Clinical Use of Molecular Biomarkers in Canine and Feline Oncology: Current and Future
Abstract
:Simple Summary
Abstract
1. Introduction
2. Molecular Alterations in Cancer
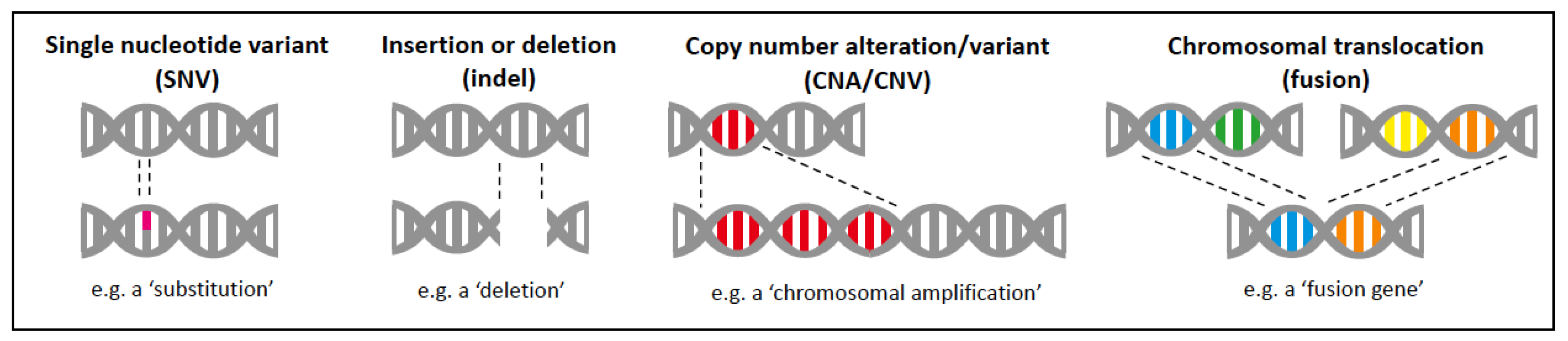
2.1. Types of Molecular Alterations
2.2. Somatic versus Germline Alterations
2.3. Tissues/Fluids for Molecular Analysis
2.4. Tools for Molecular Analysis
- Sample type (tissue versus body fluid, formalin-fixed versus fresh)
- Nucleic acids being analysed (DNA versus RNA)
- Type of genomic aberration being detected (SNVs, MNVs, CNAs, methylation)
- Cost (use of expensive equipment and highly specialised personnel/staff)
- Availability of technical expertise (computational analyses and interpretations)
3. Classes of Biomarkers
3.1. Diagnostic Molecular Biomarkers
3.2. Prognostic Molecular Biomarkers
3.3. Predictive Molecular Biomarkers
3.4. Screening Molecular Biomarkers
