Denaturing Gradient Gel Electrophoresis Approach for Microbial Shift Analysis in Thermophilic and Mesophilic Anaerobic Digestions
Abstract
:1. Introduction

2. Results and Discussion
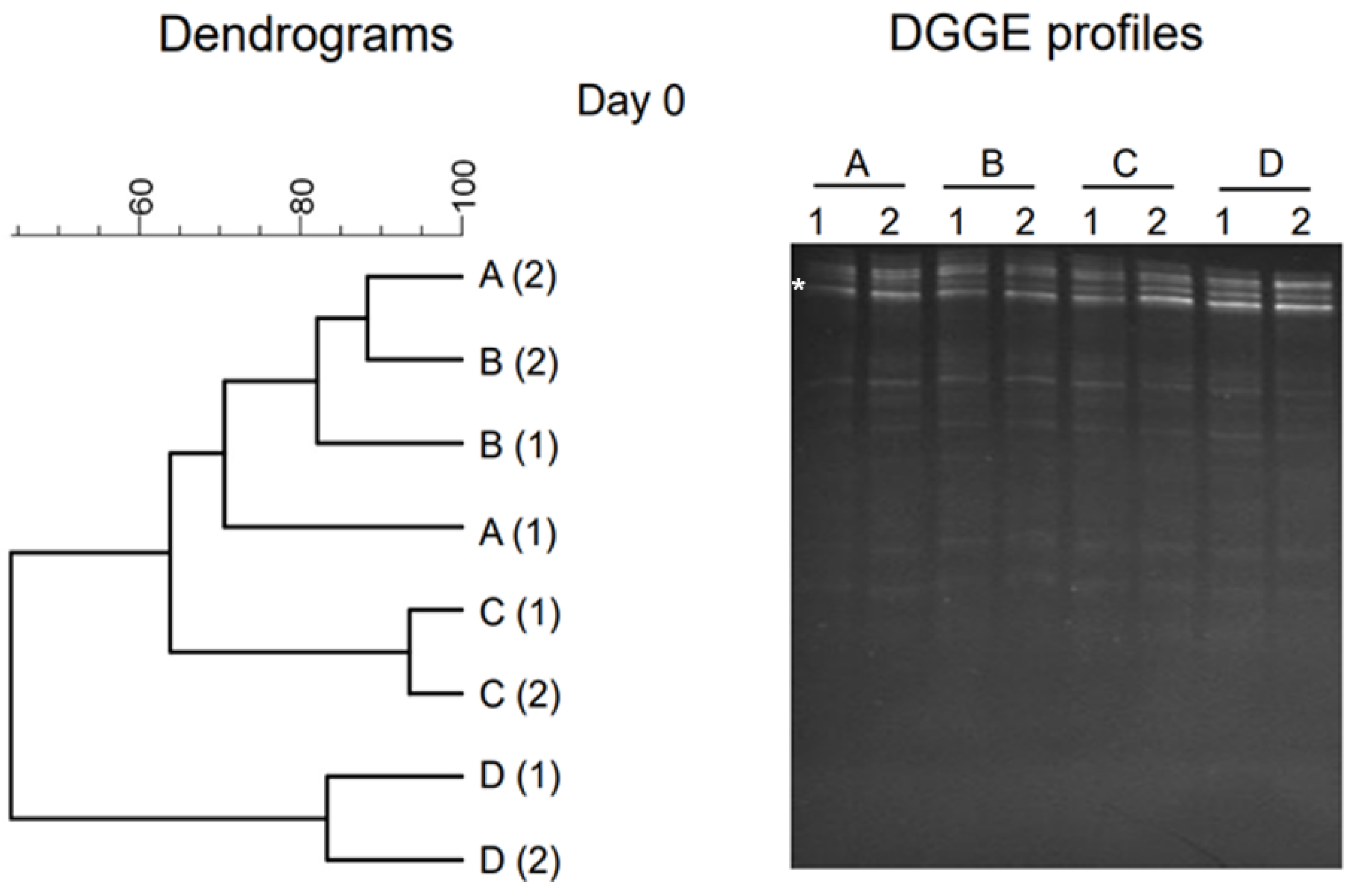
2.1. PCR-DGGE Analysis of Initial Samples
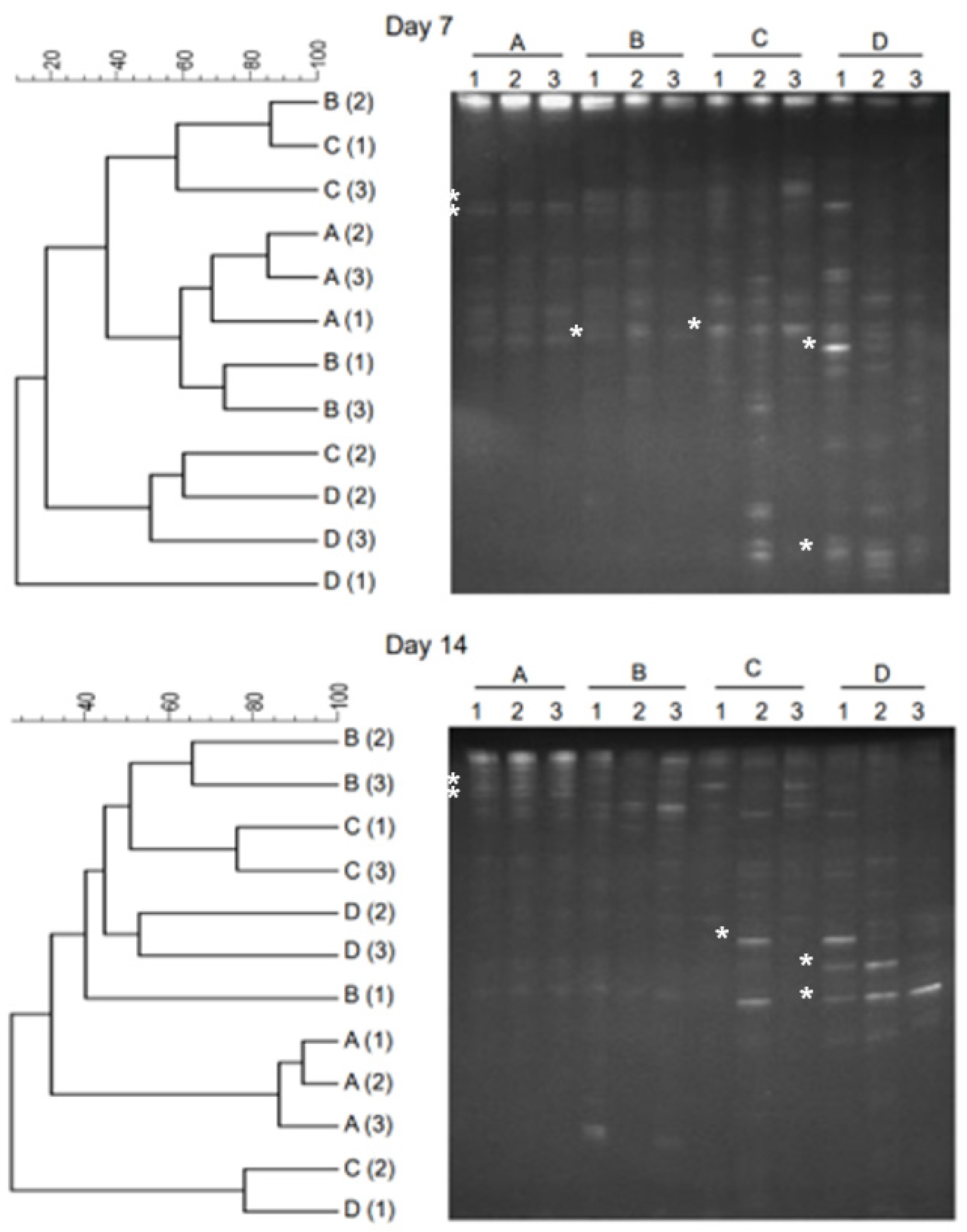
2.2. Microbial Dynamics in Intermediate Stages of Anaerobic Processes
2.3. Microbial Dynamics in Late Stages of Anaerobic Processes
2.4. DGGE Profiles of Microbial Community Dynamics over Various Incubation Periods
2.5. Comparison of Bands, Sequence Sizes, Closest Relatives, and Alignment Similarities
| Incubation Time | |||||
|---|---|---|---|---|---|
| Day 0 | Day 7 | Day 14 | Day 30 | Day 60 | |
| Temperature | Number of extracted bands # | ||||
| 28 °C | 1 | 2 | 2 | 0 | 3 |
| 36 °C | 1 | 1 | 0 | 1 | 1 |
| 44 °C | 1 | 1 | 1 | 0 | 1 |
| 52 °C | 1 | 2 | 2 | 4 | 4 |
| Temperature | Sequence size (bp) | ||||
| 28 °C | 195 | 195 | 169, 172 | - | 188, 194, 195 |
| 36 °C | 195 | 195 | - | 194 | 194 |
| 44 °C | 195 | 170 | 171 | - | 194 |
| 52 °C | 195 | 169, 194 | 170 | 172 | 170, 172 |
| Temperature | Accession number | ||||
| 28 °C | MK872367 | MK872368 | MK872373, MK872374 | - | MK872383, MK872384, MK872385 |
| 36 °C | MK872367 | MK872369 | - | MK872378 | MK872386 |
| 44 °C | MK872367 | MK872370 | MK872375 | - | MK872387 |
| 52 °C | MK872367 | MK872371, MK872372 | MK872376, MK872377 | MK872379, MK872380, MK872381, MK872382 | MK872388, MK872389, MK872390, MK872391 |
| Temperature | Closest relatives (alignment similarity %) | ||||
| 28 °C | Acinetobacter beijerinckii (97%), Acinetobacter bouvetii (97%), Acinetobacter venetianus (97%) | Acinetobacter beijerinckii (97–98%), Acinetobacter bouvetii (97–98%), Acinetobacter venetianus (97–98%) | Clostridium sporosphaeroides, Clostridium jeddahense, Phocea massiliensis, Intestinimonas butyriciproducens, Ercella succinigenes, Papillibacter cinnamivorans | Galbibacter mesophilus (87%), Zeaxanthinibacter enoshimensis (87%), Sphaerochaeta pleomorpha (91%), Sphaerochaeta associate (90%), Sphaerochaeta globose (90%), Syntrophomonas sapovorans (94%), Syntrophomonas curvata (93%), Thermosyntropha tengcongensis (93%) | |
