Shiftless Is a Novel Member of the Ribosome Stress Surveillance Machinery That Has Evolved to Play a Role in Innate Immunity and Cancer Surveillance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Plasmid Transfections
2.2. shRNA Knockdowns
2.3. Generation of CRISPR Knockout Cell Lines
2.4. Growth Curve of SFL-/- HEK293T Cells
2.5. Preparation of Reporter Plasmids
2.6. Dual Luciferase Assays of -1 PRF
2.7. Bifluorescence Assays of -1 PRF
2.8. qRT PCR and RT PCR Methods
2.9. Bioinformatics Analysis of SFL Expression in Cancer Cells
3. Results
3.1. SFL Is Constitutively Expressed in Human-Derived Cells
3.2. First-Generation Dual Luciferase Reporters, Not the CCR5 -1 PRF Element, Promote Off-Target mRNA Splicing
3.3. SFL Overexpression or Knockdown Results in 2-Fold Reciprocal Effects on -1 PRF
3.4. SFL Is Not Limited to -1 PRF Signals
3.5. Disrupting SFL Homeostasis Reduces Reporter Protein Activity
3.6. Disruption of SFL Homeostasis Decreases mRNA Steady State Abundances
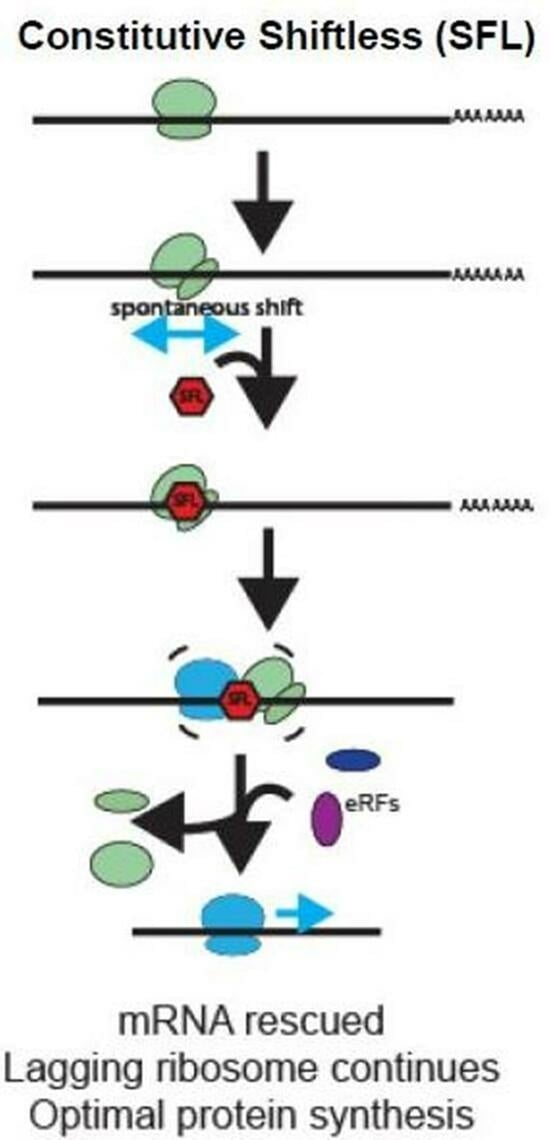
3.7. Genetic Analysis of SFL in the Context of Ribosome-Associated Quality Control (RQC)
3.8. SFL Expression Is Significantly Reduced in Many Common Cancers and Correlates with Worse Clinical Outcomes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warner, J.R. The Economics of Ribosome Biosynthesis in Yeast. Trends Biochem. 1999, 24, 437–440. [Google Scholar] [CrossRef]
- Rudra, D.; Warner, J.R. What Better Measure than Ribosome Synthesis? Genes Dev. 2004, 18, 2431–2436. [Google Scholar] [CrossRef]
- Calamita, P.; Gatti, G.; Miluzio, A.; Scagliola, A.; Biffo, S. Translating the Game: Ribosomes as Active Players. Front. Genet. 2018, 9, 418681. [Google Scholar] [CrossRef]
- Kurland, C.G. Translational Accuracy In Vitro. Cell 1982, 28, 201–202. [Google Scholar] [CrossRef]
- Belcourt, M.F.; Farabaugh, P.J. Ribosomal Frameshifting in the Yeast Retrotransposon Ty: TRNAs Induce Slippage on a 7 Nucleotide Minimal Site. Cell 1990, 62, 339–352. [Google Scholar] [CrossRef]
- Dinman, J.D.; Icho, T.; Wickner, R.B. A -1 Ribosomal Frameshift in a Double-Stranded RNA Virus Forms a Gag-Pol Fusion Protein. Proc. Natl. Acad. Sci. USA 1991, 88, 174–178. [Google Scholar] [CrossRef]
- Parker, J. Errors and Alternatives in Reading the Universal Genetic-Code. Microbiol. Rev. 1989, 53, 273–298. [Google Scholar] [CrossRef]
- Kurland, C.G. Translational Accuracy and the Fitness of Bacteria. Annu. Rev. Genet. 1992, 26, 29–50. [Google Scholar] [CrossRef]
- Gu, J.; Yan, H.; Huang, Y.; Zhu, Y.; Sun, H.; Qiu, Y.; Chen, S. Heightened Protein-Translation Activities in Mammalian Cells and the Disease/Treatment Implications. Natl. Sci. Rev. 2020, 7, 1851–1855. [Google Scholar] [CrossRef]
- Bang, M.L.; Centner, T.; Fornoff, F.; Geach, A.J.; Gotthardt, M.; McNabb, M.; Witt, C.C.; Labeit, D.; Gregorio, C.C.; Granzier, H.; et al. The Complete Gene Sequence of Titin, Expression of an Unusual ≈700-KDa Titin Isoform, and Its Interaction with Obscurin Identify a Novel Z-Line to I-Band Linking System. Circ. Res. 2001, 89, 1065–1072. [Google Scholar] [CrossRef]
