1. Introduction
Hyperspectral remote sensors can collect data simultaneously in hundreds of narrow, adjacent spectral bands covering the visible to near-infrared wavelengths. The resulting hyperspectral images contain a wealth of high-resolution spectral information permitting a wide range of remote sensing applications, such as environment monitoring, geological surveying, and surveillance. However, visualizing the information content of this high-dimensional data space is difficult and often involves trade-offs between the complexity of information being conveyed, similarity of the output to the human visual system, and speed of visualization display.
Hyperspectral image visualization has been part of remote sensing for decades in order to provide a quick overview and understanding of a hyperspectral data set. Visualization is implemented by mapping the original high-dimensional data into a three-dimensional color space and showing the mapped data on a tristimulus-based display device (e.g., a standard computer screen). There is inevitable loss of information in the process of visualization. However, in order to effectively preserve the information contained in hyperspectral imagery and to intuitively provide a discriminative color representation, some objective visualization criteria have been proposed, including pairwise distance preservation [
1–
3] and feature separability (or contrast) [
4,
5]. The former tries to maintain pairwise distances in hyperspectral imagery and in the final color image while the latter, introduced by Cui
et al. [
4], aims to display the differences between pixels as distinctively as possible. It is difficult to simultaneously satisfy these two criteria since the smaller differences between pixels cannot be well distinguished when all pairwise distances are exactly preserved in the final color image. From the subjective perspective, the visualization aim is to generate a trichromatic composite image with perceptual appeal [
6–
8] suitable for interpretation by the human visual system, referred to as naturalness. However, it is challenging to display the full information content in mapped data on devices designed for natural images.
In the literature, many visualization methods have been proposed for hyperspectral images and can be roughly classified into four categories: band selection, linear spectral combination, data projection, and convex optimization.
Band selection refers to techniques that select three representative bands as color components. The simplest scheme is to pick three bands with wavelengths closest to the natural wavelengths of red, green and blue. More sophisticated rule based approaches, such as one-bit transform (1BT) [
9], normalized information (NI) [
6], minimum redundancy maximum relevance (mRMR) [
10], minimum estimated abundance covariance (MEAC) and linear prediction [
11], have also been introduced for the selection of triplet bands. However, this kind of techniques usually loses a large amount of information contained in the data space not represented in the three bands.
Another kind of visualization method is linear spectral combination. The idea is to create the final image from a weighted sum of pixels across the available sampled wavelength spectrum. The design of the weights involved is typically related to the visualization effect. In [
3], a set of fixed spectral weighting envelopes are derived from color matching functions, designed to mimic how human photopic (daylight) vision works. In [
12], a bilateral filtering scheme is presented for retaining the minor details, while, more recently, a multiobjective optimization-based approach has been proposed to provide the solution of weights by taking some desired properties of the resultant image into account [
7]. Relatively speaking, the fused images are more informative due to the process of combining information from all bands.
The third family of visualization methods is composed of data transformations that aim to project the original hyperspectral image into a low-dimensional space. The classical example is principal component analysis (PCA), which has been extensively used for visualization purposes. In general, PCA and its variants are explored as an
N-to-3 projection into the color space with the first three principal components [
13–
15]. Further PCA methods have been investigated based on perceptual attributes [
16] and class separability [
17]. An alternative way,
i.e., segmented PCA, is to perform PCA on three subgroups to extract principal components used for visualization [
18,
19]. Additionally, regarding the nonlinear characteristics of hyperspectral data [
20], a straightforward but time-consuming extension is to employ nonlinear projection methods (e.g., local linear embedding) for dimensionality reduction into three bands with the goal of preserving the local distances between a pixel and its neighbors.
A new class of visualization methods, designed to preserve some desired properties, was recently proposed from the convex optimization perspective by formulating the related criteria into the underlying objective function. In [
4], Cui
et al. exploited pairwise distance preservation and feature separability in a two-step dimensionality-reduction framework. In their approach, the pairwise distance preservation criterion is explicitly set as a three-dimensional mapping problem decomposed into two steps: a two-dimensional projection as a partial solution using classical PCA and a linear programming method to solve for the one-dimensional coordinate in the third dimension with the separability of features criterion as a set of constraints. From a different perspective, Mignotte proposed a bicriteria optimization for color display model (BCOCDM) method [
5] by combing these two contradictory criteria with an internal parameter, thus making this algorithm express the contribution of two criteria for a specific application, besides only focusing on the criterion of pairwise distance preservation in [
21]. This kind of visualization method provides interesting and promising results to some extent.