4. Somatic Molecular Alterations Currently Tested in the Clinical Setting
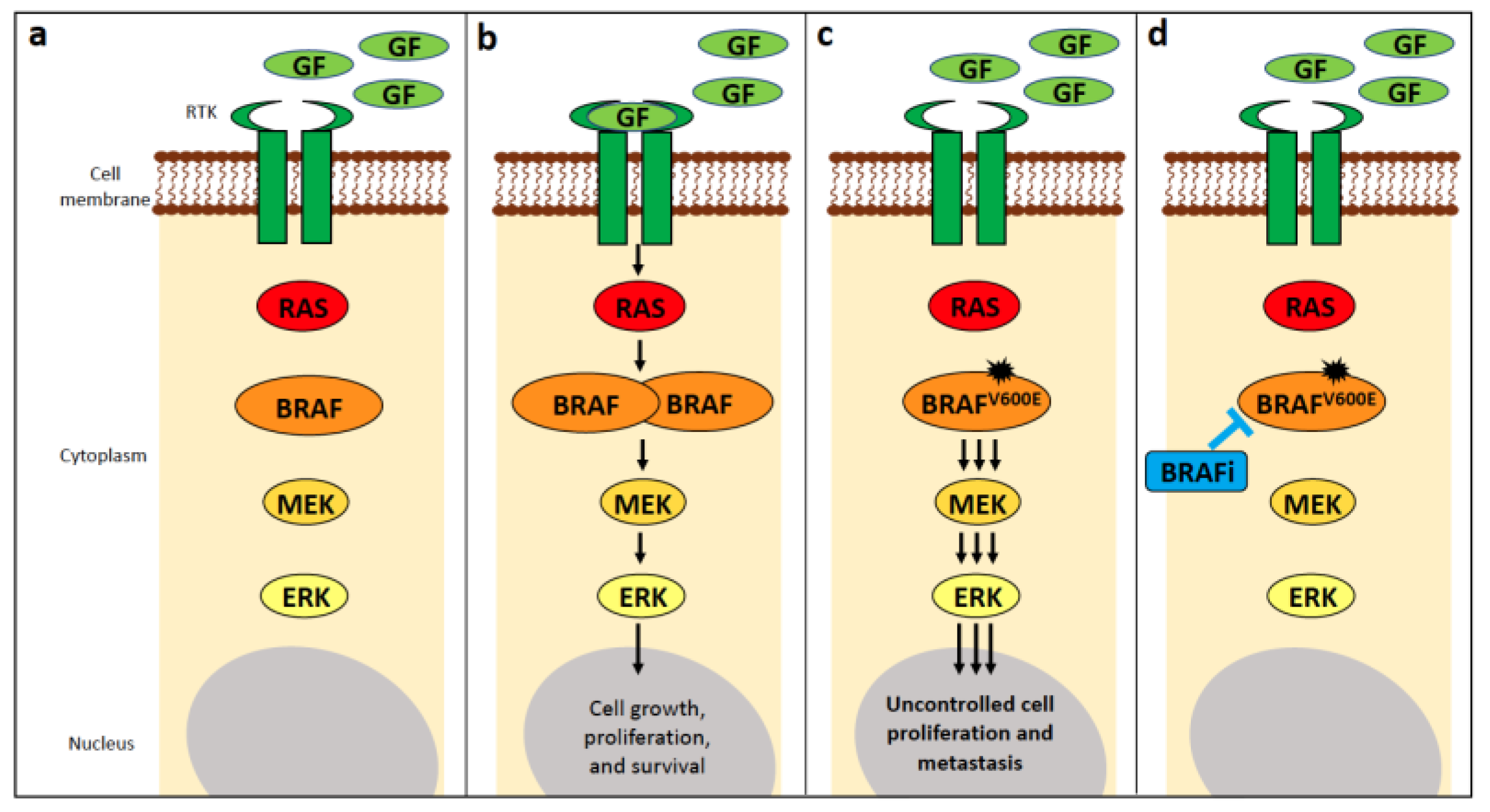
4.1. BRAF
4.1.1. BRAF Alterations in Human Cancer
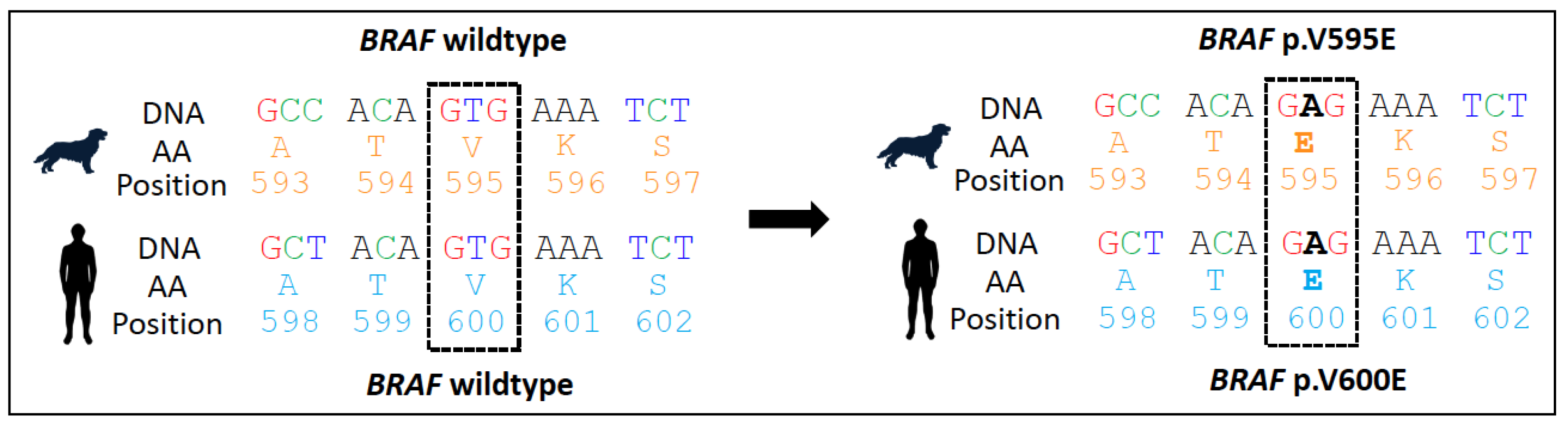
4.1.2. BRAF Alterations in Canine Cancer
4.1.3. Use as a Diagnostic Biomarker in Canines
4.1.4. Use as a Prognostic Biomarker in Canines
4.1.5. Use as a Predictive Biomarker in Canines
4.1.6. BRAF Alterations in Feline Cancer
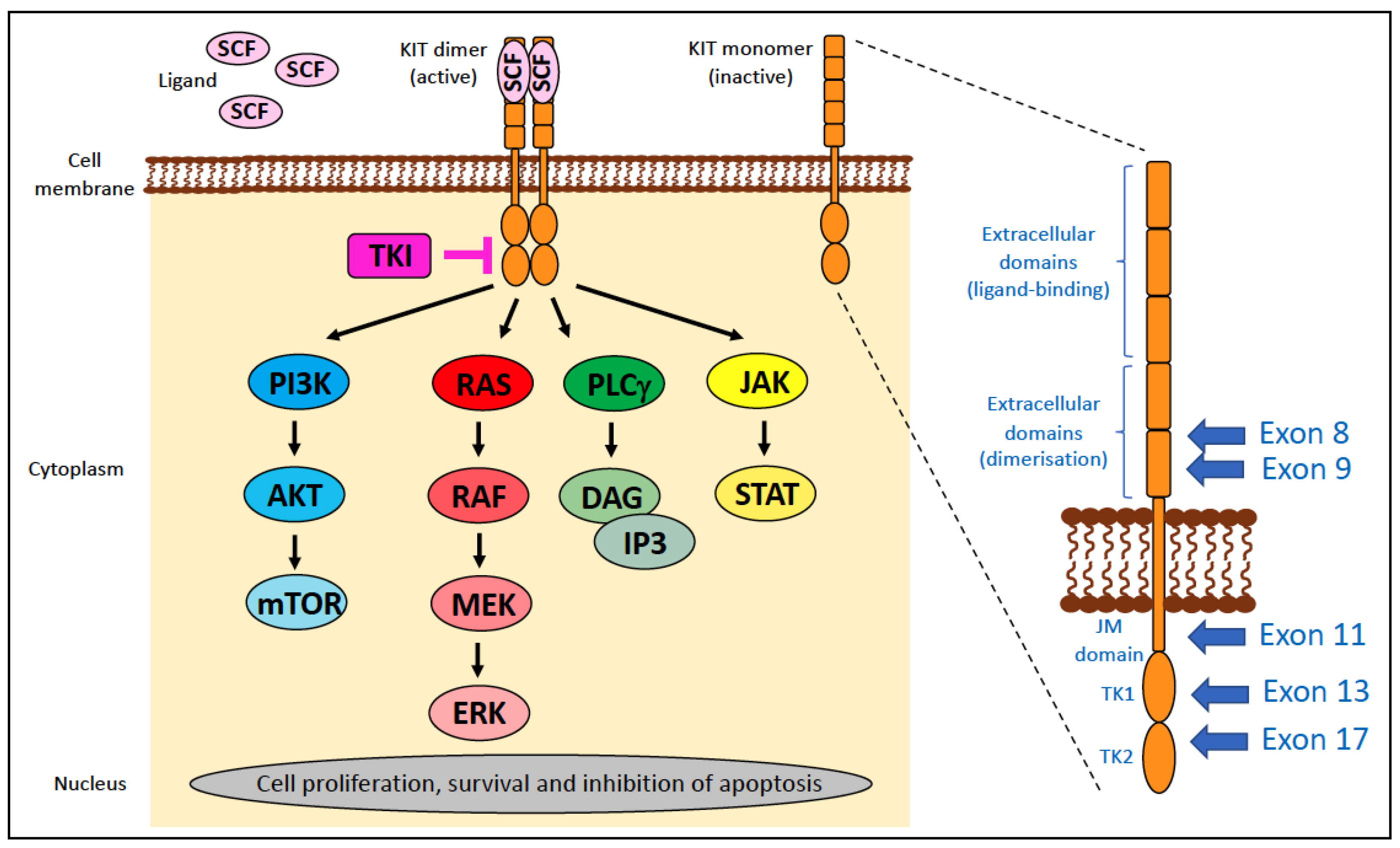
4.2. KIT
- ‘regulatory’ type—affects portions of the molecule that regulate kinase activity, such as releasing the inhibitory regulation of ligand-unoccupied c-Kit, resulting in constitutive activation. These include mutations in exons 8, 9, and 11.
- ‘enzymatic pocket’ type—affects the activation loop at the entrance to the enzymatic ‘pocket’. These include mutations in exons 13–21, typically exon 17.