- Chang, Y.F.; Imam, J.S.; Wilkinson, M.F. The Nonsense-Mediated Decay RNA Surveillance Pathway. Annu. Rev. Biochem. 2007, 76, 51–74. [Google Scholar] [CrossRef]
- Dever, T.E.; Dinman, J.D.; Green, R. Translation Elongation and Recoding in Eukaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10, a032649. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.H.; Brierley, I. Structural and Functional Insights into Viral Programmed Ribosomal Frameshifting. Annu. Rev. Virol. 2023, 10, 217–242. [Google Scholar] [CrossRef]
- Atkins, J.F.; Loughran, G.; Bhatt, P.R.; Firth, A.E.; Baranov, P.V. Ribosomal Frameshifting and Transcriptional Slippage: From Genetic Steganography and Cryptography to Adventitious Use. Nucleic Acids Res. 2016, 44, 7007–7078. [Google Scholar] [CrossRef]
- Penn, W.D.; Mukhopadhyay, S. Abracadabra, One Becomes Two: The Importance of Context in Viral -1 Programmed Ribosomal Frameshifting. MBio 2022, 13, e02468-21. [Google Scholar] [CrossRef] [PubMed]
- Dinman, J.D.; Wickner, R.B. Ribosomal Frameshifting Efficiency and Gag/Gag-Pol Ratio Are Critical for Yeast M1 Double-Stranded RNA Virus Propagation. J. Virol. 1992, 66, 3669–3676. [Google Scholar] [CrossRef] [PubMed]
- Dinman, J.D. Programmed Ribosomal Frameshifting Goes Beyond Viruses: Organisms from All Three Kingdoms Use Frameshifting to Regulate Gene Expression, Perhaps Signaling a Paradigm Shift. Microbe 2006, 1, 521–527. [Google Scholar] [PubMed]
- Belew, A.T.; Meskauskas, A.; Musalgaonkar, S.; Advani, V.M.; Sulima, S.O.; Kasprzak, W.; Shapiro, B.A.; Dinman, J.D. Ribosomal Frameshifting in the CCR5 MRNA Is Regulated by MiRNAs and the NMD Pathway. Nature 2014, 512, 265–269. [Google Scholar] [CrossRef]
- Riegger, R.J.; Caliskan, N. Thinking Outside the Frame: Impacting Genomes Capacity by Programmed Ribosomal Frameshifting. Front. Mol. Biosci. 2022, 9, 842261. [Google Scholar] [CrossRef] [PubMed]
- Loughran, G.; Howard, M.T.; Firth, A.E.; Atkins, J.F. Avoidance of Reporter Assay Distortions from Fused Dual Reporters. RNA 2017, 23, 1285–1289. [Google Scholar] [CrossRef]
- Dinman, J.D. Translational Recoding Signals: Expanding the Synthetic Biology Toolbox. J. Biol. Chem. 2019, 294, 7537–7545. [Google Scholar] [CrossRef]
- Kelly, J.; Woodside, M.; Dinman, J. Programmed -1 Ribosomal Frameshifting in Coronaviruses: A Therapeutic Target. Virology 2021, 554, 75–82. [Google Scholar] [CrossRef]
- Dinman, J.D.; Ruiz-Echevarria, M.J.; Peltz, S.W. Translating Old Drugs into New Treatments: Ribosomal Frameshifting as a Target for Antiviral Agents. Trends Biotechnol. 1998, 16, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Hung, M.; Patel, P.; Davis, S.; Green, S.R. Importance of Ribosomal Frameshifting for Human Immumodeficiency Virus Type 1 Assembly and Replication. J. Virol. 1998, 72, 4819–4824. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xuan, Y.; Han, Y.; Ding, X.; Ye, K.; Yang, F.; Gao, P.; Goff, S.P.; Gao, G. Regulation of HIV-1 Gag-Pol Expression by Shiftless, an Inhibitor of Programmed-1 Ribosomal Frameshifting. Cell 2019, 176, 625–635.e14. [Google Scholar] [CrossRef] [PubMed]