| 36 °C | Acinetobacter (97%) | Syntrophomonas zehnderi (92%), Syntrophomonas sapovorans (91%), Syntrophomonas palmitatica (91%) | Sphaerochaeta pleomorpha (91%), Sphaerochaeta associata (90%), Sphaerochaeta globosa (90%), | ||
| 44 °C | Acinetobacter (97%) | Coprothermobacter proteolyticus (100%), Coprothermobacter platensis (95%) | Thermoclostridium caenicola (94%), Intestinimonas butyriciproducens (94%), Hungateiclostridium thermocellum (94%) | Dielma fastidiosa (86%), Acholeplasma parvum (86%), Acholeplasma vituli (86%) | |
| 52 °C | Acinetobacter (97%) | Tepidimicrobium ferriphilum (100%), Tepidimicrobium xylanilyticum (100%), Symbiobacterium turbinis (93%), Symbiobacterium terraclitae (92%), Caldibacillus debilis (89%) | Coprothermobacter proteolyticus (100%), Coprothermobacter platensis (99%), Hungateiclostridium thermocellum (100%) | Caldicoprobacter faecalis (97%), Caldicoprobacter oshimai (97%), Caldicoprobacter guelmensis (96%), Caldicoprobacter oshimai (96%), Caldicoprobacter guelmensis (96%), Hungateiclostridium straminisolvens (98%) | Coprothermobacter proteolyticus (99%), Coprothermobacter platensis (95%), Caldicoprobacter faecalis (97%), Caldicoprobacter oshimai (97%), Caldicoprobacter guelmensis (97%), Caldicoprobacter faecalis (97%) |
3. Conclusions
4. Material and Methods
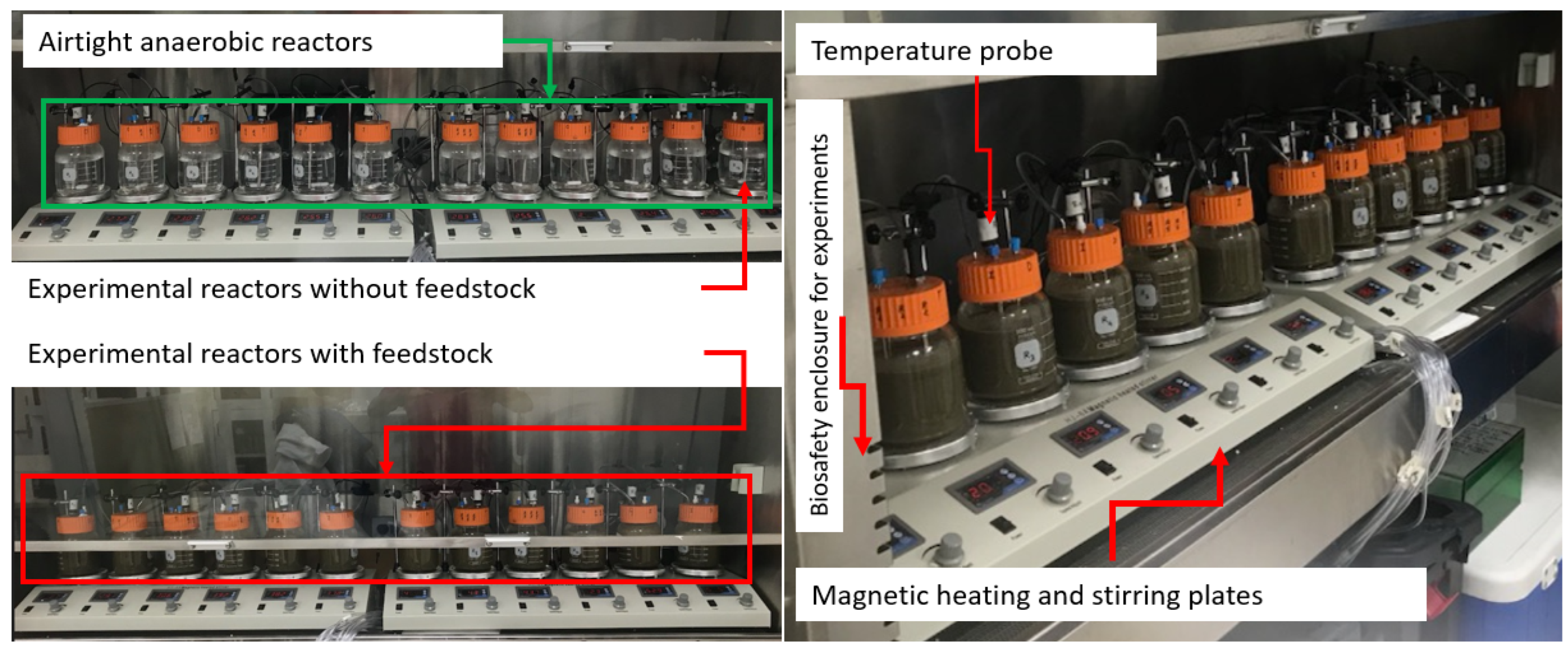
4.1. Anaerobic Experiment and Sample Collection
4.2. PCR Amplification of 16S rRNA V3 Region and DGGE
4.3. Statistical Analysis of DGGE Images
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Forney, L.J.; Zhou, X.; Brown, C.J. Molecular microbial ecology: Land of the one-eyed king. Curr. Opin. Microbiol. 2004, 7, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Strathdee, F.; Free, A. Denaturing gradient gel electrophoresis (DGGE). Methods Mol. Biol. 2013, 1054, 145–157. [Google Scholar]
- Fernández-Gómez, M.J.; Nogales, R.; Insam, H.; Romero, E.; Goberna, M. Use of DGGE and COMPOCHIP for investigating bacterial communities of various vermicomposts produced from different wastes under dissimilar conditions. Sci. Total Environ. 2012, 414, 664–671. [Google Scholar] [CrossRef]
- Festa, M.; Abbruscato, P.; Manachini, B. Plant-Nematode Interactions; Springer: Berlin/Heidelberg, Germany, 2024; pp. 247–255. [Google Scholar]
