1
Department of Botany, Jomo Kenyatta University of Agriculture and Technology, Nairobi P.O. Box 62000-02000, Kenya
2
Department of Food and Nutritional Sciences, Jomo Kenyatta University of Agriculture and Technology, Nairobi P.O. Box 62000-02000, Kenya
3
International Centre of Insect Physiology and Ecology (icipe), Nairobi P.O. Box 30772-00100, Kenya
Microorganisms 2021, 9(12), 2417; https://doi.org/10.3390/microorganisms9122417 - 23 Nov 2021
Cited by 53 | Viewed by 5957
Abstract
Globally, the broad-spectrum antimicrobial activity of chitin and chitosan has been widely documented. However, very little research attention has focused on chitin and chitosan extracted from black soldier fly pupal exuviae, which are abundantly present as byproducts from insect-farming enterprises. This study presents
[...] Read more.
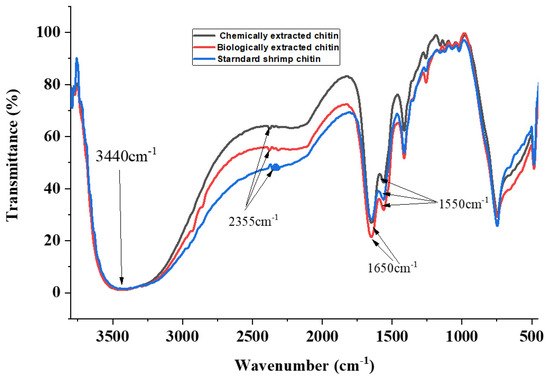
Globally, the broad-spectrum antimicrobial activity of chitin and chitosan has been widely documented. However, very little research attention has focused on chitin and chitosan extracted from black soldier fly pupal exuviae, which are abundantly present as byproducts from insect-farming enterprises. This study presents the first comparative analysis of chemical and biological extraction of chitin and chitosan from BSF pupal exuviae. The antibacterial activity of chitosan was also evaluated. For chemical extraction, demineralization and deproteinization were carried out using 1 M hydrochloric acid at 100 °C for 2 h and 1 M NaOH for 4 h at 100 °C, respectively. Biological chitin extraction was carried out by protease-producing bacteria and lactic-acid-producing bacteria for protein and mineral removal, respectively. The extracted chitin was converted to chitosan via deacetylation using 40% NaOH for 8 h at 100 °C. Chitin characterization was done using FTIR spectroscopy, while the antimicrobial properties were determined using the disc diffusion method. Chemical and biological extraction gave a chitin yield of 10.18% and 11.85%, respectively. A maximum chitosan yield of 6.58% was achieved via chemical treatment. From the FTIR results, biological and chemical chitin showed characteristic chitin peaks at 1650 and 1550 cm−1—wavenumbers corresponding to amide I stretching and amide II bending, respectively. There was significant growth inhibition for Escherichia coli, Bacillus subtilis,Pseudomonas aeruginosa,Staphylococcus aureus, and Candida albicans when subjected to 2.5 and 5% concentrations of chitosan. Our findings demonstrate that chitosan from BSF pupal exuviae could be a promising and novel therapeutic agent for drug development against resistant strains of bacteria.
Full article
(This article belongs to the Special Issue Microbial Biocontrol in the Agri-Food Industry)
▼
Show Figures