4.2.1. KIT Alterations in Human Cancer
4.2.2. KIT Alterations in Canine Cancer
4.2.3. Use as a Diagnostic Biomarker in Canine Cancer
4.2.4. Use as a Prognostic Biomarker in Canine Cancer
4.2.5. Use as a Predictive Biomarker in Canine Cancer
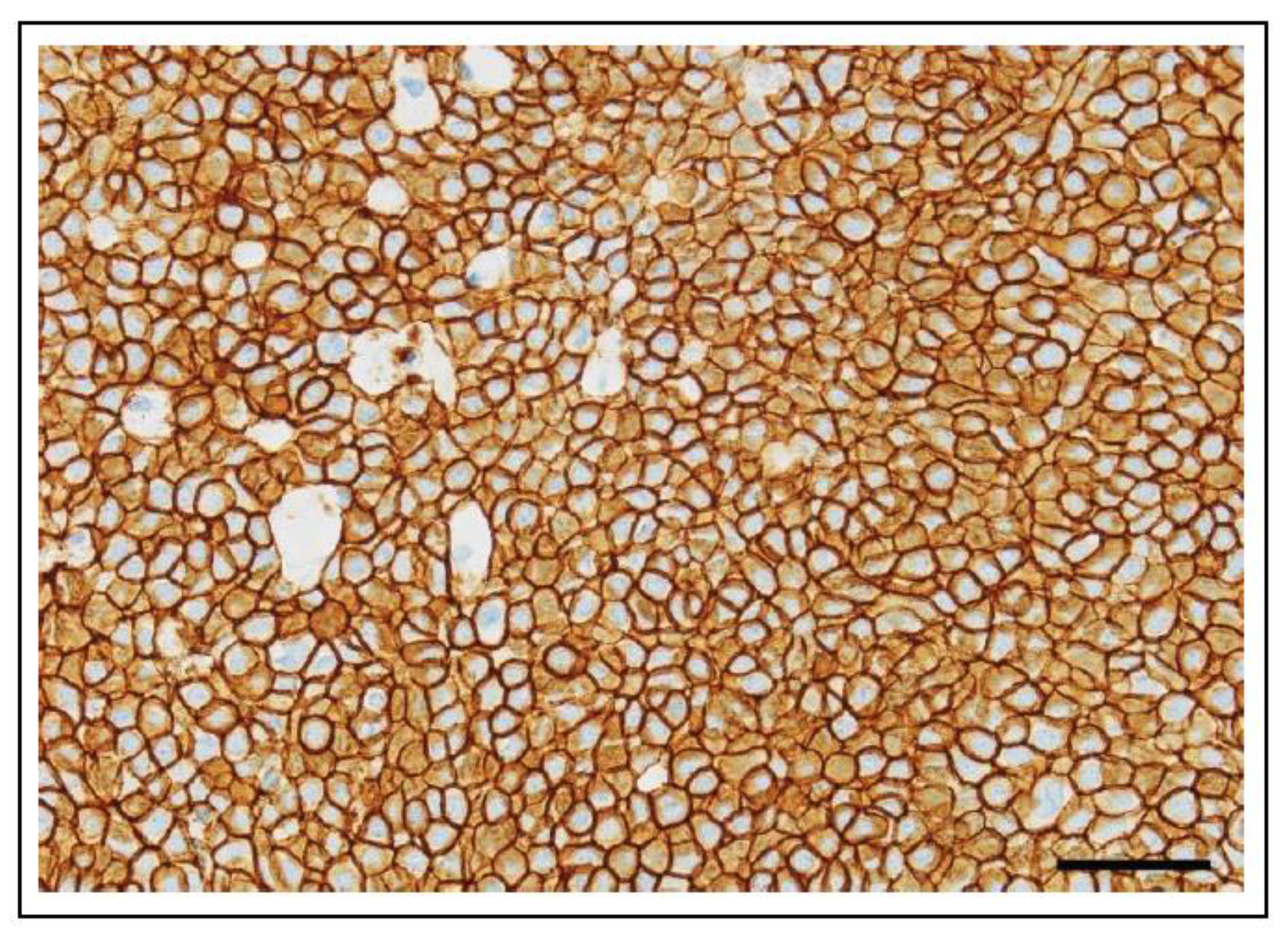
4.2.6. KIT Mutations in Feline Cancer
4.2.7. Use as a Diagnostic Biomarker in Feline Cancer
4.2.8. Use as a Prognostic Biomarker in Feline Cancer
4.2.9. Use as a Predictive Biomarker in Feline Cancer
4.3. PCR for Antigen Receptor Rearrangement in Dogs and Cats
4.4. Multi-Gene Panels
4.4.1. Clinical Use of Multi-Gene Panels for Human Cancer Patients
4.4.2. Clinical Use of Multi-Gene Panels for Veterinary Cancer Patients
4.4.3. Use of Multi-Gene Panels for Cancer Screening in Dogs
5. Germline Alterations Currently Tested in the Veterinary Clinical Setting
5.1. Canine Hereditary Multifocal Renal Cystadenocarcinoma and Nodular Dermatofibrosis
5.2. Familial Follicular Thyroid Carcinoma in German Longhaired Pointer Dogs
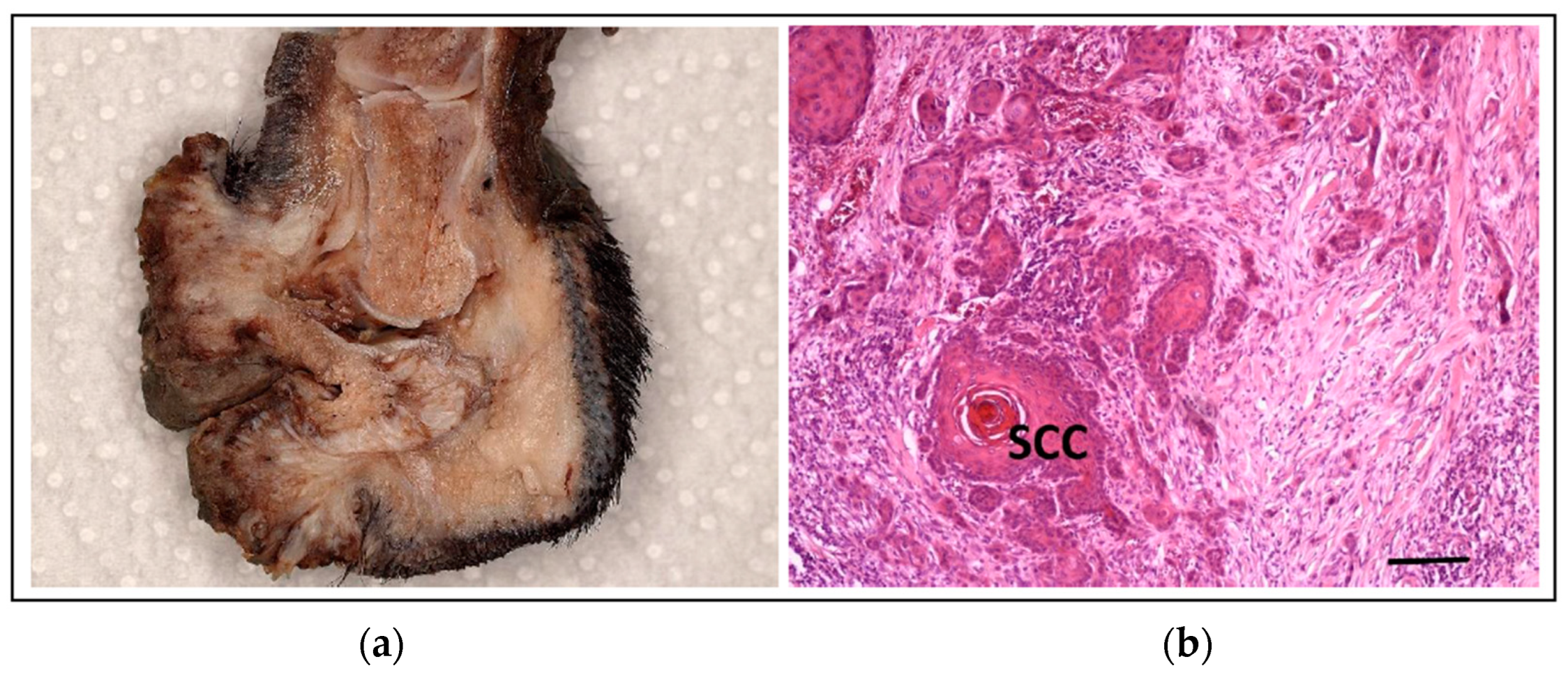
5.3. Squamous Cell Carcinoma of the Digit in Black Poodles and Black Giant Schnauzers
5.4. Histiocytic Sarcoma in the Bernese Mountain Dog
- Index A: The individual has 4× the chance of not developing HS
- Index B: Neutral index
- Index C: The individual has 4× the risk of developing HS
6. Emerging Molecular Alterations Showing Promise for Use as Biomarkers in the Clinic
6.1. Potential Somatic Molecular Biomarkers in Dogs and Cats
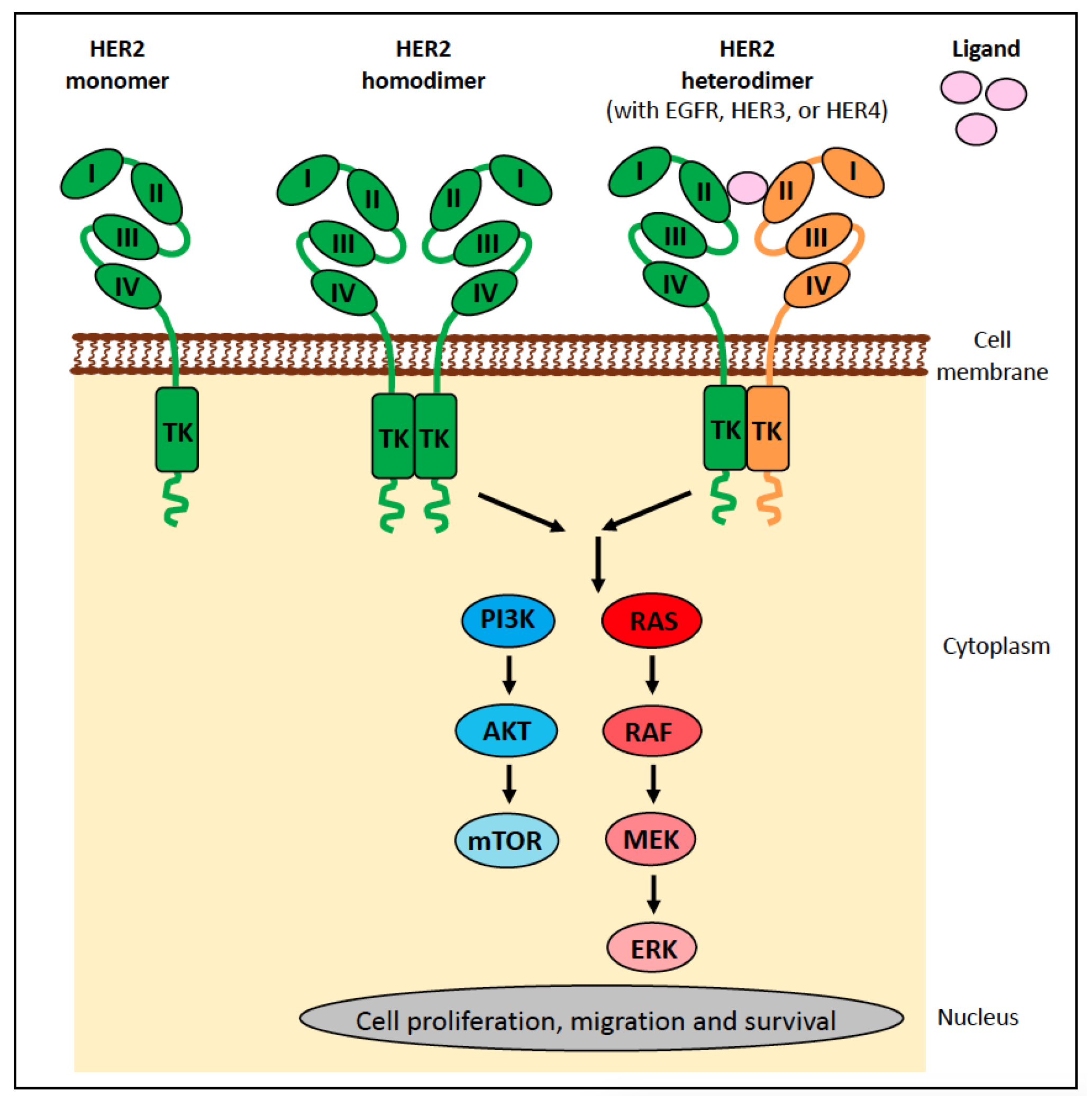
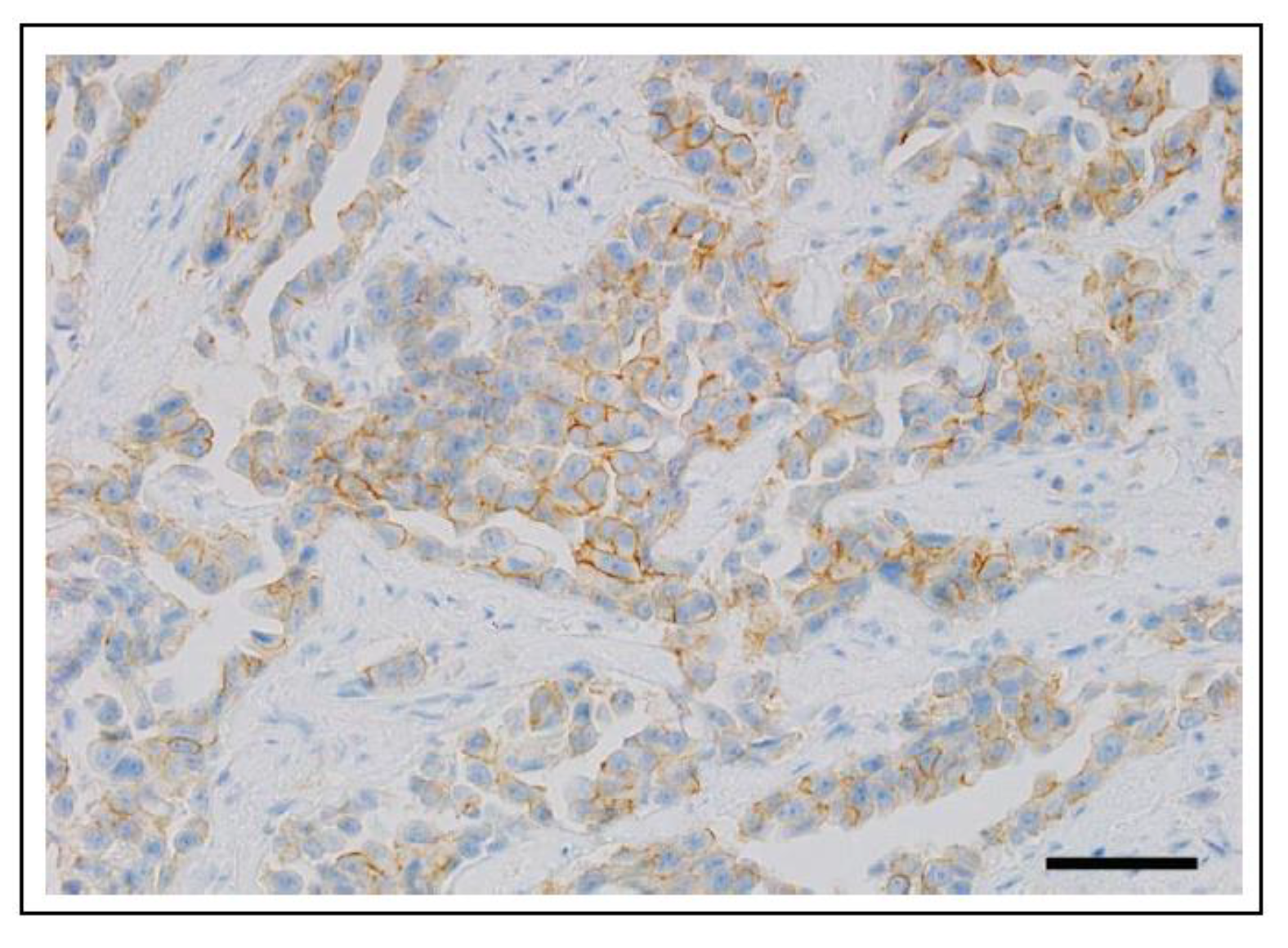
6.1.1. ERBB2 (HER2) in Human Cancer
6.1.2. ERBB2 (HER2) in Canine Cancer
6.1.3. ERBB2 (HER2) in Feline Cancer
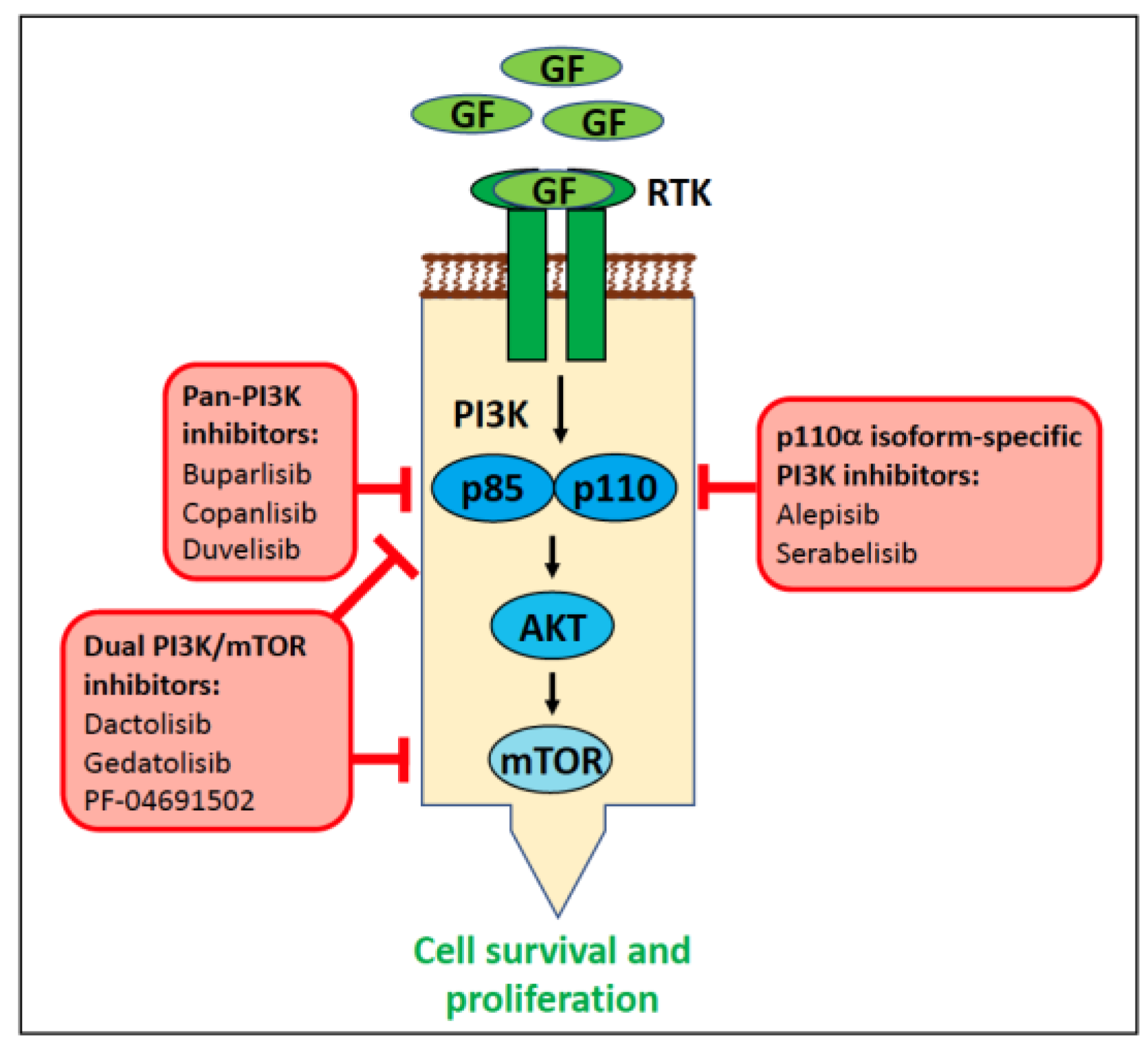
6.1.4. PIK3CA in Human Cancer
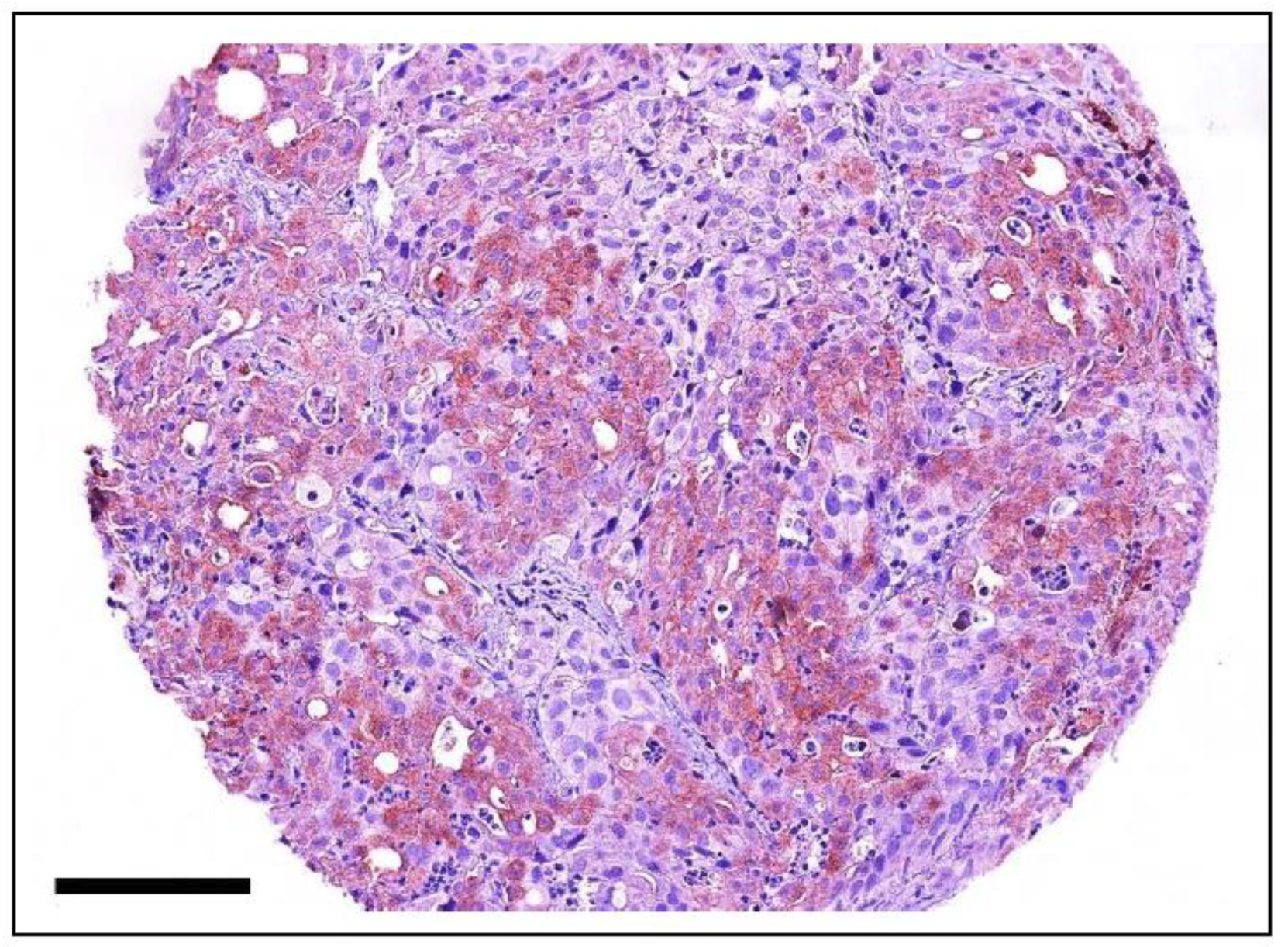
6.1.5. PIK3CA in Canine Cancer
6.1.6. PI3KCA in Feline Cancer
6.2. Potential Germline Molecular Biomarkers in Dogs and Cats
6.2.1. Potential Molecular Diagnostic Biomarkers in Dogs
6.2.2. Potential Molecular Diagnostic Biomarkers in Cats