These existing algorithms either favor distances preservation in high-dimensional space at the risk of low contrast or prefer high contrast for achieving the good features separability in the low-dimensional space but allow excessive distortion of the pairwise-distances or sometimes unwanted loss of the local details. More importantly, the intrinsic properties of the pairwise measured space for high-dimensional hyperspectral data, such as large-scale dynamic range of pairwise distances and concentration phenomenon, make it difficult for them to yield a natural visual representation.
In this paper, we propose a new hyperspectral image visualization strategy with double mappings for improved balancing of the trade-off between pairwise distance preservation and feature separability. Our contributions are three-fold:
In order to decrease the dynamic range of pairwise distances and uniformly distribute energy in the three color components as that in natural color images, the principle of equal variance is adapted to divide the hyperspectral image bands into three subgroups prior to further mapping.
Two different mapping methods are proposed for normal pixels and outlier pixels respectively. This can achieve good contrast with minimal distortion of the pairwise distances while preserving local fine information.
In the mapping case of outlier pixels, an objective function, based on the weighting of pairwise distances, is designed to adaptively inhibit/enhance the large/small pairwise-distances for the purpose of preserving local topology in the hyperspectral data space.
The reminder of this paper is organized as follows. Section 2 analyzes the characteristics of pairwise distances in hyperspectral imagery and discusses the display problems of color space mapped data. Section 3 describes the pairwise-distances-analysis-driven visualization strategy (PDADVS) with double mappings. Experimental results and comparisons with existing methods are presented in Section 4, followed by concluding remarks in Section 6.