- Muyzer, G.; Smalla, K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 1998, 73, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Giangeri, G.; Tsapekos, P.; Gaspari, M.; Ghofrani-Isfahani, P.; Treu, L.; Kougias, P.; Campanaro, S.; Angelidaki, I. A bioaugmentation strategy to recover methane production under sulfate-stressed conditions: Highlights on targeted sulfate-reducing bacteria and DIET-related species. Appl. Energy 2024, 362, 122940. [Google Scholar] [CrossRef]
- Luo, Y.; Shen, J.; Wang, X.; Xiao, H.; Yaser, A.Z.; Fu, J. Recent advances in research on microbial community in the composting process. Biomass Convers. Biorefinery 2023, 1–15. [Google Scholar] [CrossRef]
- Üstüntürk-Onan, M.; Tüccar, T.; Ilhan-Sungur, E. Occurrence of sulfate-reducing bacteria in well water: Identification of anaerobic sulfidogenic bacterial enrichment cultures. J. Water Health 2024, 22, 746–756. [Google Scholar] [CrossRef]
- García-Ruíz, M.J.; Castellano-Hinojosa, A.; Armato, C.; González-Martínez, A.; González-López, J.; Osorio, F. Biogas production and microbial community structure in a stable-stage of a two-stage anaerobic digester. AIChE J. 2020, 66, e16807. [Google Scholar] [CrossRef]
- Hu, J.; Zhou, D.; Zhong, R.; Zhao, C.; Li, Q. Response of microbial community diversity and subsoil properties to cow manure amendment in mollisols. Arch. Agron. Soil Sci. 2023, 69, 785–802. [Google Scholar] [CrossRef]
- Wang, Y.; Pandey, P.K.; Kuppu, S.; Pereira, R.; Aly, S.; Zhang, R. Degradation of antibiotic resistance genes and mobile gene elements in dairy manure anerobic digestion. PLoS ONE 2021, 16, e0254836. [Google Scholar] [CrossRef]
- Ozbayram, E.G.; Ince, O.; Ince, B.; Harms, H.; Kleinsteuber, S. Comparison of Rumen and Manure Microbiomes and Implications for the Inoculation of Anaerobic Digesters. Microorganisms 2018, 6, 15. [Google Scholar] [CrossRef]
- Zhi, W.; Ge, Z.; He, Z.; Zhang, H. Methods for understanding microbial community structures and functions in microbial fuel cells: A review. Bioresour. Technol. 2014, 171, 461–468. [Google Scholar] [CrossRef]
- Bio-Rad. D GENE Denaturing Gel Electrophoresis System, Instruction Manual and Application Guide. Code System for DGGE (Catalog Numbers 170-9000). 2024. BIO-RAD Life science, Hercules, California 94547, USA. Available online: https://www.bio-rad.com/webroot/web/pdf/lsr/literature/4000039b.pdf (accessed on 14 May 2024).
