Identification of Chromosomal Regions and Candidate Genes for Round leaf Locus in Cucumis melo L.
Abstract
:1. Introduction
2. Results
2.1. The Round leaf in MR-1 Is Controlled by a Single Recessive Locus
2.2. Mapping of the Cmrl Locus into an 80.27-kb Region
2.3. Pentatricopeptide Repeat-Containing Family Protein Is the Candidate Gene for Cmrl Locus
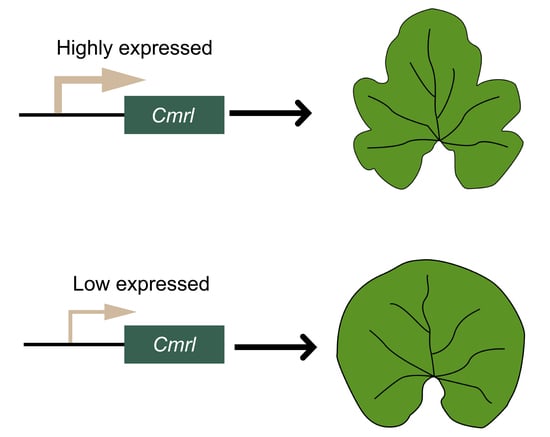
2.4. Cmrl Encodes a DYW-PPR Protein
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. BSA-Seq and Initial Mapping
4.3. Fine Mapping
4.4. Gene Annotation and Cloning
4.5. RNA Extraction and Gene Expression Analysis
4.6. Subcellular Localization
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feng, X.; Li, X.; Yang, X.; Zhu, P. Fine Mapping and Identification of the Leaf Shape Gene BoFL in Ornamental Kale. Theor. Appl. Genet. 2020, 133, 1303–1312. [Google Scholar] [CrossRef] [PubMed]
- Peppe, D.J.; Royer, D.L.; Cariglino, B.; Oliver, S.Y.; Newman, S.; Leight, E.; Enikolopov, G.; Fernandez-Burgos, M.; Herrera, F.; Adams, J.M.; et al. Sensitivity of leaf size and shape to climate: Global patterns and paleoclimatic applications. New Phytol. 2011, 190, 724–739. [Google Scholar] [CrossRef] [PubMed]
- Zuniga, A. Next generation limb development and evolution: Old questions, new perspectives. Development 2015, 142, 3810–3820. [Google Scholar] [CrossRef]
- Runions, A.; Tsiantis, M. The shape of things to come: From typology to predictive models for leaf diversity. Am. J. Bot. 2017, 104, 1437–1441. [Google Scholar] [CrossRef] [PubMed]
- Kierzkowski, D.; Runions, A.; Vuolo, F.; Strauss, S.; Lymbouridou, R.; Routier-Kierzkowska, A.L.; Wilson-Sánchez, D.; Jenke, H.; Galinha, C.; Mosca, G.; et al. A Growth-Based Framework for Leaf Shape Development and Diversity. Cell 2019, 177, 1405–1418. [Google Scholar] [CrossRef] [PubMed]
- Kucypera, K.; Lipowczan, M.; Piekarska-Stachowiak, A.; Nakielski, J. A Method to Generate the Surface Cell Layer of the 3D Virtual Shoot Apex from Apical Initials. Plant Methods 2017, 13, 110. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Kong, F.; Zhou, C. From Genes to Networks: The Genetic Control of Leaf Development. J. Integr. Plant Biol. 2021, 63, 1181–1196. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.; Southam, P.; Pantin, F.; Kennaway, R.; Robinson, S.; Castorina, G.; Sánchez-Corrales, Y.E.; Sablowski, R.; Chan, J.; Grieneisen, V.; et al. Spatiotemporal coordination of cell division and growth during organ morphogenesis. PLoS Biol. 2018, 16, e2005952. [Google Scholar] [CrossRef]
- Nicotra, A.B.; Leigh, A.; Boyce, C.K.; Jones, C.S.; Niklas, K.J.; Royer, D.L.; Tsukaya, H. The Evolution and Functional Significance of Leaf Shape in the Angiosperms. Funct. Plant Biol. 2011, 38, 535–552. [Google Scholar] [CrossRef]
- Sisó, S.; Camarero, J.; Gil-Pelegrín, E. Relationship between Hydraulic Resistance and Leaf Morphology in Broadleaf Quercus Species: A New Interpretation of Leaf Lobation. Trees-Struct. Funct. 2001, 15, 341–345. [Google Scholar] [CrossRef]
- Smith, D.D.; Sperry, J.S.; Adler, F.R. Convergence in leaf size versus twig leaf area scaling: Do plants optimize leaf area partitioning? Ann. Bot. 2017, 119, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Lusk, C.H.; Grierson, E.R.P.; Laughlin, D.C. Large leaves in warm, moist environments confer an advantage in seedling light interception efficiency. New Phytol. 2019, 223, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Royer, D.L.; Wilf, P. Why do toothed leaves correlate with cold climates? Gas exchange at leaf margins provides new insights into a classic paleotemperature proxy. Int. J. Plant Sci. 2006, 167, 11–18. [Google Scholar] [CrossRef]
