Physiological Studies and Transcriptomic Analysis Reveal the Mechanism of Saline-Alkali Stress Resistance of Malus sieversii f. niedzwetzkyan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Saline-Alkali Treatment
2.2. Physiological Indexes
2.3. RNA Extraction and Library Construction
2.4. Quality Control, Alignment, and Analysis of Sequencing Data
2.5. Differentially Expressed Gene (DEG), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis
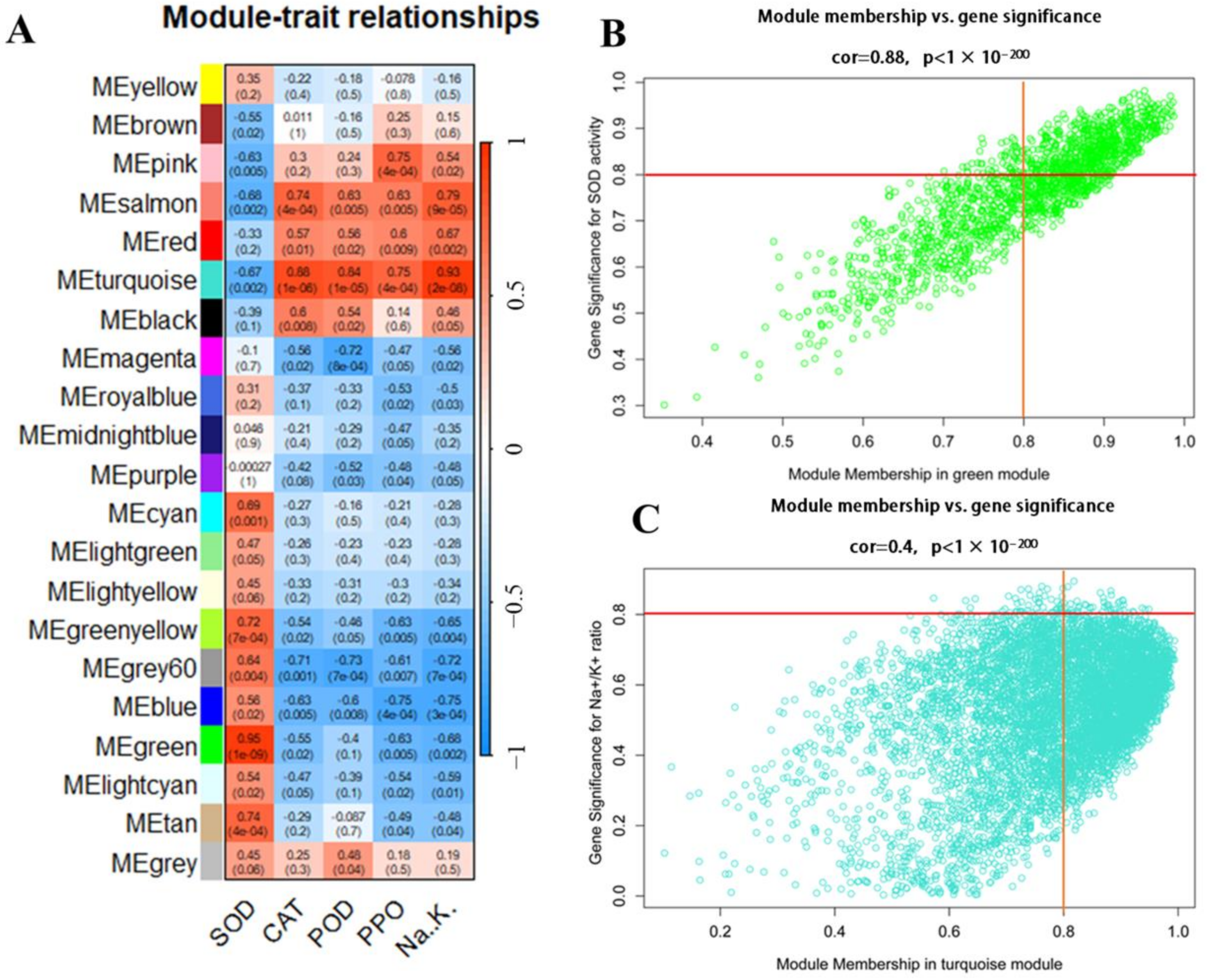
2.6. Weighted Gene Co-Expression Network Analysis (WGCNA)
2.7. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR) Validation of RNA Sequencing (RNA-Seq) Data
2.8. Data Accessibility
2.9. Statistical Analysis
3. Results
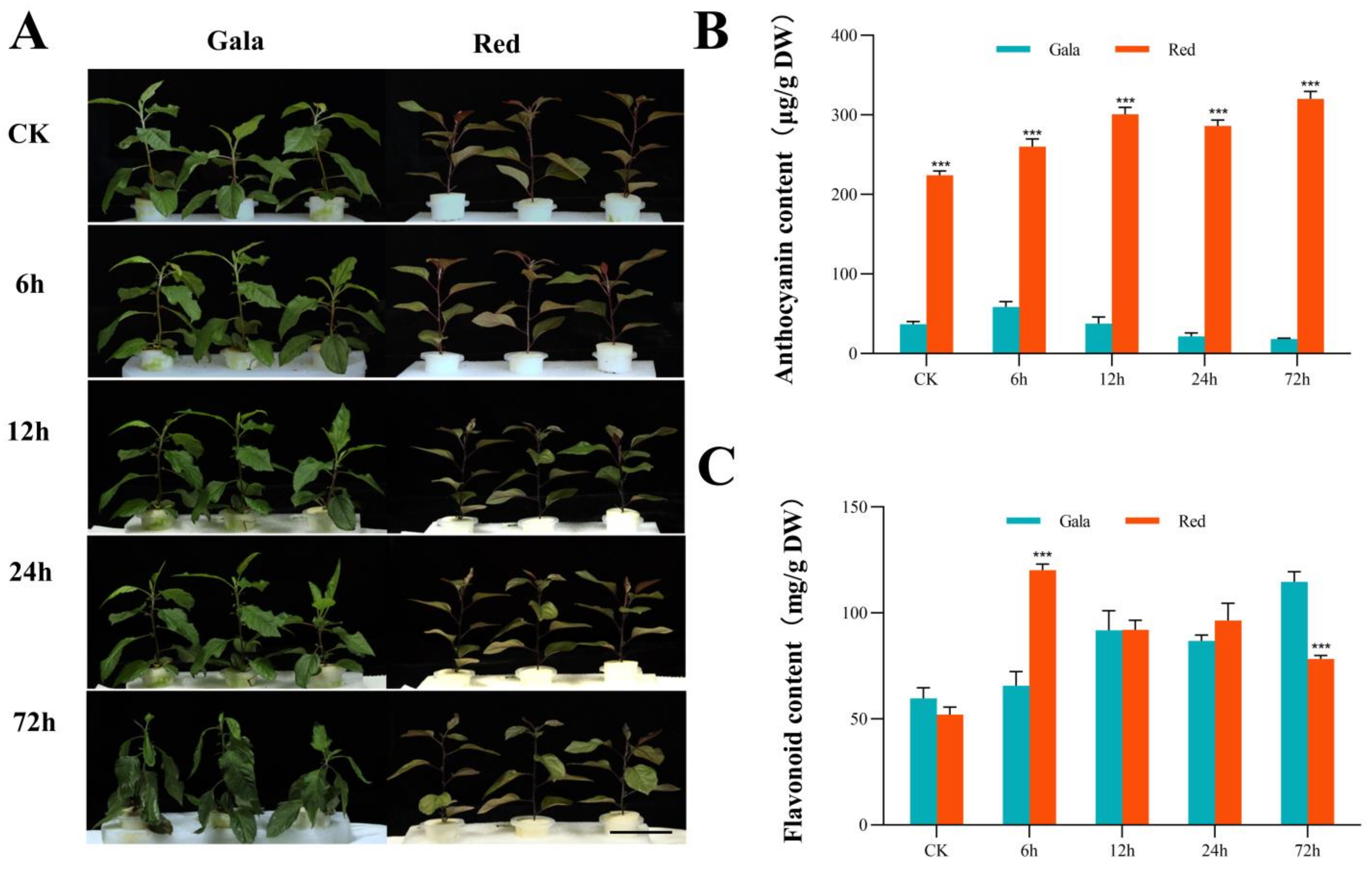
3.1. Phenotypic Changes under Saline-Alkali Treatment
3.2. Activity of Antioxidant Enzymes under Saline-Alkali Treatment
3.3. Metal Ion and PRO Content under Saline-Alkali Treatment
3.4. RNA-Seq Analysis
3.5. DEGs Involved in the Response to Saline-Alkali Stress
3.6. GO Analysis of DEGs Involved in Saline-Alkali Stress
3.7. KEGG Pathway Enrichment Analysis of DEGs Involved in Saline-Alkali Stress
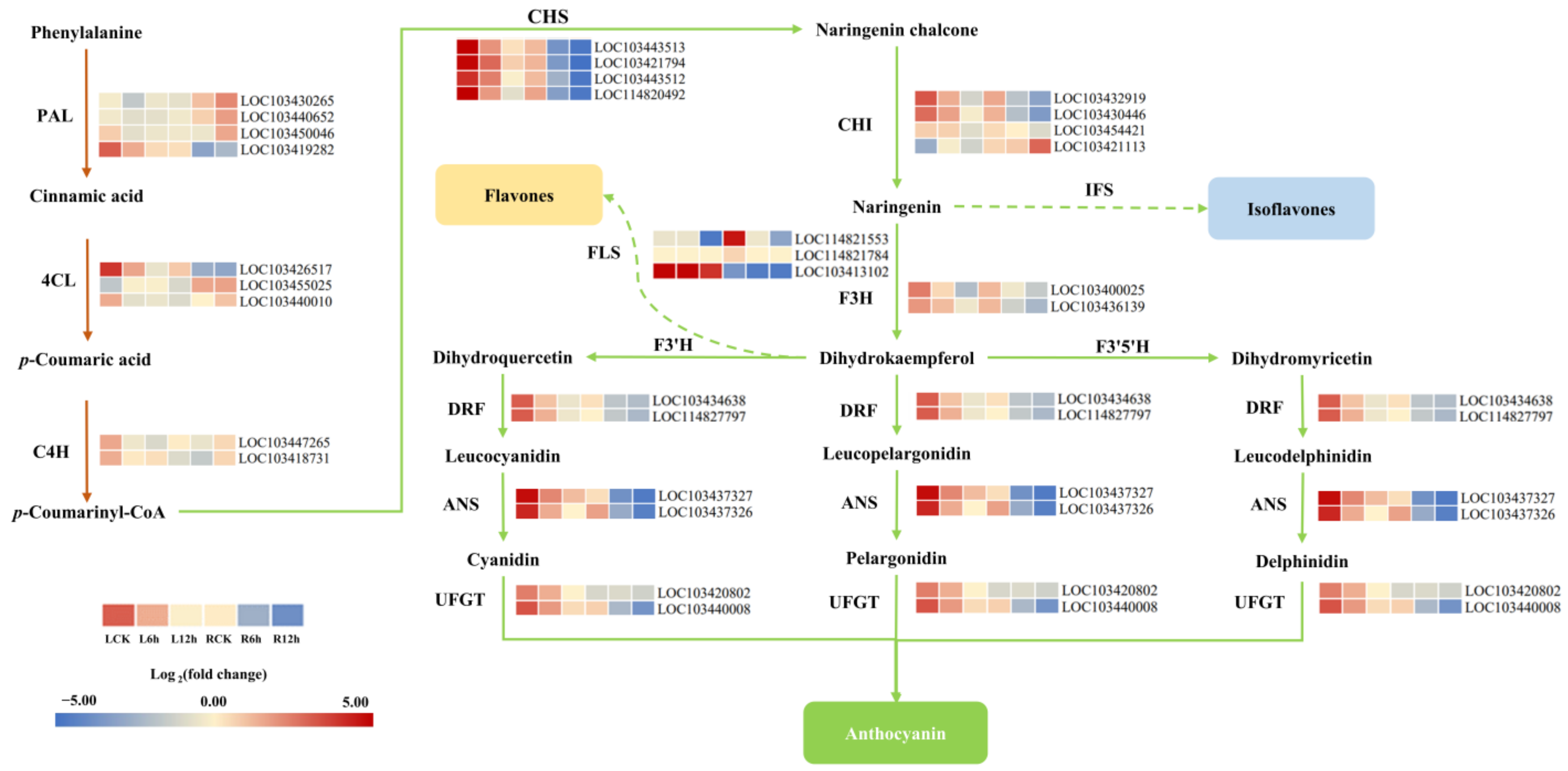
3.8. DEGs Involved in the Anthocyanin Synthesis Pathway Mediate the Response to Saline-Alkali Stress
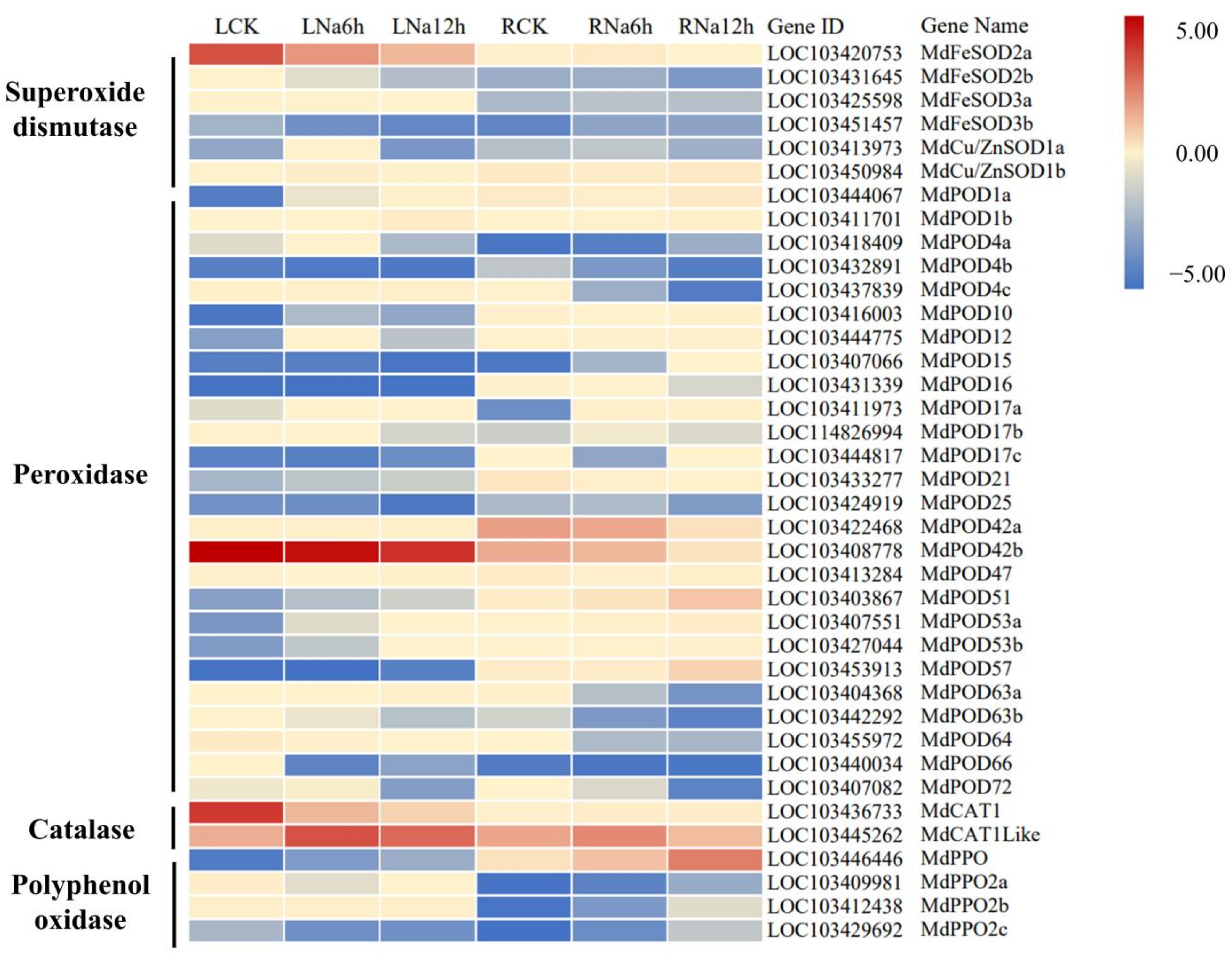
3.9. DEGs Encoding Antioxidant Enzymes Involved in the Response to Saline-Alkali Stress
3.10. Ion Transporters in Response to Saline-Alkali Stress in DEGs
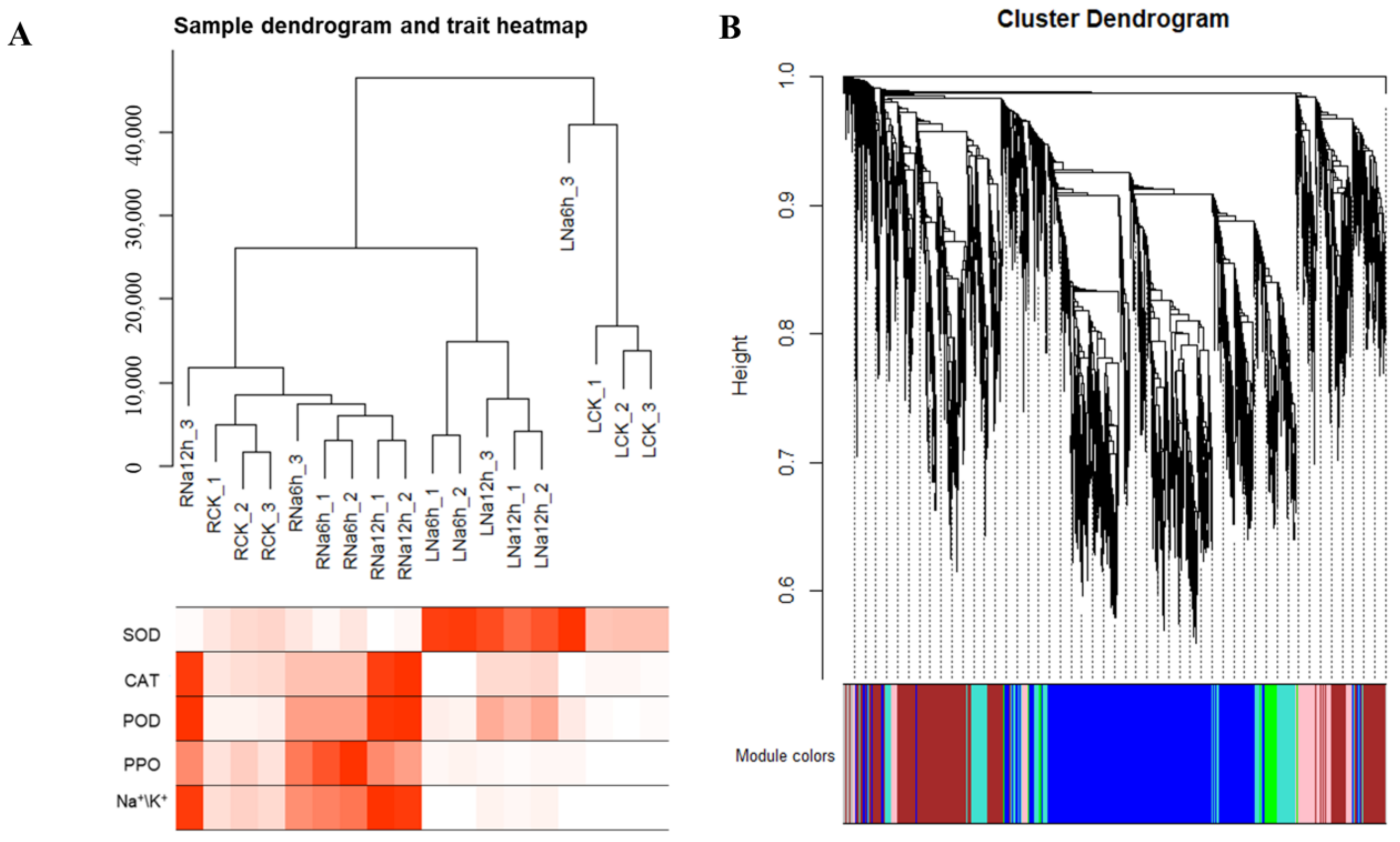
3.11. WGCNA
3.12. RT-qPCR Validation of Candidate Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jesus, J.; Castro, F.; Niemelä, A.; Borges, M.T.; Danko, A.S. Evaluation of the Impact of Different Soil Salinization Processes on Organic and Mineral Soils. Water Air Soil Pollut. 2015, 226, 12. [Google Scholar] [CrossRef]
- Nachshon, U. Cropland Soil Salinization and Associated Hydrology: Trends, Processes and Examples. Water 2018, 10, 1030. [Google Scholar] [CrossRef]
- Li, J.G.; Pu, L.J.; Han, M.F.; Zhu, M.; Zhang, R.S.; Xiang, Y.Z. Soil salinization research in China: Advances and prospects. J. Geogr. Sci. 2014, 24, 943–960. [Google Scholar] [CrossRef]