2. Problem Statement
Despite the progress of visualization methods, intrinsic measure phenomena of the high-dimensional hyperspectral data space are often overlooked. As such, this section presents important characteristics of pairwise distances in hyperspectral images and highlights how these properties may affect the visualization performance.
2.1. Analysis of Pairwise Distances in Hyperspectral Images
Visualization of hyperspectral imagery attempts to reduce the data dimensionality while retaining the information content of the full data space. Toward this goal, pairwise distance preservation is a good foundation for hyperspectral visualization because keeping pairwise distances in the visualized data space indicates a good mapping with high faithfulness of a resultant color image to raw hyperspectral data.
Before proceeding to the analysis of pairwise distances, we first define the notations used in what follows. Let I be a hyperspectral image of w × h pixels with r bands in a tensor space Tr ∈ ℝw×h×r, and
be spectral vector (also is corresponding to a hyperspectral image pixel) with the subscript denoting the spatial location and the superscript denoting the range of bands. A subgroup of hyperspectral image is referred to as Ik1~k2, whose spectral vector is
. The pairwise distance
between two spectral vectors
and
is defined as
, which can be straightforwardly extended to the full-band case with k1 = 1 and k2 = r.
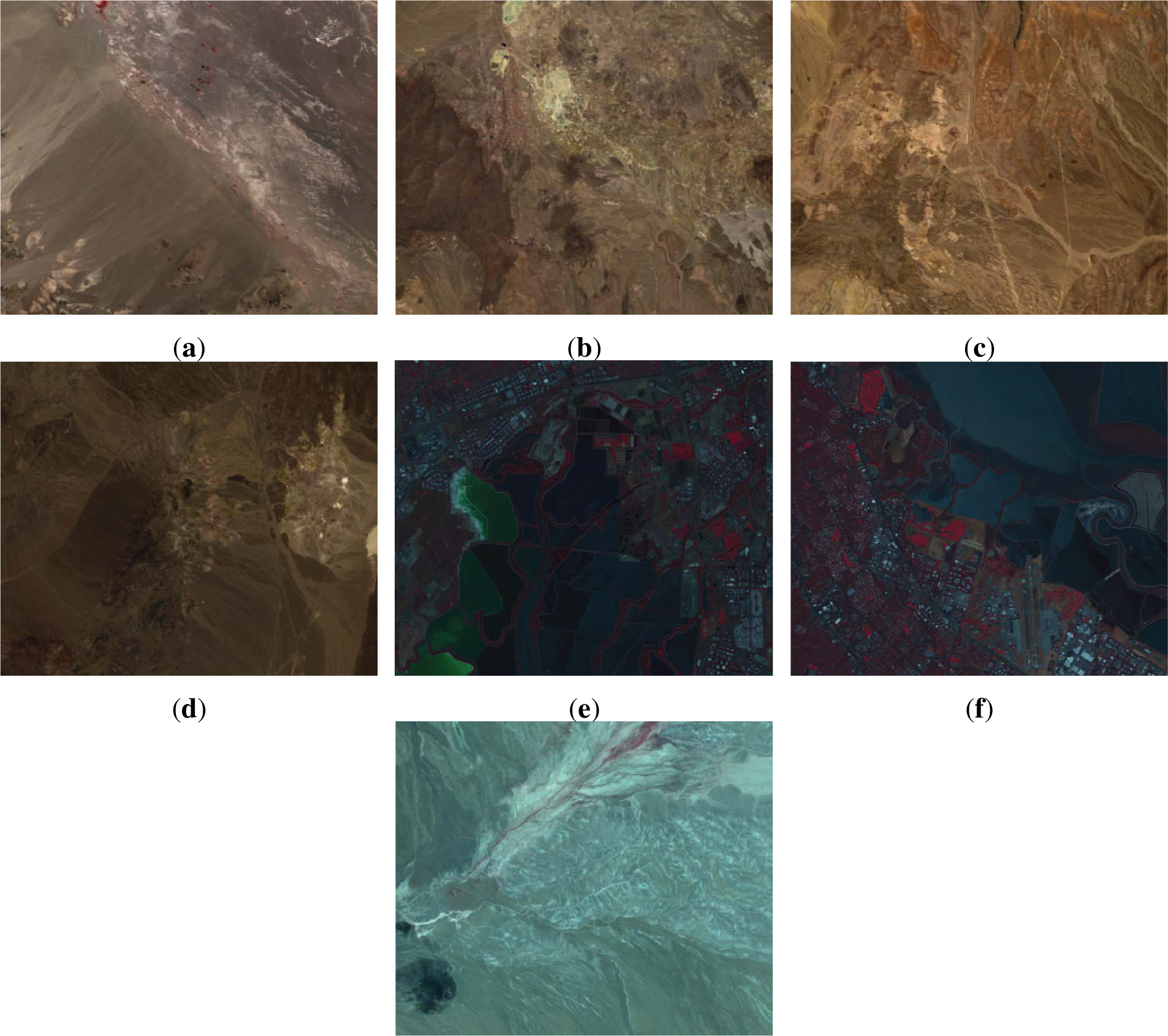
The images used later for experimental validation are taken as examples to illustrate the characteristics of pairwise distances in hyperspectral images, whose false-color composites are presented in
Figure 1. In all cases, the black and noise bands are first removed as outlined in Section 3.2.
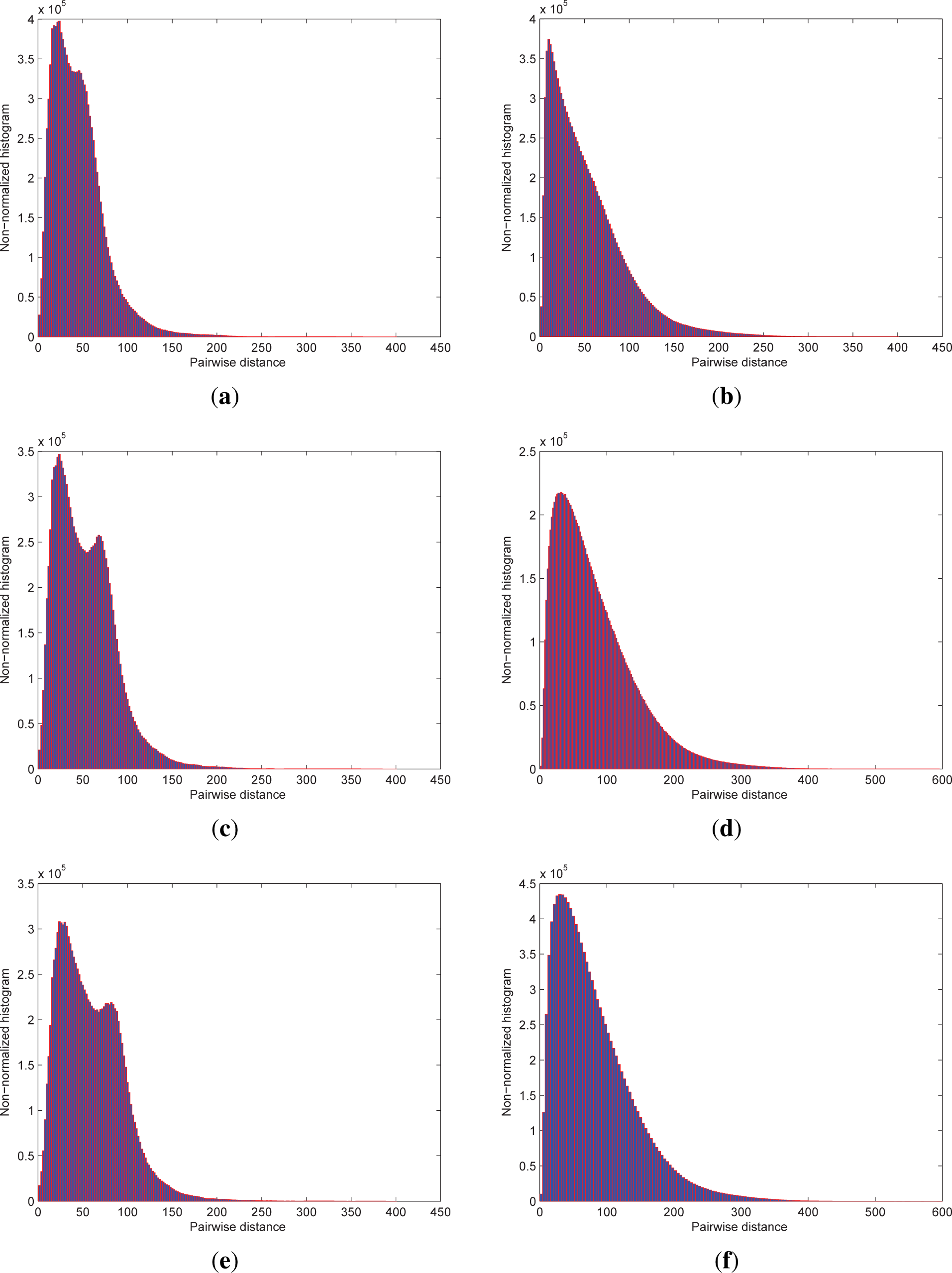
Table 1 lists the measures of pairwise-distance variation for different subgroups of seven hyperspectral images, while
Figure 2 shows the non-normalized histograms of pairwise distances of