- Elsharkawy, M.M.; Kuno, S.; Hyakumachi, M.; Mostafa, Y.S.; Alamri, S.A.; Alrumman, S.A. PCR-DGGE Analysis Proves the Suppression of Rhizoctonia and Sclerotium Root Rot Due to Successive Inoculations. J. Fungi 2022, 8, 133. [Google Scholar] [CrossRef]
- Engelen, B.; Nguyen, T.; Heyerhoff, B.; Kalenborn, S.; Sydow, K.; Tabai, H.; Peterson, R.N.; Wegener, G.; Teske, A. Microbial Communities of Hydrothermal Guaymas Basin Surficial Sediment Profiled at 2 Millimeter-Scale Resolution. Front. Microbiol. 2021, 12, 710881. [Google Scholar] [CrossRef] [PubMed]
- Fontana, C.; Vignolo, G.; Cocconcelli, P.S. PCR–DGGE analysis for the identification of microbial populations from Argentinean dry fermented sausages. J. Microbiol. Methods 2005, 63, 254–263. [Google Scholar] [CrossRef]
- Gee, H.Y.; Kim, C.K.; Kim, S.W.; Lee, J.H.; Kim, J.-H.; Kim, K.H.; Lee, M.G. The L441P mutation of cystic fibrosis transmembrane conductance regulator and its molecular pathogenic mechanisms in a Korean patient with cystic fibrosis. J. Korean Med. Sci. 2010, 25, 166. [Google Scholar] [CrossRef] [PubMed]
- Green, S.J.; Leigh, M.B.; Neufeld, J.D. Denaturing gradient gel electrophoresis (DGGE) for microbial community analysis. In Hydrocarbon and Lipid Microbiology Protocols: Microbial Quantitation, Community Profiling and Array Approaches; Springer: Berlin/Heidelberg, Germany, 2017; pp. 77–99. [Google Scholar]
- Larsson, M.E.; Bramucci, A.R.; Collins, S.; Hallegraeff, G.; Kahlke, T.; Raina, J.B.; Seymour, J.R.; Doblin, M.A. Mucospheres produced by a mixotrophic protist impact ocean carbon cycling. Nat. Commun. 2022, 13, 1301. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, H.; Muyzer, G. Denaturing gradient gel electrophoresis in marine microbial ecology. Methods Microbiol. 2001, 30, 425–468. [Google Scholar]
- Leung, K.; Topp, E. Bacterial community dynamics in liquid swine manure during storage: Molecular analysis using DGGE/PCR of 16S rDNA. FEMS Microbiol. Ecol. 2001, 38, 169–177. [Google Scholar] [CrossRef]
- Rasolofo, E.A.; LaPointe, G.; Roy, D. Assessment of the bacterial diversity of treated and untreated milk during cold storage by T-RFLP and PCR-DGGE methods. Dairy Sci. Technol. 2011, 91, 573–597. [Google Scholar] [CrossRef]