7. Barriers to Clinical Translation of Biomarkers
7.1. Study Design
7.2. Clinical Trials
7.2.1. Good Clinical Practice
7.2.2. Legal Considerations
7.2.3. Ethical Considerations
7.2.4. Sponsorship
7.3. Tumour Heterogeneity
7.4. Complexity of Cellular Oncogenic Mechanisms
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADC | antibody-drug conjugate |
| AI | artificial intelligence |
| AKT | protein kinase B |
| BMD | Bernese Mountain dog |
| BRAFi | BRAF inhibitors |
| cfDNA | cell-free DNA |
| ctDNA | circulating tumour DNA |
| CFA | Canis familiaris chromosome |
| CNA | copy number alteration |
| CNG | copy number gain |
| CNV | copy number variation |
| ddPCR | droplet digital PCR |
| DSCC | squamous cell carcinoma of the digit |
| EMA | European Medicines Agency |
| ERBB2 | erb-b2 receptor tyrosine kinase 2 |
| ERK | extracellular signal-regulated kinase |
| ESS | English Springer Spaniel |
| FCR | Flat-coated Retriever |
| FDA | United States Food and Drug Administration |
| FFPE | formalin-fixed paraffin-embedded |
| FFTC | familial follicular thyroid carcinoma |
| FISS | Feline injection-site sarcoma |
| FTC | follicular thyroid carcinoma |
| GCP | Good Clinical Practice |
| GEP | gene expression profile |
| GOF | gain of function |
| GIST | gastrointestinal tumour |
| GR | Golden Retriever |
| GS | Giant Schnauzer |
| GSD | German Shepherd dog |
| GWAS | genome-wide association study |
| HE | hematoxylin and eosin |
| HS | histiocytic sarcoma |
| HSA | hemangiosarcoma |
| ICH | International Council on Harmonization |
| IgH | immunoglobulin heavy chain |
| IHC | immunohistochemistry |
| ITD | internal tandem duplications |
| ITH | intratumoural heterogeneity |
| JAK | Janus kinase |
| JM | juxtamembrane |
| MAPK | mitogen-activated protein kinase |
| MCED | multi-cancer early detection |
| MCT | mast cell tumour |
| MEK | mitogen-activated protein kinase kinase |
| MNV | multi-nucleotide variant |
| MSKCC | Memorial Sloan Kettering Cancer Center |
