- Article
Engineering the Future of Heart Failure Therapeutics: Integrating 3D Printing, Silicone Molding, and Translational Development for Implantable Cardiac Devices
- Carleigh Eagle,
- Aarti Desai and
- Rohan Goswami
- + 5 authors
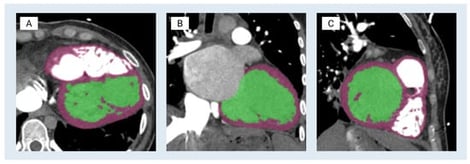
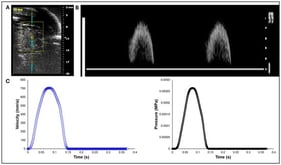
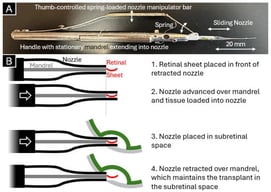
Three-dimensional (3D) anatomic modeling derived from high-resolution medical imaging, such as computed tomography (CT) and magnetic resonance imaging (MRI), has been increasingly adopted in preclinical testing and device development. This white paper describes a cardiac-specific workflow that integrates 3D printing and silicone molding for support device development and procedural simulation. Patient-derived computed tomography angiography data were segmented using FDA-cleared medical modeling software to isolate the left ventricular anatomy and were further processed in computer-aided design (CAD) to ensure accurate physiological wall thickness and structural fidelity. Material jetting 3D printing was performed on a Stratasys J750 using material distributions designed to mimic the mechanical properties of myocardium, thereby approximating myocardial compliance. In parallel, stereolithography apparatus molds were designed from the left ventricle CAD model to cast transparent, pliable left ventricular models in Sorta-Clear™ 18 silicone. The 3D-printed models preserved intricate morphological detail and were suitable for mechanical manipulation and device deployment studies, whereas silicone models offered tunable mechanical properties, transparency for visualization, and durability for repeated use. Together, these complementary modalities provided rapid manufacturing capability and application-relevant physical representation. Case-specific parameters, strengths, and limitations of both models in enhancing patient care and device testing are highlighted, with relevance to heart failure applications. Current knowledge gaps, workflow and integration challenges, and future opportunities are identified, positioning this work as a reference framework for continued innovation in anatomic modeling. Within the collaborative framework of Mayo Clinic’s Anatomic Modeling Unit and Simulation Center, this integrated modeling workflow demonstrates the value of multidisciplinary collaboration between engineers and clinicians. Clinically, these patient-specific left ventricular models may enable pre-procedural device sizing and positioning and may support simulation of mechanical circulatory support (MCS) deployment while identifying possible anatomic constraints prior to intervention. This workflow has direct applicability in advanced heart failure patients undergoing MCS support, such as the Impella axillary MCS device or the durable LVAD, with potential to reduce procedural uncertainty while reducing complications and improving peri-procedural outcomes. Additionally, these models also serve as high-accuracy educational tools, enabling trainees and multidisciplinary care teams to visualize and possibly rehearse procedural steps while gaining hands-on experience in a risk-free environment.
8 February 2026