- Wang, N.; Zheng, Y.; Duan, N.; Zhang, Z.; Ji, X.; Jiang, S.; Sun, S.; Yang, L.; Bai, Y.; Fei, Z. Comparative transcriptomes analysis of red-and white-fleshed apples in an F1 population of Malus sieversii f. niedzwetzkyana crossed with M. domestica ‘Fuji’. PLoS ONE 2015, 10, e0133468. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Bai, Y.; Sun, H.; Wang, N.; Ma, Y.; Li, M.; Wang, X.; Jiao, C.; Legall, N.; Mao, L.; et al. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat. Commun. 2017, 8, 249. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.Y.; Zhao, X.Y.; Zhao, Q.; Wang, G.L.; Li, Y.Y.; Hao, Y.J. MdDREB2A in apple is involved in the regulation of multiple abiotic stress responses. Hortic. Plant J. 2021, 7, 197–208. [Google Scholar] [CrossRef]
- Mei, C.; Yang, J.; Mei, Q.L.; Jia, D.F.; Yan, P.; Feng, B.B.; Mamat, A.; Gong, X.Q.; Guan, Q.M.; Mao, K.; et al. MdNAC104 positively regulates apple cold tolerance via CBF-dependent and CBF-independent pathways. Plant Biotechnol. J. 2023, 21, 2057–2073. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.R.; Yang, Y.Y.; Zhao, Q.; Zhang, T.E.; Wang, C.K.; Hao, Y.J.; You, C.X. MdCIB1, an apple bHLH transcription factor, plays a positive regulator in response to drought stress. Environ. Exp. Bot. 2021, 188, 11. [Google Scholar] [CrossRef]
- Li, J.; Yang, Y.Q. How do plants maintain pH and ion homeostasis under saline-alkali stress? Front. Plant Sci. 2023, 14, 1217193. [Google Scholar] [CrossRef]
- Zhang, J.-T.; Mu, C.-S. Effects of saline and alkaline stresses on the germination, growth, photosynthesis, ionic balance and anti-oxidant system in an alkali-tolerant leguminous forage Lathyrus quinquenervius. Soil Sci. Plant Nutr. 2009, 55, 685–697. [Google Scholar] [CrossRef]
- Guo, R.; Shi, L.X.; Yang, Y.F. Germination, growth, osmotic adjustment and ionic balance of wheat in response to saline and alkaline stresses. Soil Sci. Plant Nutr. 2009, 55, 667–679. [Google Scholar] [CrossRef]