Moffet02_
igm and
Cuprite02 with different band coverage for the intuitive analysis.
As shown in
Table 1 and
Figure 2, pairwise distances are distributed in a very wide range and exhibit heavy-tailed characteristics. In addition, the distributions show both increased variance and dynamic range as the dimensionality of the subgroups increase.
Figure 2 reveals that the majority of pairwise distances populate the smaller distance values, which is consistent with the phenomenon of measure concentration in high-dimensional space [
22–
24]. Furthermore, the average and maximum quantities of pairwise distances together with their ratios are presented in
Table 2 for all available bands of seven hyperspectral images. In general, the mean is significantly less than the corresponding maximum value for each image. As a result, the dissimilarities represented by smaller pairwise distances become less meaningful because of their poor discernibility. On the contrary, a few larger pairwise distances indicate that some pixels are considerably dissimilar from the normal ones and are considered outliers as in [
4,
25,
26], whose ranking and detection will be discussed in Section 3.3.
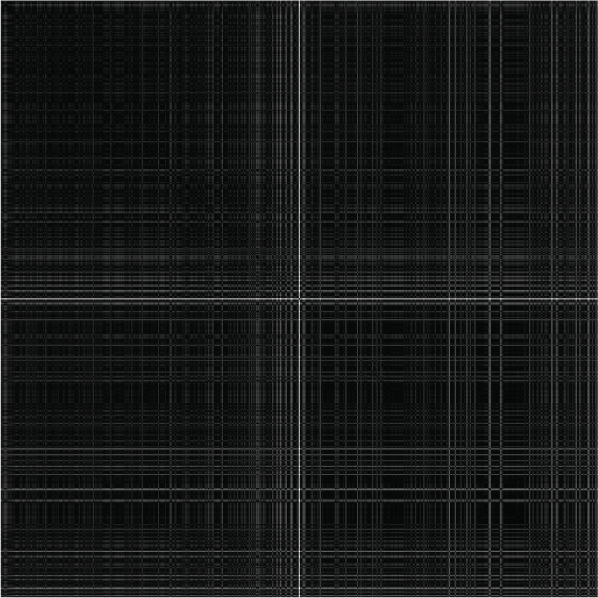
In order to further demonstrate the inconsistency of outlier pixels, the normalized pairwise-distance matrix image of an outlier located in (183, 211) and 512 randomly selected normal pixels extracted from
Cuprite02 image is shown in
Figure 3. To construct this matrix, the outlier pixel is positioned in the median x-/y-coordinates. In this illustration, with white points denoting the high degree of dissimilarity, it is clear that large distances between this outlier and the normal pixels are much larger than that calculated among the normal pixels. Large pairwise distances is another consequence of the existence of outlier pixels, besides high dimensionality of hyperspectral images. Therefore, careful handling is necessary for the optimal visualization of hyperspectral data.