- Hanners, N.W.; Mar, K.B.; Boys, I.N.; Eitson, J.L.; Cruz-Rivera, P.C.D.L.; Richardson, R.B.; Fan, W.; Wight-Carter, M.; Schoggins, J.W. Shiftless Inhibits Flavivirus Replication in Vitro and Is Neuroprotective in a Mouse Model of Zika Virus Pathogenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2111266118. [Google Scholar] [CrossRef]
- Rodriguez, W.; Muller, M. Shiftless, a Critical Piece of the Innate Immune Response to Viral Infection. Viruses 2022, 14, 1338. [Google Scholar] [CrossRef]
- Balinsky, C.A.; Schmeisser, H.; Wells, A.I.; Ganesan, S.; Jin, T.; Singh, K.; Zoon, K.C. IRAV (FLJ11286), an Interferon-Stimulated Gene with Antiviral Activity against Dengue Virus, Interacts with MOV10. J. Virol. 2017, 91, e01606-16. [Google Scholar] [CrossRef]
- Snieckute, G.; Genzor, A.V.; Vind, A.C.; Ryder, L.; Stoneley, M.; Chamois, S.; Dreos, R.; Nordgaard, C.; Sass, F.; Blasius, M.; et al. Ribosome Stalling Is a Signal for Metabolic Regulation by the Ribotoxic Stress Response. Cell Metab. 2022, 34, 2036–2046.e8. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Harger, J.W.; Dinman, J.D. An in Vivo Dual-Luciferase Assay System for Studying Translational Recoding in the Yeast Saccharomyces Cerevisiae. RNA 2003, 9, 1019–1024. [Google Scholar] [CrossRef]
- Jacobs, J.L.; Dinman, J.D. Systematic Analysis of Bicistronic Reporter Assay Data. Nucleic Acids Res. 2004, 32, e160. [Google Scholar] [CrossRef] [PubMed]
- Munshi, S.; Neupane, K.; Ileperuma, S.M.; Halma, M.T.J.; Kelly, J.A.; Halpern, C.F.; Dinman, J.D.; Loerch, S.; Woodside, M.T. Identifying Inhibitors of −1 Programmed Ribosomal Frameshifting in a Broad Spectrum of Coronaviruses. Viruses 2022, 14, 177. [Google Scholar] [CrossRef]
- Lee, B.T.; Barber, G.P.; Benet-Pagès, A.; Casper, J.; Clawson, H.; DIekhans, M.; Fischer, C.; Gonzalez, J.N.; Hinrichs, A.S.; Lee, C.M.; et al. The UCSC Genome Browser Database: 2022 Update. Nucleic Acids Res. 2022, 50, D1115–D1122. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Khan, Y.A.; Loughran, G.; Steckelberg, A.-L.; Brown, K.; Baranov, P.V.; Kieft, J.S.; Firth, A.E.; Atkins, J.F. Evaluating the Evidence for −1 Frameshifting in Immune-Functioning C-C Chemokine Receptor (CCR5)—The HIV1 Co-Receptor. Nature 2019, 604, E16–E23. [Google Scholar] [CrossRef] [PubMed]
- Napthine, S.; Hill, C.H.; Nugent, H.C.M.; Brierley, I. Modulation of Viral Programmed Ribosomal Frameshifting and Stop Codon Readthrough by the Host Restriction Factor Shiftless. Viruses 2021, 13, 1230. [Google Scholar] [CrossRef] [PubMed]