- Ondrasek, G.; Rathod, S.; Manohara, K.K.; Gireesh, C.; Anantha, M.S.; Sakhare, A.S.; Parmar, B.; Yadav, B.K.; Bandumula, N.; Raihan, F.; et al. Salt Stress in Plants and Mitigation Approaches. Plants 2022, 11, 717. [Google Scholar] [CrossRef] [PubMed]
- Janku, M.; Luhová, L.; Petrivalsky, M. On the Origin and Fate of Reactive Oxygen Species in Plant Cell Compartments. Antioxidants 2019, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lu, J.H.; Zhang, J.D.; Liu, D.K.; Wang, Y.; Niu, Q.D.; Huang, D.D. Transcriptome revealed the molecular mechanism of Glycyrrhiza inflata root to maintain growth and development, absorb and distribute ions under salt stress. BMC Plant Biol. 2021, 21, 599. [Google Scholar] [CrossRef] [PubMed]
- Chrysargyris, A.; Papakyriakou, E.; Petropoulos, S.A.; Tzortzakis, N. The combined and single effect of salinity and copper stress on growth and quality of Mentha spicata plants. J. Hazard. Mater. 2019, 368, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Ganapati, R.K.; Naveed, S.A.; Zafar, S.; Wang, W.; Xu, J. Saline-Alkali Tolerance in Rice: Physiological Response, Molecular Mechanism, and QTL Identification and Application to Breeding. Rice Sci. 2022, 29, 412–434. [Google Scholar] [CrossRef]
- Martínez-Cuenca, M.-R.; Legaz, F.; Forner-Giner, M.Á.; Primo-Millo, E.; Iglesias, D.J. Bicarbonate blocks iron translocation from cotyledons inducing iron stress responses in Citrus roots. J. Plant Physiol. 2013, 170, 899–905. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) Pathway: Established and Emerging Roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Lin, H.; Yang, Y.; Quan, R.; Mendoza, I.; Wu, Y.; Du, W.; Zhao, S.; Schumaker, K.S.; Pardo, J.M.; Guo, Y. Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 Protein Kinase Stabilizes Their Protein Complex and Regulates Salt Tolerance in Arabidopsis. Plant Cell 2009, 21, 1607–1619. [Google Scholar] [CrossRef]
- Qiu, Q.-S.; Guo, Y.; Dietrich, M.A.; Schumaker, K.S.; Zhu, J.-K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 2002, 99, 8436–8441. [Google Scholar] [CrossRef]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.T.; Chen, Q.; Xiang, N.; Liu, Y.Y.; Kong, X.X.; Yang, Y.P.; Hu, X.Y. SIP1, a novel SOS2 interaction protein, is involved in salt-stress tolerance in Arabidopsis. Plant Physiol. Biochem. 2018, 124, 167–174. [Google Scholar] [CrossRef]