| NGS | next-generation sequencing |
| OS | overall survival |
| OSA | osteosarcoma |
| PARR | PCR for antigen receptor rearrangement |
| PC | prostate carcinoma |
| PCR | polymerase chain reaction |
| PI3K | Phosphatidylinositol 3-kinase |
| PFS | progression-free survival |
| PLCγ | phospholipase C-γ |
| PODS | Precision Oncology Decision Support |
| RAF | rapidly accelerated fibrosarcoma |
| RCND | renal cystadenocarcinoma and nodular dermatofibrosis |
| RTK | receptor tyrosine kinase |
| SCC | squamous cell carcinoma |
| SCF | stem cell factor |
| SNV | single nucleotide variation |
| SS | Standard Schnauzer |
| STAT | signal transducer and activator of transcription |
| STPO | standard poodle |
| TCR | T-cell receptor |
| TGS | targeted sequencing |
| TSG | tumour suppressor gene |
| TK | tyrosine kinase |
| TKI | tyrosine kinase inhibitor |
| UC | urothelial carcinoma |
| VICH | International Cooperation on Harmonisation of Technical Requirements for Registration of Veterinary Medicinal Products’ |
| VUS | variants of unknown significance |
| WES | whole exome sequencing |
| WGS | whole genome sequencing |
References
- Sosinsky, A.; Ambrose, J.; Cross, W.; Turnbull, C.; Henderson, S.; Jones, L.; Hamblin, A.; Arumugam, P.; Chan, G.; Chubb, D.; et al. Insights for precision oncology from the integration of genomic and clinical data of 13,880 tumors from the 100,000 Genomes Cancer Programme. Nat. Med. 2024, 30, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Takeshima, H.; Ushijima, T. Accumulation of genetic and epigenetic alterations in normal cells and cancer risk. NPJ Precis. Oncol. 2019, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of somatic mutation in human cancer genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef]
- Miller, D.G. On the nature of susceptibility to cancer. The presidential address. Cancer 1980, 46, 1307–1318. [Google Scholar] [CrossRef]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef]