- Sack, L.; Scoffoni, C.; McKown, A.D.; Frole, K.; Rawls, M.; Havran, J.C.; Tran, H.; Tran, T. Developmentally based scaling of leaf venation architecture explains global ecological patterns. Nat. Commun. 2012, 15, 837. [Google Scholar] [CrossRef] [PubMed]
- Field, T.S.; Sage, T.L.; Czerniak, C.; Iles, W.J.D. Hydathodal leaf teeth of Chloranthus japonicus (Chloranthaceae) prevent guttation-induced flooding of the mesophyll. Plant Cell Environ. 2005, 28, 1179–1190. [Google Scholar] [CrossRef]
- Dkhar, J.; Pareek, A. What Determines a Leaf’s Shape? Evodevo 2014, 5, 47. [Google Scholar] [CrossRef] [PubMed]
- Lurin, C.; Andrés, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyère, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-Wide Analysis of Arabidopsis Pentatricopeptide Repeat Proteins Reveals Their Essential Role in Organelle Biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef]
- Cheng, S.; Gutmann, B.; Zhong, X.; Ye, Y.; Fisher, M.F.; Bai, F.; Castleden, I.; Song, Y.; Song, B.; Huang, J.; et al. Redefining the Structural Motifs That Determine RNA Binding and RNA Editing by Pentatricopeptide Repeat Proteins in Land Plants. Plant J. 2016, 85, 532–547. [Google Scholar] [CrossRef]
- Wang, X.; An, Y.; Xu, P.; Xiao, J. Functioning of PPR Proteins in Organelle RNA Metabolism and Chloroplast Biogenesis. Front. Plant Sci. 2021, 12, 627501. [Google Scholar] [CrossRef]
- Yuan, Y.W.; Liu, C.; Marx, H.E.; Olmstead, R.G. The Pentatricopeptide Repeat (PPR) Gene Family, a Tremendous Resource for Plant Phylogenetic Studies. New Phytol. 2009, 182, 272–283. [Google Scholar] [CrossRef]
- Ren, R.C.; Wang, L.L.; Zhang, L.; Zhao, Y.J.; Wu, J.W.; Wei, Y.M.; Zhang, X.S.; Zhao, X.Y. DEK43 Is a P-Type Pentatricopeptide Repeat (PPR) Protein Responsible for the Cis-Splicing of Nad4 in Maize Mitochondria. J. Integr. Plant Biol. 2020, 62, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Cheng, Y.; Yap, A.; Chateigner-Boutin, A.L.; Delannoy, E.; Hammani, K.; Small, I.; Huang, J. The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 2009, 58, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Van Rooijen, R.; Kruijer, W.; Boesten, R.; Van Eeuwijk, F.A.; Harbinson, J.; Aarts, M.G.M. Natural Variation of YELLOW SEEDLING1 Affects Photosynthetic Acclimation of Arabidopsis thaliana. Nat. Commun. 2017, 8, 1421. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.B.; Jiang, Y.; Chong, K.; Yang, Z.N. AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J. 2009, 59, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, W.Y.; Liao, J.C.; Chang, C.Y.; Harrison, T.; Boucher, C.; Hsieh, M.H. The SLOW GROWTH3 Pentatricopeptide Repeat Protein Is Required for the Splicing of Mitochondrial NADH Dehydrogenase Subunit7 Intron 2 in Arabidopsis. Plant Physiol. 2015, 168, 490–501. [Google Scholar] [CrossRef] [PubMed]
- Doniwa, Y.; Ueda, M.; Ueta, M.; Wada, A.; Kadowaki, K.I.; Tsutsumi, N. The Involvement of a PPR Protein of the P Subfamily in Partial RNA Editing of an Arabidopsis Mitochondrial Transcript. Gene 2010, 454, 39–46. [Google Scholar] [CrossRef]