- Wu, H.; Shabala, L.; Zhou, M.; Su, N.; Wu, Q.; Ul-Haq, T.; Zhu, J.; Mancuso, S.; Azzarello, E.; Shabala, S. Root vacuolar Na+ sequestration but not exclusion from uptake correlates with barley salt tolerance. Plant J. 2019, 100, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Hong, P.; Luo, L.; Xia, T. Overexpression of AtNHX1, a Vacuolar Na+/H+ Antiporter from Arabidopsis thalina, in Petunia hybrida Enhances Salt and Drought Tolerance. J. Plant Biol. 2009, 52, 453–461. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Zhang, H.M.; Liu, Z.H.; Li, H.C.; Guo, X.L.; Li, G.A. The wheat NHX antiporter gene TaNHX2 confers salt tolerance in transgenic alfalfa by increasing the retention capacity of intracellular potassium. Plant Mol. Biol. 2015, 87, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef]
- Farooq, M.A.; Niazi, A.K.; Akhtar, J.; Saifullah; Farooq, M.; Souri, Z.; Karimi, N.; Rengel, Z. Acquiring control: The evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol. Biochem. 2019, 141, 353–369. [Google Scholar] [CrossRef]
- Pan, Y.; Wu, L.J.; Yu, Z.L. Effect of salt and drought stress on antioxidant enzymes activities and SOD isoenzymes of liquorice (Glycyrrhiza uralensis Fisch). Plant Growth Regul. 2006, 49, 157–165. [Google Scholar] [CrossRef]
- Fini, A.; Guidi, L.; Ferrini, F.; Brunetti, C.; Di Ferdinando, M.; Biricolti, S.; Pollastri, S.; Calamai, L.; Tattini, M. Drought stress has contrasting effects on antioxidant enzymes activity and phenylpropanoid biosynthesis in Fraxinus ornus leaves: An excess light stress affair? J. Plant Physiol. 2012, 169, 929–939. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Dong, Q.L.; Liu, D.D.; An, X.H.; Hu, D.G.; Yao, Y.X.; Hao, Y.J. MdVHP1 encodes an apple vacuolar H(+)-PPase and enhances stress tolerance in transgenic apple callus and tomato. J. Plant Physiol. 2011, 168, 2124–2133. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.B.; Song, H.F.; Zhang, L.Y. New Insight into Plant Saline-Alkali Tolerance Mechanisms and Application to Breeding. Int. J. Mol. Sci. 2022, 23, 16048. [Google Scholar] [CrossRef] [PubMed]