- Wittebolle, L.; Boon, N.; Vanparys, B.; Heylen, K.; De Vos, P.; Verstraete, W. Failure of the ammonia oxidation process in two pharmaceutical wastewater treatment plants is linked to shifts in the bacterial communities. J. Appl. Microbiol. 2005, 99, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Marzorati, M.; Wittebolle, L.; Boon, N.; Daffonchio, D.; Verstraete, W. How to get more out of molecular fingerprints: Practical tools for microbial ecology. Environ. Microbiol. 2008, 10, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Bier, R.L.; Bernhardt, E.S.; Boot, C.M.; Graham, E.B.; Hall, E.K.; Lennon, J.T.; Nemergut, D.R.; Osborne, B.B.; Ruiz-Gonzalez, C.; Schimel, J.P. Linking microbial community structure and microbial processes: An empirical and conceptual overview. FEMS Microbiol. Ecol. 2015, 91, fiv113. [Google Scholar] [CrossRef]
- Duan, N.; Kougias, P.G.; Campanaro, S.; Treu, L.; Angelidaki, I. Evolution of the microbial community structure in biogas reactors inoculated with seeds from different origin. Sci. Total Environ. 2021, 773, 144981. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.A. Microbial community structure and its functional implications. Nature 2009, 459, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.M.; Park, S.K.; Ha, J.H.; Park, J.M. Microbial community structure in a thermophilic aerobic digester used as a sludge pretreatment process for the mesophilic anaerobic digestion and the enhancement of methane production. Bioresour. Technol. 2013, 145, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Ju, F.; Lau, F.; Zhang, T. Linking microbial community, environmental variables, and methanogenesis in anaerobic biogas digesters of chemically enhanced primary treatment sludge. Environ. Sci. Technol. 2017, 51, 3982–3992. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Kang, H.-J.; Lee, Y.H.; Lee, T.J.; Han, K.; Choi, Y.; Park, H.-D. Monitoring bacterial community structure and variability in time scale in full-scale anaerobic digesters. J. Environ. Monit. 2012, 14, 1893–1905. [Google Scholar] [CrossRef]
- Rastogi, G.; Sani, R.K. Microbes and Microbial Technology: Agricultural and Environmental Applications; Springer: Berlin/Heidelberg, Germany, 2011; pp. 29–57. [Google Scholar]