- Wei, C.; Chen, X.; Wang, Z.; Liu, Q.; Li, H.; Zhang, Y.; Ma, J.; Yang, J.; Zhang, X. Genetic Mapping of the LOBED LEAF 1 (ClLL1) Gene to a 127.6-Kb Region in Watermelon (Citrullus lanatus L.). PLoS ONE 2017, 12, e0180741. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Gao, M.; Amanullah, S.; Guo, Y.; Bao, X.; Duan, Y.; Liu, X.; Liu, J.; Gao, Y.; Luan, F. Fine Genetic Mapping Confers a Major Gene Controlling Leaf Shape Variation in Watermelon. Euphytica 2023, 219, 92. [Google Scholar] [CrossRef]
- Bo, K.; Duan, Y.; Qiu, X.; Zhang, M.; Shu, Q.; Sun, Y.; He, Y.; Shi, Y.; Weng, Y.; Wang, C. Promoter Variation in a Homeobox Gene, CpDll, Is Associated with Deeply Lobed Leaf in Cucurbita pepo L. Theor. Appl. Genet. 2022, 135, 1223–1234. [Google Scholar] [CrossRef]
- Gao, X.; Ning, X.; Wang, Y.; Wang, X.; Yan, W.; Zhang, Z.; Li, G. Fine Mapping of a Gene That Confers Palmately Lobed Leaf (Pll) in Melon (Cucumis melo L.). Euphytica 2014, 200, 337–347. [Google Scholar] [CrossRef]
- Herrington, M.E.; Brown, P.J. Inheritance of leaf and fruit characteristics in Cucurbita maxima Duch. cv. Queensland blue × Cucurbita ecuadorensis cutler and whitaker. Qld. J. Agric. Animal Sci. 1988, 45, 45–48. [Google Scholar]
- Dyutin, K.E. Spontaneous mutant of Cucurbita maxima Duch. with lobed leaves. Genetika 1980, 16, 176–178. [Google Scholar]
- Wang, X.; Ando, K.; Wu, S.; Reddy, U.K.; Tamang, P.; Bao, K.; Hammar, S.A.; Grumet, R.; McCreight, J.D.; Fei, Z. Genetic Characterization of Melon Accessions in the U.S. National Plant Germplasm System and Construction of a Melon Core Collection. Mol. Hortic. 2021, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Vollbrecht, E.; Veit, B.; Sinha, N.; Hake, S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 1991, 350, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Tsiantis, M. KNOX Genes: Versatile Regulators of Plant Development and Diversity. Development 2010, 137, 3153–3165. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Fan, M.; Zhao, M.; Jiang, X.; Liu, Q. Overexpression of LtKNOX1 from Lilium tsingtauense in Nicotiana benthamiana affects the development of leaf morphology. Plant Signal. Behav. 2022, 17, 2031783. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.F.; Fu, L.M.; Deng, L.; Liu, M.F.; Gan, Z.M.; Zhou, H.; Hu, S.F.; Hu, C.G.; Zhang, J.Z. CiKN1 and CiKN6 Are Involved in Leaf Development in Citrus by Regulating CimiR164. Plant J. 2022, 110, 828–848. [Google Scholar] [CrossRef]
- Müller, K.J.; Romano, N.; Gerstner, O.; Garcia-Maroto, F.; Pozzi, C.; Salamini, F.; Rohde, W. The barley Hooded mutation caused by a duplication in a homeobox gene intron. Nature 1995, 374, 727–730. [Google Scholar] [CrossRef]
- Lincoln, C.; Long, J.; Yamaguchi, J.; Serikawa, K.; Hake, S. A Knotted1-like Homeobox Gene in Arabidopsis Is Expressed in the Vegetative Meristem and Dramatically Alters Leaf Morphology When Overexpressed in Transgenic Plants. Plant Cell 1994, 6, 1859–1876. [Google Scholar]
- Spinelli, S.V.; Martin, A.P.; Viola, I.L.; Gonzalez, D.H.; Palatnik, J.F. A Mechanistic Link between STM and CUC1 during Arabidopsis Development. Plant Physiol. 2011, 156, 1894–1904. [Google Scholar] [CrossRef]