2.2. Display Problems of The Mapped Data in Color Space
To visualize hyperspectral images, the mapped data will be displayed on a standard tristimulus device, whose gamut is restricted to a triangle in chromaticity space and three primary colors generally have fixed quantization levels. Under the criterion of pairwise distance preservation, the characteristics of pairwise distances in the color space are similar to that of pairwise distances in the original high-dimensional space. Then the dynamic range, as that in the original high-dimensional space, will become large accordingly with the increase of dimensionality. Naturally, each color value will represent a larger pairwise-distance range. In addition, due to the concentration of measure phenomenon, the quantizing levels in each component of three primaries, associated with the majority of normal pixels, only account for a small central part {a + 1, a + 2,…, b − 1} of the whole gray level set G = {0, 1, 2,…, L}, while the top and bottom parts of this set, i.e., {0, 1, 2,…, a} and {b, b + 1,…, L}, are used to indicate a small number of the outlier pixels. As a result, the differences between normal pixels, represented by small pairwise distances, are difficult to distinguish in the color image and has poor contrast. Meanwhile, like the outliers originally measured in the high-dimensional space, their counterparts in the color space are few and far way from other color pixels. This makes it difficult to illustrate the pixel diversity in the mapped color space.
These difficulties have been explored by previous research and many techniques have been proposed to achieve a preferable visual effect. In this simplest case, these pixels, whose duplicates in the color image are so large that they suppress the brightness of other pixels, can be removed before dimensionality reduction [
27]. In [
3], Jacobson
et al. proposed to render no more than 2% of pixels by clipping their out-of-gamut color values to the gamut in sRGB after fixed integration envelopes are applied. PCA2% used in ENVI linearly stretches each of three display bands so that 2% of pixels at both ends of each color channel are at the minimum and maximum display value. However, in these cases, some pixels (
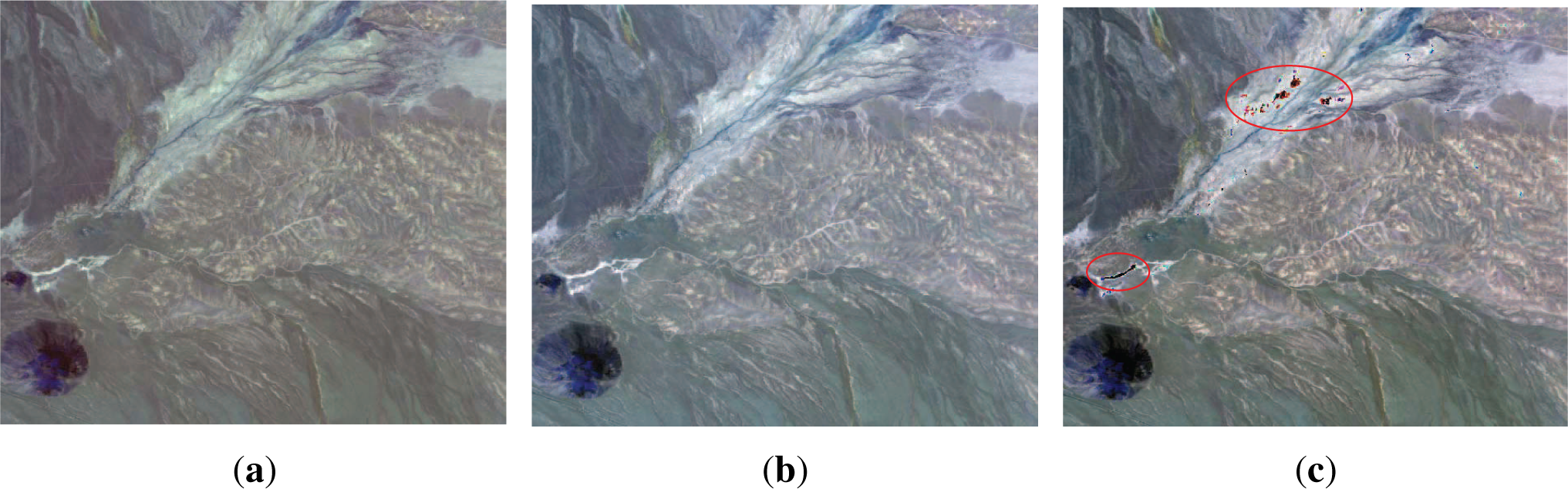
i.e., outliers) regarded as a nuisance which impede the contrast enhancement tend to be eliminated. Although this creates a vivid contrast in some regions, the dissimilarities between outliers, which contain useful information, are lost. Furthermore, the local contrast where these outliers exist cannot be reflected in the visualized color image even though they have topological structure in a local region as shown in
Figure 4. Good visualization cannot be achieved unless the visualization method is designed specifically to take these outliers, as well as the more general properties of hyperspectral images, into account. This is the motivation of our proposed method described in next section.