- Schimel, J.P.; Gulledge, J. Microbial community structure and global trace gases. Glob. Change Biol. 1998, 4, 745–758. [Google Scholar] [CrossRef]
- Sun, H.; Deng, S.; Raun, W. Bacterial community structure and diversity in a century-old manure-treated agroecosystem. Appl. Environ. Microbiol. 2004, 70, 5868–5874. [Google Scholar] [CrossRef]
- Wang, X.; Feng, J.; Ao, G.; Qin, W.; Han, M.; Shen, Y.; Liu, M.; Chen, Y.; Zhu, B. Globally nitrogen addition alters soil microbial community structure, but has minor effects on soil microbial diversity and richness. Soil Biol. Biochem. 2023, 179, 108982. [Google Scholar] [CrossRef]
- Carballa, M.; Smits, M.; Etchebehere, C.; Boon, N.; Verstraete, W. Correlations between molecular and operational parameters in continuous lab-scale anaerobic reactors. Appl. Microbiol. Biotechnol. 2011, 89, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Dongsu, B.; Tao, Z.; Xintong, J.; Ming, C.; Siqi, W.; Zheng, S.; Yalei, Z. Anaerobic co-digestion of three commercial bio-plastic bags with food waste: Effects on methane production and microbial community structure. Sci. Total Environ. 2023, 859, 159967. [Google Scholar] [CrossRef]
- Regueiro, L.; Veiga, P.; Figueroa, M.; Alonso-Gutierrez, J.; Stams, A.J.; Lema, J.M.; Carballa, M. Relationship between microbial activity and microbial community structure in six full-scale anaerobic digesters. Microbiol. Res. 2012, 167, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Pagliano, G.; Ventorino, V.; Panico, A.; Romano, I.; Robertiello, A.; Pirozzi, F.; Pepe, O. The effect of bacterial and archaeal populations on anaerobic process fed with mozzarella cheese whey and buttermilk. J. Environ. Manag. 2018, 217, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Toumi, J.; Miladi, B.; Farhat, A.; Nouira, S.; Hamdi, M.; Gtari, M.; Bouallagui, H. Microbial ecology overview during anaerobic codigestion of dairy wastewater and cattle manure and use in agriculture of obtained bio-fertilisers. Bioresour. Technol. 2015, 198, 141–149. [Google Scholar] [CrossRef]
- Yong, X.; Cui, Y.; Chen, L.; Ran, W.; Shen, Q.; Yang, X. Dynamics of bacterial communities during solid-state fermentation using agro-industrial wastes to produce poly-γ-glutamic acid, revealed by real-time PCR and denaturing gradient gel electrophoresis (DGGE). Appl. Microbiol. Biotechnol. 2011, 92, 717–725. [Google Scholar] [CrossRef]