- Matsufuji, S.; Matsufuji, T.; Miyazaki, Y.; Murakami, Y.; Atkins, J.F.; Gesteland, R.F.; Hayashi, S. Autoregulatory Frameshifting in Decoding Mammalian Ornithine Decarboxylase Antizyme. Cell 1995, 80, 51–60. [Google Scholar] [CrossRef]
- Firth, A.E.; Wills, N.M.; Gesteland, R.F.; Atkins, J.F. Stimulation of Stop Codon Readthrough: Frequent Presence of an Extended 3′ RNA Structural Element. Nucleic Acids Res. 2011, 39, 6679–6691. [Google Scholar] [CrossRef] [PubMed]
- Strauss, E.G.; Rice, C.M.; Strauss, J.H. Sequence Coding for the Alphavirus Nonstructural Proteins Is Interrupted by an Opal Termination Codon. Proc. Natl. Acad. Sci. USA 1983, 80, 5271–5275. [Google Scholar] [CrossRef]
- Kendra, J.A.; de la Fuente, C.; Brahms, A.; Woodson, C.; Bell, T.M.; Chen, B.; Khan, Y.A.; Jacobs, J.L.; Kehn-Hall, K.; Dinman, J.D. Ablation of Programmed −1 Ribosomal Frameshifting in Venezuelan Equine Encephalitis Virus Results in Attenuated Neuropathogenicity. J. Virol. 2017, 91, e01766-16. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.-C.; Peterson, A.; Zinshteyn, B.; Regot, S.; Green, R. Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 2020, 182, 404–416.e14. [Google Scholar] [CrossRef]
- Meydan, S.; Guydosh, N.R. Disome and Trisome Profiling Reveal Genome-Wide Targets of Ribosome Quality Control. Mol. Cell 2020, 79, 588–602.e6. [Google Scholar] [CrossRef] [PubMed]
- Dinman, J.D. Scaring Ribosomes Shiftless. Biochemistry 2019, 58, 1831–1832. [Google Scholar] [CrossRef]
- Hickey, K.L.; Dickson, K.; Cogan, J.Z.; Replogle, J.M.; Schoof, M.; D’Orazio, K.N.; Sinha, N.K.; Hussmann, J.A.; Jost, M.; Frost, A.; et al. GIGYF2 and 4EHP Inhibit Translation Initiation of Defective Messenger RNAs to Assist Ribosome-Associated Quality Control. Mol. Cell 2020, 79, 950–962.e6. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, Y.; Ikeuchi, K.; Saeki, Y.; Iwasaki, S.; Schmidt, C.; Udagawa, T.; Sato, F.; Tsuchiya, H.; Becker, T.; Tanaka, K.; et al. Ubiquitination of Stalled Ribosome Triggers Ribosome-Associated Quality Control. Nat. Commun. 2017, 8, 159. [Google Scholar] [CrossRef]
- Garzia, A.; Jafarnejad, S.M.; Meyer, C.; Chapat, C.; Gogakos, T.; Morozov, P.; Amiri, M.; Shapiro, M.; Molina, H.; Tuschl, T.; et al. The E3 Ubiquitin Ligase and RNA-Binding Protein ZNF598 Orchestrates Ribosome Quality Control of Premature Polyadenylated MRNAs. Nat. Commun. 2017, 8, 16056. [Google Scholar] [CrossRef]
- D’Orazio, K.N.; Green, R. Ribosome States Signal RNA Quality Control. Mol. Cell 2021, 81, 1372–1383. [Google Scholar] [CrossRef]
- Amrani, N.; Ganesan, R.; Kervestin, S.; Mangus, D.A.; Ghosh, S.; Jacobson, A. A Faux 3′-UTR Promotes Aberrant Termination and Triggers Nonsense-Mediated MRNA Decay. Nature 2004, 432, 112–118. [Google Scholar] [CrossRef]
- Howlader, N.; Noone, A.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review, 1975–2017; National Cancer Institute: Bethesda, MD, USA, 2020. Available online: https://seer.cancer.gov/statistics/interactive.html (accessed on 28 February 2022).