3. Proposed Visualization Technique
In this section, a new visualization technique for hyperspectral imagery is proposed,
i.e., the pairwise-distances-analysis-driven visualization strategy (PDADVS). In this approach, normal pixels and outliers are treated separately using two different mapping methods for enhancing global contrast while avoiding the high risk of sacrificing local details and excessively distorting pairwise distances. For clarity, the flowchart of PDADVS is shown in
Figure 5 and detailed descriptions of each step are provided in Sections 3.2 through 3.5.
3.1. Partitioning of Hyperspectral Bands
3.2. Preprocessing
A hyperspectral image generally contains some bad bands (e.g., water absorption and low SNR bands), which impact the following analysis. Therefore, preprocessing is a necessary part of the visualization process. Because of the high-spectral correlation, two adjacent bands in hyperspectral imagery tend to be highly correlated, which conversely provides us a probability to perform the removal of bad bands. Specifically, those bands, whose correlation coefficients to their neighboring bands are less than a given threshold
η = 0.8, are removed. Additionally, hyperspectral imagery is affected by additive noise that impairs information extraction and scene interpretation. To reduce this noise, the wavelet hard thresholding approach [
28] is applied to each band. Finally, bands with SNR values less than 15dB are excluded.
According to the analysis of pairwise distances in Section 2, we partition all available hyperspectral bands into three subgroups for decreasing the dynamic range of pairwise distances. As such, in terms of the consistency in distance preservation, the relatively smaller dynamic range of pairwise distances in the hyperspectral image space will lead to the smaller gamut of pixels in the color space correspondingly. This is conducive to the feature separability in the sense that the differences between pixels can be better reflected by three color components corresponding to different subgroups respectively rather than these corresponding to the whole band.
To this end, the principle of equal variance is proposed to partition all hyperspectral bands into three subgroups prior to mapping. Different from the equal subgroups technique used in [
5,
21], equal variance can guarantee that three components of the final color image are correlated (
i.e., if the intensity changes, all three components will change accordingly) and the energy is distributed uniformly as in natural color images [
29], such that the visualization result is optimal for human visual system. To provide perceptual visualization, three subgroups are then mapped to the device-independent and approximately perceptually uniform CIE (Coherent Infrared Energy)
L∗a∗b∗ color space.
To achieve the partitioning, we minimize the following objective function
where
σ denotes the variance, and
is a
row vector for an image of size
w ×
h, corresponding to the Euclidean distances
of pairs of spectral vectors in
Ik1∼k2. In practice, we consider a subsampling of pairs of pixels in the image (
i.e., all pairs with horizontal or vertical displacements of 2
p pixels for
p ≥ 3) to compute the vector of pairwise distances for reducing computational complexity. As is well known, there is high relevance between the adjacent-band images in hyperspectral imagery. So each subgroup should consist of contiguous bands in order to keep as much of the common information as possible contained within the subgroup during the mapping. On the other hand, it can be seen from
Tables 1 and
2 that both mean and variance of pairwise distances increase with the number of bands in the subgroups. Therefore, a line searching technique is introduced (summarized in
Algorithm 1) to solve
Equation (1) for the partitioning of hyperspectral bands.
Table 3 lists the number of the available bands after preprocessing for all considered experimental hyperspectral images and their partitioning results.
Algorithm 1.
Partitioning of hyperspectral bands
Algorithm 1.
Partitioning of hyperspectral bands
| Require: I - a hyperspectral image of w × h pixels with r bands; |
| Ensure: s1, s2 - two splitting points of the whole band |
| 1: | Initialize |
| 2: | k = 0; s1 = ⌊r/3⌋; s2 = 2 × ⌊r/3⌋ + 1; |
| 3: | |
| 4: | |
| 5: | Repeat |
| 6: | σAV = (U1 + U2 + U3)/3; |
| 7: |
|
| 8: |
|
| 9: |
|
| 10: |
|
| 11: |
|
| 12: |
|
| 13: |
|
| 14: |
; |
| 15: | k = k + 1; |
| 16: | until △≤10−3 |
3.3. Outlier Ranking and Detection
As described earlier in Section 2, it is difficult to deal with all pixels of hyperspectral images in the same way for simultaneous discrimination between small and large distances. To address this issue, two different mapping strategies are proposed separately for normal pixels and outlier pixels. This separate approach can achieve the better feature separability with minimum distortion of pairwise distances while avoiding the loss of spatial details.
