; Springer: Berlin, Germany, 2005; pp. 55–76. [Google Scholar]
- Biebricher, A.S.; Heller, I.; Roijmans, R.F.; Hoekstra, T.P.; Peterman, E.J.; Wuite, G.J. The impact of DNA intercalators on DNA and DNA-processing enzymes elucidated through force-dependent binding kinetics. Nat. Commun. 2015, 6, 7304. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Cattoni, D.I.; Nöllmann, M. The fluorescence properties and binding mechanism of SYTOX green, a bright, low photo-damage DNA intercalating agent. Eur. Biophys. J. 2015, 44, 337–348. [Google Scholar] [CrossRef] [PubMed]
| Experiment/Parameters | S. aureus Strains | Oxacillin, µg/mL | Lysostaphin, ng/mL | Temperature, °C | Flow Rate, mL/min | Media |
|---|---|---|---|---|---|---|
| Baseline variation | Sanger 476 | 50 | 0.7 | RT a, 30, 35 | 0.65 | MHS b |
| Chemical stressor | Sanger 476 | 50 | 0.7, 1.4 | 35 | 0.65 | MHS |
| Increased shear stress | Sanger 476 | 50 | 0.7 | RT | 0.65, 6.5 | MHS |
| Media alteration | Sanger 476 | 2 | 1.4 | 35 | 0.65 | MHS, MH c |
| Cumulative effect | Sanger 476, MW2 | 1, 3, 16 | 1.4 | 35 | 0.65 | MH |
| Optimized conditions test | MN8, Newman, NRS382 | 2 | 1.4 | 35 | 0.65 | MH |
| Experiment/Parameters | MSSA Strains | MRSA Strains | Oxacillin, µg/mL | Lysostaphin, ng/mL | Media |
|---|---|---|---|---|---|
| Lysostaphin MIC | Sanger 476 | MW2 | 0 | 0–1000 | MHS b, MH c |
| Lysostaphin effect on oxacillin MIC | Sanger 476, MN8, Newman | 0, 0.125–8 a | 0, 1.4 | MHS, MH | |
| Lysostaphin effect on oxacillin MIC | MW2, NRS382 | 0, 8–256 a | 0, 1.4 | MHS, MH |
| Oxacillin, µg/mL | 20 min | 60 min | ||||
|---|---|---|---|---|---|---|
| MRSA | MSSA | Ratio a | MRSA | MSSA | Ratio a | |
| 0 | 0 | 0 | - | 0.15 | 0.17 | 1.13 |
| 1 | 0.06 | 0.11 | 1.83 | 0.2 | 3.26 | 16.3b |
| 3 | 0.06 | 0.23 | 3.83 | 0.7 | 9.3 | 13.29b |
| 16 | 0.13 | 1.5 | 11.54b | 2.1 | 15.6 | 7.43 |
| Strain/Media Composition | MH | MH + Lyso a | MHS | MHS + Lyso a |
|---|---|---|---|---|
| Sanger 476 | 0.5 | 0.5 | 0.5 | 0.5–1 |
| MN8 | 0.25–0.5 | 0.5 | 0.25–0.5 | 0.5 |
| Newman | 0.25 | 0.25 | 0.5 | 0.25–0.5 |
| MW2 | 64 | 64–128 | 128 | 64–128 |
| NRS382 | 64 | 64 | 32 | 64 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalashnikov, M.; Lee, J.C.; Sauer-Budge, A.F. Optimization of Stress-Based Microfluidic Testing for Methicillin Resistance in Staphylococcus aureus Strains. Diagnostics 2018, 8, 24. https://doi.org/10.3390/diagnostics8020024
Kalashnikov M, Lee JC, Sauer-Budge AF. Optimization of Stress-Based Microfluidic Testing for Methicillin Resistance in Staphylococcus aureus Strains. Diagnostics. 2018; 8(2):24. https://doi.org/10.3390/diagnostics8020024
Chicago/Turabian StyleKalashnikov, Maxim, Jean C. Lee, and Alexis F. Sauer-Budge. 2018. "Optimization of Stress-Based Microfluidic Testing for Methicillin Resistance in Staphylococcus aureus Strains" Diagnostics 8, no. 2: 24. https://doi.org/10.3390/diagnostics8020024