- Rácz, G.A.; Nagy, N.; Tóvári, J.; Apáti, Á.; Vértessy, B.G. Identification of New Reference Genes with Stable Expression Patterns for Gene Expression Studies Using Human Cancer and Normal Cell Lines. Sci. Rep. 2021, 11, 19459. [Google Scholar] [CrossRef]
- Jo, J.; Choi, S.; Oh, J.; Lee, S.G.; Choi, S.Y.; Kim, K.K.; Park, C. Conventionally Used Reference Genes Are Not Outstanding for Normalization of Gene Expression in Human Cancer Research. BMC Bioinform. 2019, 20, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Chin, W.X.; Han, Q.; Ichiyama, K.; Lee, C.H.; Eyo, Z.W.; Ebina, H.; Takahashi, H.; Takahashi, C.; Tan, B.H.; et al. Characterization of RyDEN (C19orf66) as an Interferon-Stimulated Cellular Inhibitor against Dengue Virus Replication. PLoS Pathog. 2016, 12, e1005357. [Google Scholar] [CrossRef]
- Parker, M.D.; Karbstein, K. Quality Control Ensures Fidelity in Ribosome Assembly and Cellular Health. J. Cell Biol. 2023, 222, e202209115. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Q.; Zaher, H.S. Canary in a Coal Mine: Collided Ribosomes as Sensors of Cellular Conditions. Trends Biochem. Sci. 2022, 47, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Petrov, A.; Tsai, A.; O’Leary, S.E.; Puglisi, J.D. Coordinated Conformational and Compositional Dynamics Drive Ribosome Translocation. Nat. Struct. Mol. Biol. 2013, 20, 718–727. [Google Scholar] [CrossRef]
- Olson, A.N.; Dinman, J.D. Two Ribosomes Are Better Than One. Sometimes. Mol. Cell 2020, 79, 541–543. [Google Scholar] [CrossRef]
- Costa-Mattioli, M.; Walter, P. The Integrated Stress Response: From Mechanism to Disease. Science 2020, 368, eaat5314. [Google Scholar] [CrossRef]
- Lopinski, J.D.; Dinman, J.D.; Bruenn, J.A. Kinetics of Ribosomal Pausing during Programmed -1 Translational Frameshifting. Mol. Cell Biol. 2000, 20, 1095–1103. [Google Scholar] [CrossRef]
- Schuller, A.P.; Green, R. Roadblocks and Resolutions in Eukaryotic Translation. Nat. Rev. Mol. Cell Biol. 2018, 19, 526–541. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Mühlemann, O. A Comprehensive Coverage Insurance for Cells: Revealing Links between Ribosome Collisions, Stress Responses and MRNA Surveillance. RNA Biol. 2022, 19, 609–621. [Google Scholar] [CrossRef]
- Grentzmann, G.; Ingram, J.A.; Kelly, P.J.; Gesteland, R.F.; Atkins, J.F. A Dual-Luciferase Reporter System for Studying Recoding Signals. RNA 1998, 4, 479–486. [Google Scholar] [PubMed]
- Harger, J.W.; Dinman, J.D. Evidence against a Direct Role for the Upf Proteins in Frameshfiting or Nonsense Codon Readthrough. RNA 2004, 10, 1721–1729. [Google Scholar] [CrossRef] [PubMed]







| shRNA Target | Target Sequence (5′ to 3′) | Cat# |
|---|---|---|
| SFL shA | GTGTATCCAACACGGATCCTC | TRCN0000434142 |
| SFL shB | CCAAGAACTAAGTAACGATCT | TRCN0000161786 |
| SFL shC | AGCAACCCTCACATTAGCAGT | TRCN0000420530 |
| SFL shD | GAAGTTTCATGGGAAGGTATC | TRCN0000164654 |
| SFL shE | GAAGTTCTGTGGGACACATTG | TRCN0000418723 |
| ASCC3 | TGAGGAGCGAACTGGATATTT | TRCN0000296023 |
| Hbs1L | GCGATCTATTGACAAACCTTT | TRCN0000353597 |
| PELO | GCAGTGAAGACCGACAACAAA | TRCN0000163394 |
| SMG1 | GCACTGTAACTACGGCTACAA | TRCN0000037413 |