Prior to performing double mappings, we require to distinguish outliers from normal pixels with the aid of multidimensional scaling (MDS) [
30,
31]. MDS is a well-known technique for visualizing the high-dimensional data and exploring its underlying structure by analyzing the dissimilarities (or similarities) between pairs of data points, which utilizes an embedding to find coordinates in a low-dimensional space for each high-dimensional data point that preserve the given pairwise dissimilarities as faithfully as possible. Specifically, for each subgroup, the outliers are detected by the following three steps:
Step (1) Given distance matrix
with its entry
representing the pairwise distance between pixels
and
in a subgroup of hyperspectral imagery, we implement MDS to obtain the coordinates for each pixel of
Ik1∼k2 by minimizing the objective function
where
. To calculate the inner product matrix
with
and
e being a column vector of all 1’s [
31], now
Equation (2) can be reduced to
Then the solution for minimizing
Equation (3) can be found by solving an eigenvalue problem, given by
Xk1∼k2 = Λ
1/2VT, where Λ is the top
q eigenvalues of (
Ik1∼k2)
T Ik1∼k2 and
V is the eigenvectors of (
Ik1∼k2)
T Ik1∼k2 corresponding to the top
q eigenvalues.
Step (2) Since the coordinates are computed on the premise of distance preservation, they can be used to measure the degree of deviation of pixels in original high-dimensional space. With the results in Step (1), we define the rank for each pixel of
Ik1∼k2,
i.e.,
Step (3) Finally, we sort Ik1∼k2 according to their ranks, and then the pixels whose ranks are in the top ξ percent and in the bottom ξ percent should be identified as outliers.
3.4. The Mapping of Normal Pixels
For the hyperspectral image visualization, it is of importance to provide a quick overview for the existing materials and their distribution in the final color image. Moreover, as mentioned in Section 2, the existence of outliers has a negative effect on the feature separability due to their large pairwise distances with normal pixels. As for normal pixels dominating hyperspectral data, it is desirable to keep pairwise distance among them in dimensionality reduction to yield a faithful representation for hyperspectral images.
In terms of the requirements on low computational complexity and the preservation of pairwise distance, we prefer to utilize MDS for the mapping of normal pixels. Here the results is directly extracted from that obtained by MDS in Section 3.3, instead of recalculating the embedded points by defining a new objective function of MDS only involving normal pixels. In such a way, besides simplicity, it is of importance to maintain the consistency of the whole pairwise distances, since normal pixels and outliers will be simultaneously involved in the mapping of outliers in next subsection.
3.5. The Mapping of Outliers
In this subsection, a novel mapping method is proposed for the outliers. This method takes pairwise distance preservation and feature separability into account, and, unlike MDS, adaptively deals with the small and large pairwise distances.
Suppose we have the set
ψ of outliers in a subgroup, the objective function for each outlier
is designed as follows
Where
are the pairwise distances between current outlier
and its neighbors (other outliers or normal pixels), and
are the distances between the embedded point of
in color component P and the neighboring points.
, Ns denotes a square fixed-size neighborhood centered around an outlier, and λ is a positive internal parameter balancing the contribution of pairwise distance preservation and feature separability.
The first term
is used to achieve distance preservation by minimizing the difference of pairwise distances between
in hyperspectral image and
in the color image. The involving weight
is the inverse of the square root of pairwise distance and is designed to enhance the importance of small distances among the neighborhood of outlier
in the objective function. Correspondingly, small distances are emphasized and local dissimilarity can be preserved in the neighborhood of
in the color component. Meanwhile, due to large pairwise distances between outliers and normal pixels, a second term is introduced in order to prevent the embeddings of outliers in three color components from departing too far from those that correspond to their respective neighbors. There are two points worth being mentioned about this dimensionality reduction:
The constraint term in BCOCDM tries to keep the dissimilarity between color pixels. In comparison, the second term of the proposed objective function aims to reduce the deviation between an outlier and its neighbors (especially normal pixels in its neighborhood). This results in a reduction of the color component range. As such, as mentioned in Section 2, more color levels will be used for describing the embedded points associated with normal pixels, which account for the overwhelming majority.
In order to get good contrast not only in different local regions but also in the full image, the presented method does not simply eliminate or clip the outliers, like the previous algorithms (e.g., PCA2%, and Cui’s [
4]). Instead, it adjusts their values in color components based on visualization criteria of pairwise distance preservation and feature separability. As a result, local pairwise distances between outliers and their respective neighbors are preserved, and local information around outliers is also kept.
Algorithm 2.