- Forni, C.; Duca, D.; Glick, B.R. Mechanisms of plant response to salt and drought stress and their alteration by rhizobacteria. Plant Soil 2017, 410, 335–356. [Google Scholar] [CrossRef]
- Yu, Z.P.; Duan, X.B.; Luo, L.; Dai, S.J.; Ding, Z.J.; Xia, G.M. How Plant Hormones Mediate Salt Stress Responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Yang, J.; Li, W.; Guo, X.; Chen, P.; Cheng, Y.; Mao, K.; Ma, F. Cation/Ca2+ Exchanger 1 (MdCCX1), a Plasma Membrane-Localized Na+ Transporter, Enhances Plant Salt Tolerance by Inhibiting Excessive Accumulation of Na+ and Reactive Oxygen Species. Front. Plant Sci. 2021, 12, 746189. [Google Scholar] [CrossRef]
- Yin, Y.-C.; Zhang, X.-D.; Gao, Z.-Q.; Hu, T.; Liu, Y. The Research Progress of Chalcone Isomerase (CHI) in Plants. Mol. Biotechnol. 2019, 61, 32–52. [Google Scholar] [CrossRef]
- Jiang, W.; Yin, Q.; Wu, R.; Zheng, G.; Liu, J.; Dixon, R.A.; Pang, Y. Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 7165–7179. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.H.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef]
- Zhang, M.J.; Liu, Y.L.; Han, G.L.; Zhang, Y.; Wang, B.S.; Chen, M. Salt tolerance mechanisms in trees: Research progress. Trees 2021, 35, 717–730. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, X.L. Recent advances in polyphenol oxidase-mediated plant stress responses. Phytochemistry 2021, 181, 112588. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Meng, S.; Xiang, Z.; Yang, G.; He, N. An R1R2R3 MYB Transcription Factor, MnMYB3R1, Regulates the Polyphenol Oxidase Gene in Mulberry (Morus notabilis). Int. J. Mol. Sci. 2019, 20, 2602. [Google Scholar] [CrossRef] [PubMed]
- Plett, D.C.; Moller, I.S. Na plus transport in glycophytic plants: What we know and would like to know. Plant Cell Environ. 2010, 33, 612–626. [Google Scholar] [CrossRef]
- Munns, R.; Gilliham, M. Salinity tolerance of crops-what is the cost? New Phytol. 2015, 208, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Wakeel, A.; Farooq, M.; Qadir, M.; Schubert, S. Potassium Substitution by Sodium in Plants. Crit. Rev. Plant Sci. 2011, 30, 401–413. [Google Scholar] [CrossRef]