- Schneider, K.; Zelley, K.; Nichols, K.E.; Garber, J. Li-Fraumeni Syndrome. In GeneReviews(®); Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2019. [Google Scholar]
- Kehl, A.; Aupperle-Lellbach, H.; de Brot, S.; van der Weyden, L. Review of Molecular Technologies for Investigating Canine Cancer. Animals 2024, 14, 769. [Google Scholar] [CrossRef]
- Steiert, T.A.; Parra, G.; Gut, M.; Arnold, N.; Trotta, J.R.; Tonda, R.; Moussy, A.; Gerber, Z.; Abuja, P.M.; Zatloukal, K.; et al. A critical spotlight on the paradigms of FFPE-DNA sequencing. Nucleic Acids Res. 2023, 51, 7143–7162. [Google Scholar] [CrossRef]
- Mathieson, W.; Thomas, G.A. Why Formalin-fixed, Paraffin-embedded Biospecimens Must Be Used in Genomic Medicine: An Evidence-based Review and Conclusion. J. Histochem. Cytochem. 2020, 68, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Bayle, A.; Peyraud, F.; Belcaid, L.; Brunet, M.; Aldea, M.; Clodion, R.; Dubos, P.; Vasseur, D.; Nicotra, C.; Geraud, A.; et al. Liquid versus tissue biopsy for detecting actionable alterations according to the ESMO Scale for Clinical Actionability of molecular Targets in patients with advanced cancer: A study from the French National Center for Precision Medicine (PRISM). Ann. Oncol. 2022, 33, 1328–1331. [Google Scholar] [CrossRef]
- Chibuk, J.; Flory, A.; Kruglyak, K.M.; Leibman, N.; Nahama, A.; Dharajiya, N.; van den Boom, D.; Jensen, T.J.; Friedman, J.S.; Shen, M.R.; et al. Horizons in Veterinary Precision Oncology: Fundamentals of Cancer Genomics and Applications of Liquid Biopsy for the Detection, Characterization, and Management of Cancer in Dogs. Front. Vet. Sci. 2021, 8, 664718. [Google Scholar] [CrossRef] [PubMed]
- Sarhadi, V.K.; Armengol, G. Molecular Biomarkers in Cancer. Biomolecules 2022, 12, 1021. [Google Scholar] [CrossRef] [PubMed]
- FDA-NIH Biomarker Working Group. BEST (Biomarkers, EndpointS, and Other Tools) Resource; Food and Drug Administration: Silver Spring, MD, USA; National Institutes of Health: Bethesda, MD, USA, 2016.