SD algorithm for solving Equation (5)
| Require: - initial mapping value of outlier; ϵ - tolerance; |
| |Ns| - the size of neighborhood window; Itermax - the maximum number of iterations; |
| Ensure: - the output mapping value of outlier; |
| 1: | Initialize k = 0 |
| 2: | repeat |
| 3: | Compute the gradient direction:
|
| 4: | Compute the search direction:
; |
| 5: | Compute the step length:
; |
| 6: | Compute the (k + 1)th iterative value of
; |
| 7: | k = k + 1. |
| 8: | Until |
To find the solution of
Equation (5), the method of steepest descent (SD) is suggested, for which a quantity, calculated from multiplying the mean of pairwise distances between the outlier and its neighbors by a small scaling factor, is introduced as the step length for improving the convergence speed. This step length will be further adjusted step by step as the solving procedure reaches the best point, so that it fluctuates smoothly and avoids oscillations often arisen in the gradient descent algorithm. The details of SD algorithm for solving
Equation (5) are given in
Algorithm 2.
5. Discussion
In terms of visualization effectiveness, our proposed PDADVS is superior to other state-of-the-art algorithms for all the hyperspectral images, whose good performance mainly comes from the separate treatment of outliers. By doing so, the gap between color pixels in the mapped color space corresponding to outliers and normal pixels can be decreased such that the pixel diversity is achieved, on the premise of preserving pairwise distances. As such, it is very suitable for handling the hyperspectral images with more complicated geographical features. On the contrary, a possible limitation of the proposed method is that it is not too effective in the hyperspectral images whose scenes are relative simple. For example, from
Table 6 we can see that the proposed PDADVS with the same parameter settings yields the higher
δ values for
Moffett02_
igm,
Moffett03_
igm and
Cuprite02 than
Lun.lake01,
Cuprite03,
Cuprite04, and
Lun.lake_igm.
As further remarks, it is worth noting that, although the above experimental analysis is performed on the portions of the original hyperspectral images, this technique can be used to the large images. To illustrate the applicability, we test the performance of PDADVS on the original images of
Moffett02_
igm and
Moffett03_
igm, respectively, having 2032×614 and 1924×753 pixels. It allows achieving
ρ = 0.981,
δ = 61.3 for
Moffett02_
igm and
ρ = 0.980,
δ = 50.3 for
Moffett03_
igm. The running times are 56.4 and 56.2 seconds for
Moffett02_
igm and
Moffett03_
igm, respectively.
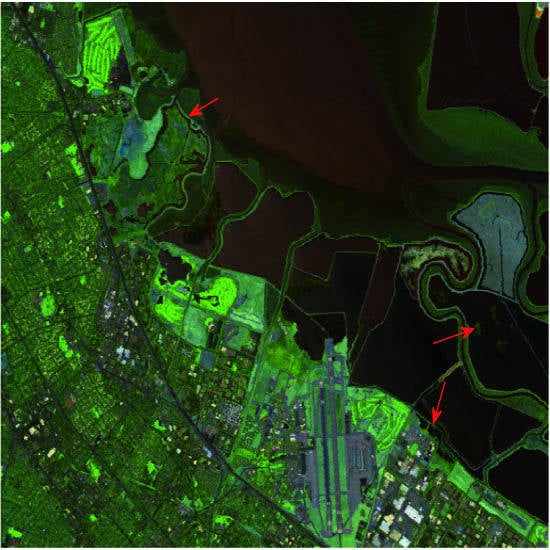
Figure 11 shows their corresponding visualization results, which provide evidence for good performance of PDADVS. From
Figure 11, it is interesting to observe that the different regions as well as man-made structures and objects are still clearly identifiable in these two images.
In addition, we discuss the computational complexity of the PDADVS algorithm. Specifically, the embedded points corresponding to normal pixels are obtained through calculating eigenvalues and eigenvectors, whose computational time is less than a millisecond. So we don’t care about the running time of the mapping of normal pixels. On the other hand, the mapping of outliers is a spring system [
24]. Each outlier is linked to its neighborhood through springs whose lengths correspond to the pairwise distances between the outlier and its neighbors in the original space. In this way, we place the outliers in the output space and keep the pairwise distances between the embedded points of outliers and their respective neighbors as similar as possible to those in the original space. The implementation of the mapping for the outliers would require the computational complexity
O(|
NS| × Itermax × ON) (
ON is the number of outliers). As mentioned above, the number of outliers takes a very small proportion of pixels in hyperspectral image (e.g., 0.5 even less). Compared with other algorithms using nonlinear mapping for all pixels such as BCOCDM, the computational complexity of the mapping in our algorithm is greatly reduced. For example, if the outliers are about 0.5% of all pixels, the running time of our method should reduce to about 0.5% of the BCOCDM’s.