- Article
Transcriptome-Based Selection and Validation of Reference Genes for Gene Expression Analysis in Roegneria ciliaris ‘Liao Sheng’ Across Various Tissues and Under Drought Stress
- Qianyun Luo,
- Yue Liu and
- Sijia Liu
- + 7 authors
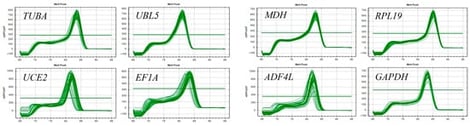
Backgrounds: Roegneria ciliaris is a perennial tetraploid wild relative of wheat that is widely distributed in China. It can be used both as a forage crop and ecological grass (the grasses specifically bred for ecological restoration) due to its strong stress tolerance, early green-up, vigorous seedling growth in spring, and great palatability. Methods: It is necessary to select and validate appropriate reference genes (RGs) for gene expression normalization by qRT-PCR in order to decipher the stress tolerance mechanism of this grass species. Therefore, eight candidate RGs were identified from transcriptome data of R. ciliaris ‘Liao sheng’ in response to drought stress. The expression stability of these RGs was evaluated by five algorithms (∆Ct, geNorm, NormFinder, Bestkeeper and ReFinder) using samples from different tissues and drought stress. Results: The results showed that MDH and RPL19 were the most stable RGs among all samples, while GAPDH and TUBA presented the lowest expression stability. These representative RGs were further used to normalize the expression level of the pyrroline-5-carboxylate synthase (P5CS) and protein phosphatase 2C (PP2C) genes in different tissues and under drought stress. The results of P5CS and PP2C expression were consistent with transcriptome data. Conclusion: Our study provided the first systematic evaluation of the most stable RG selection for qRT-PCR normalization in R. ciliaris, which will promote further research on its tissue-specific gene expression and mechanism of drought tolerance.
14 February 2026



![Schematic representation of the discontinuous (top) and continuous (bottom) selection strategies used to generate sequential SA628 mutant strains. The strains were exposed to streptomycin (800 mg/L) or rifampicin (1000 mg/L). Discontinuous selection resulted in a stepwise evolutionary trajectory (SA628 mut1 → SA628 mut3), whereas continuous selection led to the independent emergence of SA628 mut2. The figure was created with BioGDP [28].](https://mdpi-res.com/cdn-cgi/image/w=281,h=192/https://mdpi-res.com/genes/genes-17-00235/article_deploy/html/images/genes-17-00235-g001-550.jpg)