- Oldenhuis, C.N.; Oosting, S.F.; Gietema, J.A.; de Vries, E.G. Prognostic versus predictive value of biomarkers in oncology. Eur. J. Cancer 2008, 44, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Alowais, S.A.; Alghamdi, S.S.; Alsuhebany, N.; Alqahtani, T.; Alshaya, A.I.; Almohareb, S.N.; Aldairem, A.; Alrashed, M.; Bin Saleh, K.; Badreldin, H.A.; et al. Revolutionizing healthcare: The role of artificial intelligence in clinical practice. BMC Med. Educ. 2023, 23, 689. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Yang, D.; Arikawa, K.; Bai, C. Application of artificial intelligence in modern medicine. Clin. eHealth 2023, 6, 130–137. [Google Scholar] [CrossRef]
- Corti, C.; Cobanaj, M.; Dee, E.C.; Criscitiello, C.; Tolaney, S.M.; Celi, L.A.; Curigliano, G. Artificial intelligence in cancer research and precision medicine: Applications, limitations and priorities to drive transformation in the delivery of equitable and unbiased care. Cancer Treat. Rev. 2023, 112, 102498. [Google Scholar] [CrossRef] [PubMed]
- Colomer, R.; Mondejar, R.; Romero-Laorden, N.; Alfranca, A.; Sanchez-Madrid, F.; Quintela-Fandino, M. When should we order a next generation sequencing test in a patient with cancer? EClinicalMedicine 2020, 25, 100487. [Google Scholar] [CrossRef] [PubMed]
- Hanrahan, A.J.; Chen, Z.; Rosen, N.; Solit, D.B. BRAF—A tumour-agnostic drug target with lineage-specific dependencies. Nat. Rev. Clin. Oncol. 2024, 21, 224–247. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- The AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar] [CrossRef] [PubMed]
- Wan, P.T.; Garnett, M.J.; Roe, S.M.; Lee, S.; Niculescu-Duvaz, D.; Good, V.M.; Jones, C.M.; Marshall, C.J.; Springer, C.J.; Barford, D.; et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004, 116, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Poulikakos, P.I.; Sullivan, R.J.; Yaeger, R. Molecular Pathways and Mechanisms of BRAF in Cancer Therapy. Clin. Cancer Res. 2022, 28, 4618–4628. [Google Scholar] [CrossRef] [PubMed]
- Atkins, M.B.; Lee, S.J.; Chmielowski, B.; Tarhini, A.A.; Cohen, G.I.; Truong, T.G.; Moon, H.H.; Davar, D.; O’Rourke, M.; Stephenson, J.J.; et al. Combination Dabrafenib and Trametinib Versus Combination Nivolumab and Ipilimumab for Patients with Advanced BRAF-Mutant Melanoma: The DREAMseq Trial-ECOG-ACRIN EA6134. J. Clin. Oncol. 2023, 41, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Bouffet, E.; Hansford, J.R.; Garrè, M.L.; Hara, J.; Plant-Fox, A.; Aerts, I.; Locatelli, F.; van der Lugt, J.; Papusha, L.; Sahm, F.; et al. Dabrafenib plus Trametinib in Pediatric Glioma with BRAF V600 Mutations. N. Engl. J. Med. 2023, 389, 1108–1120. [Google Scholar] [CrossRef] [PubMed]
- Hargrave, D.R.; Terashima, K.; Hara, J.; Kordes, U.R.; Upadhyaya, S.A.; Sahm, F.; Bouffet, E.; Packer, R.J.; Witt, O.; Sandalic, L.; et al. Phase II Trial of Dabrafenib Plus Trametinib in Relapsed/Refractory BRAF V600-Mutant Pediatric High-Grade Glioma. J. Clin. Oncol. 2023, 41, 5174–5183. [Google Scholar] [CrossRef]
- Tian, J.; Chen, J.H.; Chao, S.X.; Pelka, K.; Giannakis, M.; Hess, J.; Burke, K.; Jorgji, V.; Sindurakar, P.; Braverman, J.; et al. Combined PD-1, BRAF and MEK inhibition in BRAF(V600E) colorectal cancer: A phase 2 trial. Nat. Med. 2023, 29, 458–466. [Google Scholar] [CrossRef]
- Mochizuki, H.; Shapiro, S.G.; Breen, M. Detection of BRAF Mutation in Urine DNA as a Molecular Diagnostic for Canine Urothelial and Prostatic Carcinoma. PLoS ONE 2015, 10, e0144170. [Google Scholar] [CrossRef]
- Mochizuki, H.; Kennedy, K.; Shapiro, S.G.; Breen, M. BRAF Mutations in Canine Cancers. PLoS ONE 2015, 10, e0129534. [Google Scholar] [CrossRef] [PubMed]
- Decker, B.; Parker, H.G.; Dhawan, D.; Kwon, E.M.; Karlins, E.; Davis, B.W.; Ramos-Vara, J.A.; Bonney, P.L.; McNiel, E.A.; Knapp, D.W.; et al. Homologous Mutation to Human BRAF V600E Is Common in Naturally Occurring Canine Bladder Cancer--Evidence for a Relevant Model System and Urine-Based Diagnostic Test. Mol. Cancer Res. 2015, 13, 993–1002. [Google Scholar] [CrossRef]
- Grassinger, J.M.; Aupperle-Lellbach, H.; Erhard, H.; Merz, S.; Klopfleisch, R. Detection of BRAF mutation in canine prostatic diseases. Tierarztl. Prax. Ausg. K Kleintiere Heimtiere 2019, 47, 313–320. [Google Scholar] [CrossRef]
- Gedon, J.; Kehl, A.; Aupperle-Lellbach, H.; von Bomhard, W.; Schmidt, J.M. BRAF mutation status and its prognostic significance in 79 canine urothelial carcinomas: A retrospective study (2006–2019). Vet. Comp. Oncol. 2022, 20, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Cronise, K.E.; Das, S.; Hernandez, B.G.; Regan, D.P.; Dailey, D.D.; McGeachan, R.I.; Lana, S.E.; Page, R.L.; Gustafson, D.L.; Duval, D.L. Characterizing the molecular and immune landscape of canine bladder cancer. Vet. Comp. Oncol. 2022, 20, 69–81. [Google Scholar] [CrossRef]
- Rodrigues, L.; Watson, J.; Feng, Y.; Lewis, B.; Harvey, G.; Post, G.; Megquier, K.; White, M.E.; Lambert, L.; Miller, A.; et al. Shared hotspot mutations in oncogenes position dogs as an unparalleled comparative model for precision therapeutics. Sci. Rep. 2023, 13, 10935. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.; Abascal, F.; Ludwig, L.; Aupperle-Lellbach, H.; Grassinger, J.; Wright, C.W.; Allison, S.J.; Pinder, E.; Phillips, R.M.; Romero, L.P.; et al. Cross-species oncogenomics offers insight into human muscle-invasive bladder cancer. Genome Biol. 2023, 24, 191. [Google Scholar] [CrossRef]
- Aupperle-Lellbach, H.; Grassinger, J.; Hohloch, C.; Kehl, A.; Pantke, P. Diagnostic value of the BRAF variant V595E in urine samples, smears and biopsies from canine transitional cell carcinoma. Tierarztl. Prax. Ausg. K. Kleintiere Heimtiere 2018, 46, 289–295. [Google Scholar] [CrossRef]