- Lahmamsi, H.; Ananou, S.; Lahlali, R.; Tahiri, A. Lactic acid bacteria as an eco-friendly approach in plant production: Current state and prospects. Folia Microbiol. 2024, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Win, T.T.; Bo, B.; Kim, H.M.; Lee, J.E.; Park, Y.E.; Song, K.G. Microbial population dynamics and meta-proteomic evaluation for functional insight into an anaerobic membrane bioreactor during startup. Int. Biodeterior. Biodegrad. 2023, 178, 105547. [Google Scholar] [CrossRef]
- Wittebolle, L.; Marzorati, M.; Clement, L.; Balloi, A.; Daffonchio, D.; Heylen, K.; De Vos, P.; Verstraete, W.; Boon, N. Initial community evenness favours functionality under selective stress. Nature 2009, 458, 623–626. [Google Scholar] [CrossRef]
- Xiuheng, W.; Zhang, K.; Nanqi, R.; Nan, L.; Lijiao, R. Monitoring microbial community structure and succession of an A/O SBR during start-up period using PCR-DGGE. J. Environ. Sci. 2009, 21, 223–228. [Google Scholar]
- Zhang, W.; Long, X.; Huo, X.; Chen, Y.; Lou, K. 16S rRNA-based P CR-DGGE analysis of actinomycete communities in fields with continuous cotton cropping in Xinjiang, China. Microb. Ecol. 2013, 66, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Liang, R.; Zhang, L.; Wu, C.; Zhou, R.; Liao, X. Characterization of microbial communities in strong aromatic liquor fermentation pit muds of different ages assessed by combined DGGE and PLFA analyses. Food Res. Int. 2013, 54, 660–666. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, P.; Chowdhury, D.; Wang, Y. Denaturing Gradient Gel Electrophoresis Approach for Microbial Shift Analysis in Thermophilic and Mesophilic Anaerobic Digestions. Gels 2024, 10, 339. https://doi.org/10.3390/gels10050339
Pandey P, Chowdhury D, Wang Y. Denaturing Gradient Gel Electrophoresis Approach for Microbial Shift Analysis in Thermophilic and Mesophilic Anaerobic Digestions. Gels. 2024; 10(5):339. https://doi.org/10.3390/gels10050339
Chicago/Turabian StylePandey, Pramod, Dhrubajyoti Chowdhury, and Yi Wang. 2024. "Denaturing Gradient Gel Electrophoresis Approach for Microbial Shift Analysis in Thermophilic and Mesophilic Anaerobic Digestions" Gels 10, no. 5: 339. https://doi.org/10.3390/gels10050339





