- Article
Toward a Phenotype-Driven Continuum Model in Trigger Finger: Proposing a Sonographic Framework for Personalized Management
- Sang-Hyun Kim,
- Jihyo Hwang and
- King Hei Stanley Lam
- + 10 authors
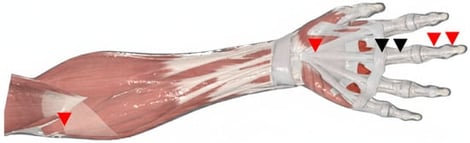
Background: The traditional A1-centric paradigm for trigger finger (TF) management does not fully capture heterogeneous pathology spanning isolated pulley stenosis, tendon degeneration, and impaired tendon–sheath gliding. Methods: A comprehensive literature synthesis (2010–2025) integrating anatomy, biomechanics, and ultrasound-guided interventions was performed to develop a testable, phenotype-driven framework. Results: A continuum model is proposed emphasizing (i) origin-to-insertion assessment of the flexor apparatus, (ii) pragmatic ultrasound phenotyping into pulley-dominant, tendon-dominant, and mixed patterns, and (iii) a stepwise, phenotype-matched management pathway incorporating conservative care, ultrasound-guided injection, selected adjuncts (e.g., hydrodissection, prolotherapy, ESWT) for tendon-dominant or mixed presentations, and percutaneous or open release when an A1 bottleneck is confirmed. Conclusions: This framework is presented as a hypothesis to guide standardized reporting, reliability testing, and phenotype-stratified comparative trials, rather than as a validated clinical guideline. This article proposes a novel, phenotype-driven clinical framework to address this limitation. Contemporary evidence is integrated to construct a model emphasizing (i) a whole-length, origin-to-insertion assessment of the flexor apparatus, (ii) sonographic phenotyping into pulley-dominant, tendon-dominant, and mixed patterns, and (iii) a stepwise treatment algorithm integrating conservative care, ultrasound-guided injections, ultrasound-guided percutaneous release, and selected adjunctive approaches such as hydrodissection (HD), prolotherapy (Prolo), and extracorporeal shockwave therapy (ESWT). While evidence supports individual modalities, the framework’s primary innovation lies in matching interventions to phenotype. This sonographic phenotyping system is presented not as a validated tool, but as a testable hypothesis designed to guide future validation studies. The proposed framework establishes research priorities, including standardized criteria, reliability testing, and comparative effectiveness research for phenotype-stratified management.
8 February 2026