- Manaa, A.; Faurobert, M.; Valot, B.; Bouchet, J.-P.; Grasselly, D.; Causse, M.; Ahmed, H.B. Effect of Salinity and Calcium on Tomato Fruit Proteome. OMICS J. Integr. Biol. 2013, 17, 338–352. [Google Scholar] [CrossRef]
- Sun, M.-H.; Ma, Q.-J.; Liu, X.; Zhu, X.-P.; Hu, D.-G.; Hao, Y.-J. Molecular cloning and functional characterization of MdNHX1 reveals its involvement in salt tolerance in apple calli and Arabidopsis. Sci. Hortic. 2017, 215, 126–133. [Google Scholar] [CrossRef]
- Sun, Z.; Li, J.; Guo, D.; Wang, T.; Tian, Y.; Ma, C.; Liu, X.; Wang, C.; Zheng, X. Melatonin enhances KCl salinity tolerance by maintaining K+ homeostasis in Malus hupehensis. Plant Biotechnol. J. 2023, 21, 2273–2290. [Google Scholar] [CrossRef] [PubMed]
- Han, D.G.; Hou, Y.J.; Wang, Y.F.; Ni, B.X.; Li, Z.T.; Yang, G.H. Overexpression of a Malus baccata WRKY transcription factor gene (MbWRKY5) increases drought and salt tolerance in transgenic tobacco. Can. J. Plant Sci. 2019, 99, 173–183. [Google Scholar] [CrossRef]
- Dong, Q.; Zheng, W.; Duan, D.; Huang, D.; Wang, Q.; Liu, C.; Li, C.; Gong, X.; Li, C.; Mao, K.; et al. MdWRKY30, a group IIa WRKY gene from apple, confers tolerance to salinity and osmotic stresses in transgenic apple callus and Arabidopsis seedlings. Plant Sci. 2020, 299, 110611. [Google Scholar] [CrossRef]
- Duan, D.; Yi, R.; Ma, Y.; Dong, Q.; Mao, K.; Ma, F. Apple WRKY transcription factor MdWRKY56 positively modulates drought stress tolerance. Environ. Exp. Bot. 2023, 212, 105400. [Google Scholar] [CrossRef]





| GO ID | GO Name | Category | LCK VS LNa6h DEGs Number | LCK VS LNa12h DEGs Number |
|---|---|---|---|---|
| GO:0016757 | transferase activity, transferring glycosyl groups | MF | 34 | 51 |
| GO:0016758 | transferase activity, transferring hexosyl groups | MF | 30 | 46 |
| GO:0020037 | heme binding | MF | 31 | 40 |
| GO:0046906 | tetrapyrrole binding | MF | 31 | 40 |
| GO:0005506 | iron ion binding | MF | 33 | 34 |
| GO:0016705 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | MF | 28 | 30 |