- Yu, C.; Yan, C.; Liu, Y.; Liu, Y.; Jia, Y.; Lavelle, D.; An, G.; Zhang, W.; Zhang, L.; Han, R.; et al. Upregulation of a KN1 Homolog by Transposon Insertion Promotes Leafy Head Development in Lettuce. Proc. Natl. Acad. Sci. USA 2020, 117, 33668–33678. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lavelle, D.; Yu, C.; Zhang, W.; Chen, J.; Wang, X.; Michelmore, R.W.; Kuang, H. The Upregulated LsKN1 Gene Transforms Pinnately to Palmately Lobed Leaves through Auxin, Gibberellin, and Leaf Dorsiventrality Pathways in Lettuce. Plant Biotechnol. J. 2022, 20, 1756–1769. [Google Scholar] [CrossRef]
- Yu, Y.H.; Li, X.F.; Yang, S.D.; Li, S.Q.; Meng, X.X.; Liu, H.N.; Pei, M.S.; Wei, T.L.; Zhang, Y.J.; Guo, D.L. Overexpression of VvPPR1, a DYW-Type PPR Protein in Grape, Affects the Phenotype of Arabidopsis thaliana Leaves. Plant Physiol. Biochem. 2021, 164, 195–204. [Google Scholar] [CrossRef]
- Bolduc, N.; Yilmaz, A.; Mejia-Guerra, M.K.; Morohashi, K.; O’Connor, D.; Grotewold, E.; Hake, S. Unraveling the KNOTTED1 Regulatory Network in Maize Meristems. Genes Dev. 2012, 26, 1685–1690. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, C.; Ma, X.; Xia, R.; Chen, J.; Liu, X.; Ying, P.; Peng, M.; Wang, J.; Shi, C.L.; et al. KNOX Protein KNAT1 Regulates Fruitlet Abscission in Litchi by Repressing Ethylene Biosynthetic Genes. J. Exp. Bot. 2020, 71, 4069–4082. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.F.; Zhou, H.; Fu, L.M.; Yan, Z.; Ye, L.X.; Hu, S.F.; Gan, Z.M.; Ai, X.Y.; Hu, C.G.; Zhang, J.Z. Two Citrus KNAT-like Genes, CsKN1 and CsKN2, Are Involved in the Regulation of Spring Shoot Development in Sweet Orange. J. Exp. Bot. 2021, 72, 7002–7019. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Thiel, T.; Kota, R.; Grosse, I.; Stein, N.; Graner, A. SNP2CAPS: A SNP and INDEL Analysis Tool for CAPS Marker Development. Nucleic Acids Res. 2004, 32, e5. [Google Scholar] [CrossRef]
- Fang, X.; Li, S.; Zhu, Z.; Zhang, X.; Xiong, C.; Wang, X.; Luan, F.; Liu, S. Clorf Encodes Carotenoid Isomerase and Regulates Orange Flesh Color in Watermelon (Citrullus lanatus L.). J. Agric. Food Chem. 2023, 71, 15445–15455. [Google Scholar] [CrossRef]
- Liu, S.; Gao, P.; Zhu, Q.; Zhu, Z.; Liu, H.; Wang, X.; Weng, Y.; Gao, M.; Luan, F. Resequencing of 297 melon accessions reveals the genomic history of improvement and loci related to fruit traits in melon. Plant Biotechnol. J. 2020, 18, 2545–2558. [Google Scholar] [CrossRef]






| Population | Plants | Deeply Lobed | Round | Expected Segregation Ratio | Actual Segregation Ratio | p Value of Chi-Square Tests |
|---|---|---|---|---|---|---|
| MR-1 | 15 | 0 | 15 | N/A | N/A | N/A |
| PI 614174 | 15 | 15 | 0 | N/A | N/A | N/A |
| F1 | 15 | 15 | 0 | N/A | N/A | N/A |
| 2021-F2 | 220 | 169 | 51 | 3:1 | 3.31:1 | 0.648 |
| 2021-F2 | 1155 | 853 | 302 | 3:1 | 2.82:1 | 0.512 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, X.; Zhu, Z.; Li, J.; Wang, X.; Wei, C.; Zhang, X.; Dai, Z.; Liu, S.; Luan, F. Identification of Chromosomal Regions and Candidate Genes for Round leaf Locus in Cucumis melo L. Plants 2024, 13, 1134. https://doi.org/10.3390/plants13081134
Fang X, Zhu Z, Li J, Wang X, Wei C, Zhang X, Dai Z, Liu S, Luan F. Identification of Chromosomal Regions and Candidate Genes for Round leaf Locus in Cucumis melo L. Plants. 2024; 13(8):1134. https://doi.org/10.3390/plants13081134
Chicago/Turabian StyleFang, Xufeng, Zicheng Zhu, Junyan Li, Xuezheng Wang, Chunhua Wei, Xian Zhang, Zuyun Dai, Shi Liu, and Feishi Luan. 2024. "Identification of Chromosomal Regions and Candidate Genes for Round leaf Locus in Cucumis melo L." Plants 13, no. 8: 1134. https://doi.org/10.3390/plants13081134