| ZNF598 | CCAACCCTCTAAAGTTGGGAA | TRCN0000222610 |
| Plasmid Name | Description |
|---|---|
| pJD2257 | Modified pSGDluc (dual luciferase with inteins) with 0-frame control sequence insert |
| pJD2256 | Modified pSGDluc (dual luciferase with inteins) with HIV-1 -1 PRF sequence insert |
| pJD2258 | Modified pSGDluc (dual luciferase with inteins) with CCR5 -1 PRF sequence insert |
| pJD2359 | Modified pSGDluc (dual luciferase with inteins) with SARS-CoV -1 PRF sequence insert |
| pJD2514 | Modified pSGDluc (dual luciferase with inteins) with SARS-CoV-2 -1 PRF sequence insert |
| pJD2450 | Modified pSGDluc (dual luciferase with inteins) with spontaneous -1 FS sequence insert |
| pJD2262 | Dual fluorescent construct with 0-frame control sequence insert |
| pJD2261 | Dual fluorescent construct with HIV-1 -1 PRF sequence insert |
| pJD2281 | Dual fluorescent construct with CCR5 -1 PRF sequence insert |
| pJD2529 | Dual fluorescent construct with SARS-CoV-2 sequence insert |
| pJD2350 | Dual fluorescent construct with OAZ1 +1 PRF sequence insert |
| pJD2455 | Dual fluorescent construct with VEEV stop codon readthrough sequence insert |
| pJD2260 | pMAX GFP monocistronic reporter |
| pJD2612 | C19orf66 ORF in pCMV-Myc |
| pJD175f | P2luci (first-generation dual luciferase) with 0-frame control sequence insert |
| pJD827 | P2luci with CCR5 -1 PRF sequence insert |
| Oligo Name | Sequence (5′ to 3′) |
|---|---|
| H_GAPDH_F | GGATGATGTTCTGGAGAGCC |
| H_GAPDH_R | CATCACCATCTTCCAGGAGC |
| GFP_qPCR_F | GGCTACGGCTTCTACCACTT |
| GFP_qPCR_R | CTCGTACTTCTCGATGCGGG |
| Firefly Luciferase qPCR Fwd | TCGCCTCTCTGATTAACGCC |
| Firefly Luciferase qPCR Rev | ATTACACCCGAGGGGGATGA |
| C19orf66_qPCR | Biorad PrimePCR qHsaCED0003572 |
| C19orf66_RT_PCR_F | CTCAGGAAGGTGTGGAGCTG |
| C19orff_RT_PCR_R | GCCACTGCTAATGTGAGGGT |
| Pelo_qPCR_F | GACCGACAACAAACTGCTCCTG |
| Pelo_qPCR_R | AGCCACAGTAGGGTCACAAAGG |
| SMG1_qPCR_F | ATGCTGGTGAGCTTCGGCAGTA |
| SMG1_qPCR_R | CGCACATACACTTCAGGGTGGT |
| CRISPR validation primer Fwd | AAATCTGGCTTCTGAACCTCCT |
| CRISPR validation primer Rev | GTGGGAGACAAAGTGGACTGAG |
| SFL gRNA Top | CACCG GGATCCGTGTTGGATACACG |
| SFL gRNA Bottom | AAAC CGTGTATCCAACACGGATCC C |
| CRISPR validation primer Fwd | AAATCTGGCTTCTGAACCTCCT |
| CRISPR validation primer Rev | GTGGGAGACAAAGTGGACTGAG |
| -1 sFS_SDM_F | AAAGAGGCTGCGGCAAAAGC |
| -1 sFS_SDM_R | GCGGCTGCTTCGGTCGAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kelly, J.A.; Dinman, J.D. Shiftless Is a Novel Member of the Ribosome Stress Surveillance Machinery That Has Evolved to Play a Role in Innate Immunity and Cancer Surveillance. Viruses 2023, 15, 2296. https://doi.org/10.3390/v15122296
Kelly JA, Dinman JD. Shiftless Is a Novel Member of the Ribosome Stress Surveillance Machinery That Has Evolved to Play a Role in Innate Immunity and Cancer Surveillance. Viruses. 2023; 15(12):2296. https://doi.org/10.3390/v15122296
Chicago/Turabian StyleKelly, Jamie A., and Jonathan D. Dinman. 2023. "Shiftless Is a Novel Member of the Ribosome Stress Surveillance Machinery That Has Evolved to Play a Role in Innate Immunity and Cancer Surveillance" Viruses 15, no. 12: 2296. https://doi.org/10.3390/v15122296