| GO:0016798 | hydrolase activity, acting on glycosyl bonds | MF | 15 | 20 |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF | 13 | 20 |
| GO:0000155 | phosphorelay sensor kinase activity | MF | 1 | 2 |
| GO:0004673 | protein histidine kinase activity | MF | 1 | 2 |
| GO:0016775 | phosphotransferase activity, nitrogenous group as acceptor | MF | 1 | 2 |
| GO ID | GO Name | Category | RCK VS RNa6h DEGs Number | RCK VS RNa12h DEGs Number |
|---|---|---|---|---|
| GO:0016758 | coenzyme binding | MF | 100 | 156 |
| GO:0015267 | tetrapyrrole binding | MF | 40 | 188 |
| GO:0046527 | heme binding | MF | 28 | 188 |
| GO:0003777 | hydrolase activity, acting on glycosyl bonds | MF | 43 | 161 |
| GO:0015631 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF | 47 | 151 |
| GO:0008017 | iron ion binding | MF | 47 | 148 |
| GO:0060089 | transferase activity, transferring hexosyl groups | MF | 21 | 172 |
| GO:0005507 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | MF | 27 | 158 |
| GO:0004553 | passive transmembrane transporter activity | MF | 91 | 61 |
| GO:0050662 | copper ion binding | MF | 101 | 41 |
| GO:0071944 | cell periphery | CC | 25 | 39 |
| GO:0005576 | extracellular region | CC | 20 | 25 |
| GO:0005618 | cell wall | CC | 16 | 20 |
| GO:0030312 | external encapsulating structure | CC | 16 | 20 |
| GO:0048046 | apoplast | CC | 16 | 20 |
| GO:0006073 | cellular carbohydrate metabolic process | BP | 26 | 64 |
| GO:0006928 | defense response | BP | 43 | 41 |
| GO:0006952 | reproduction | BP | 32 | 49 |
| GO:0051704 | reproductive process | BP | 32 | 49 |
| GO:0006270 | microtubule-based process | BP | 11 | 68 |
| GO:0044262 | cellular polysaccharide metabolic process | BP | 39 | 40 |
| GO:0008037 | pollination | BP | 30 | 47 |
| GO:0009856 | pollen-pistil interaction | BP | 30 | 47 |
| GO:0009875 | multi-multicellular organism process | BP | 30 | 47 |
| GO:0044706 | recognition of pollen | BP | 30 | 47 |
| GO:0044042 | multi-organism process | BP | 26 | 50 |
| GO:0044264 | cell recognition | BP | 26 | 47 |
| KEGGID | KEGG Pathway | LCK vs. LNa6h DEGs Number | LCK vs. LNa12h DEGs Number |
|---|---|---|---|
| mdm00480 | Glutathione metabolism | 30 | 34 |
| mdm00500 | Starch and sucrose metabolism | 33 | 50 |
| mdm00630 | Glyoxylate and dicarboxylate metabolism | 21 | 28 |
| mdm00906 | Carotenoid biosynthesis | 13 | 18 |
| mdm04712 | Circadian rhythm—plant | 21 | 30 |
| mdm01200 | Carbon metabolism | 56 | 92 |
| KEGGID | KEGG Pathway | RCK vs. RNa6h DEGs Number | RCK vs. RNa12h DEGs Number |
|---|---|---|---|
| mdm00040 | Pentose and glucuronate interconversions | 27 | 43 |
| mdm00270 | Cysteine and methionine metabolism | 41 | 65 |
| mdm00350 | Tyrosine metabolism | 18 | 29 |
| mdm00430 | Taurine and hypotaurine metabolism | 8 | 11 |
| mdm00460 | Cyanoamino acid metabolism | 23 | 29 |
| mdm00480 | Glutathione metabolism | 41 | 63 |
| mdm00592 | alpha-Linolenic acid metabolism | 22 | 38 |
| mdm00910 | Nitrogen metabolism | 16 | 28 |
| mdm00940 | Phenylpropanoid biosynthesis | 63 | 103 |
| mdm00941 | Flavonoid biosynthesis | 30 | 49 |
| mdm00950 | Isoquinoline alkaloid biosynthesis | 10 | 15 |
| mdm00960 | Tropane, piperidine, and pyridine alkaloid biosynthesis | 15 | 21 |
| mdm02010 | ABC transporters | 30 | 38 |
| mdm03030 | DNA replication | 19 | 34 |
| mdm04075 | Plant hormone signal transduction | 75 | 122 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Yang, Y.; Zhou, Z.; Chen, X. Physiological Studies and Transcriptomic Analysis Reveal the Mechanism of Saline-Alkali Stress Resistance of Malus sieversii f. niedzwetzkyan. Horticulturae 2024, 10, 510. https://doi.org/10.3390/horticulturae10050510
Jiang L, Yang Y, Zhou Z, Chen X. Physiological Studies and Transcriptomic Analysis Reveal the Mechanism of Saline-Alkali Stress Resistance of Malus sieversii f. niedzwetzkyan. Horticulturae. 2024; 10(5):510. https://doi.org/10.3390/horticulturae10050510
Chicago/Turabian StyleJiang, Lepu, Yan Yang, Zhengli Zhou, and Xuesen Chen. 2024. "Physiological Studies and Transcriptomic Analysis Reveal the Mechanism of Saline-Alkali Stress Resistance of Malus sieversii f. niedzwetzkyan" Horticulturae 10, no. 5: 510. https://doi.org/10.3390/horticulturae10050510