- Knapp, D.W.; Glickman, N.W.; Denicola, D.B.; Bonney, P.L.; Lin, T.L.; Glickman, L.T. Naturally-occurring canine transitional cell carcinoma of the urinary bladder A relevant model of human invasive bladder cancer. Urol. Oncol. 2000, 5, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Butty, E.M.; Hahn, S.; Labato, M.A. Presumptive malignant transformation of chronic polypoid cystitis into an apical transitional cell carcinoma without BRAF mutation in a young female dog. J. Vet. Intern. Med. 2021, 35, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, C.; Fonseca-Alves, C.E.; Laufer-Amorim, R. A Review on Canine and Feline Prostate Pathology. Front. Vet. Sci. 2022, 9, 881232. [Google Scholar] [CrossRef] [PubMed]
- Aupperle-Lellbach, H.; Kehl, A.; Merz, S.; Grassinger, J.; Hohloch, C.; Pantke, P. Die BRAF-Mutation V595E im Übergangszellkarzinom—Untersuchungen zur Rassedisposition bei Terriern. Kleintiermedizin 2019, 1, 30–33. [Google Scholar]
- Grassinger, J.M.; Merz, S.; Aupperle-Lellbach, H.; Erhard, H.; Klopfleisch, R. Correlation of BRAF Variant V595E, Breed, Histological Grade and Cyclooxygenase-2 Expression in Canine Transitional Cell Carcinomas. Vet. Sci. 2019, 6, 31. [Google Scholar] [CrossRef]
- Mutsaers, A.J.; Widmer, W.R.; Knapp, D.W. Canine transitional cell carcinoma. J. Vet. Intern. Med. 2003, 17, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Chambers, J.K.; Takahashi, N.; Kato, S.; Hashimoto, Y.; Goto-Koshino, Y.; Uchida, K. Diagnostic challenge in veterinary pathology: Detection of BRAF(V595E) mutation in a dog with follicular cystitis and flat urothelial lesion with atypia. Vet. Pathol. 2023, 61, 3009858231217242. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, S.G.; Raghunath, S.; Williams, C.; Motsinger-Reif, A.A.; Cullen, J.M.; Liu, T.; Albertson, D.; Ruvolo, M.; Bergstrom Lucas, A.; Jin, J.; et al. Canine urothelial carcinoma: Genomically aberrant and comparatively relevant. Chromosome Res. 2015, 23, 311–331. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, H.; Shapiro, S.G.; Breen, M. Detection of Copy Number Imbalance in Canine Urothelial Carcinoma with Droplet Digital Polymerase Chain Reaction. Vet. Pathol. 2016, 53, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Aeschlimann, L.; Kehl, A.; Guscetti, F.; Posthaus, C.; Aupperle-Lellbach, H.; Rottenberg, S.; de Brot, S. Effective detection of BRAFV595E mutation in canine urothelial and prostate carcinomas using immunohistochemistry. Vet. Comp. Oncol. 2024. Online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.M.; Pereira, E.M.; Salles, P.G.; Godrich, R.; Ceballos, R.; Kunz, J.D.; Casson, A.; Viret, J.; Chandarlapaty, S.; Ferreira, C.G.; et al. Independent real-world application of a clinical-grade automated prostate cancer detection system. J. Pathol. 2021, 254, 147–158. [Google Scholar] [CrossRef]
- Rabilloud, N.; Allaume, P.; Acosta, O.; De Crevoisier, R.; Bourgade, R.; Loussouarn, D.; Rioux-Leclercq, N.; Khene, Z.E.; Mathieu, R.; Bensalah, K.; et al. Deep Learning Methodologies Applied to Digital Pathology in Prostate Cancer: A Systematic Review. Diagnostics 2023, 13, 2676. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Wang, S.; Yang, D.M.; Xie, Y.; Chen, M.; Bishop, J.; Xiao, G. Deep learning in digital pathology for personalized treatment plans of cancer patients. Semin. Diagn. Pathol. 2023, 40, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Küchler, L.; Posthaus, C.; Jäger, K.; Guscetti, F.; van der Weyden, L.; von Bomhard, W.; Schmidt, J.M.; Farra, D.; Aupperle-Lellbach, H.; Kehl, A.; et al. Artificial Intelligence to Predict the BRAF V595E Mutation in Canine Urinary Bladder Urothelial Carcinomas. Animals 2023, 13, 2404. [Google Scholar] [CrossRef]
- Pantke, P.; Diekmann, C.; Hettig, M.; Aupperle-Lellbach, H. Erste klinische Erhebungen zur Überlebensrate von Hunden mit Übergangszellkarzinom und BRAF-Mutation V595E. Kleintierpraxis 2019, 64, 680–686. [Google Scholar] [CrossRef]
- Tagawa, M.; Tambo, N.; Maezawa, M.; Tomihari, M.; Watanabe, K.I.; Inokuma, H.; Miyahara, K. Quantitative analysis of the BRAF V595E mutation in plasma cell-free DNA from dogs with urothelial carcinoma. PLoS ONE 2020, 15, e0232365. [Google Scholar] [CrossRef]
- Kim, J.H.; Ahn, D.H.; Moon, J.S.; Han, H.J.; Bae, K.; Yoon, K.A. Longitudinal assessment of B-RAF V595E levels in the peripheral cell-free tumor DNA of a 10-year-old spayed female Korean Jindo dog with unresectable metastatic urethral transitional cell carcinoma for monitoring the treatment response to a RAF inhibitor (sorafenib). Vet. Q. 2021, 41, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Cronise, K.E.; Hernandez, B.G.; Gustafson, D.L.; Duval, D.L. Identifying the ErbB/MAPK Signaling Cascade as a Therapeutic Target in Canine Bladder Cancer. Mol. Pharmacol. 2019, 96, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Spevak, W.; Zhang, Y.; Burton, E.A.; Ma, Y.; Habets, G.; Zhang, J.; Lin, J.; Ewing, T.; Matusow, B.; et al. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature 2015, 526, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Bae, K.; Lee, J.Y.; Kim, J.H.; Han, H.J.; Yoon, H.Y.; Yoon, K.A. Establishment of Canine Transitional Cell Carcinoma Cell Lines Harboring BRAF V595E Mutation as a Therapeutic Target. Int. J. Mol. Sci. 2021, 22, 9151. [Google Scholar] [CrossRef]
- Foskett, A.; Manley, C.; Naramore, R.; Gordon, I.K.; Stewart, B.M.; Khanna, C. Tolerability of oral sorafenib in pet dogs with a diagnosis of cancer. Vet. Med. 2017, 8, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Rossman, P.; Zabka, T.S.; Ruple, A.; Tuerck, D.; Ramos-Vara, J.A.; Liu, L.; Mohallem, R.; Merchant, M.; Franco, J.; Fulkerson, C.M.; et al. Phase I/II Trial of Vemurafenib in Dogs with Naturally Occurring, BRAF-mutated Urothelial Carcinoma. Mol. Cancer Ther. 2021, 20, 2177–2188. [Google Scholar] [CrossRef]
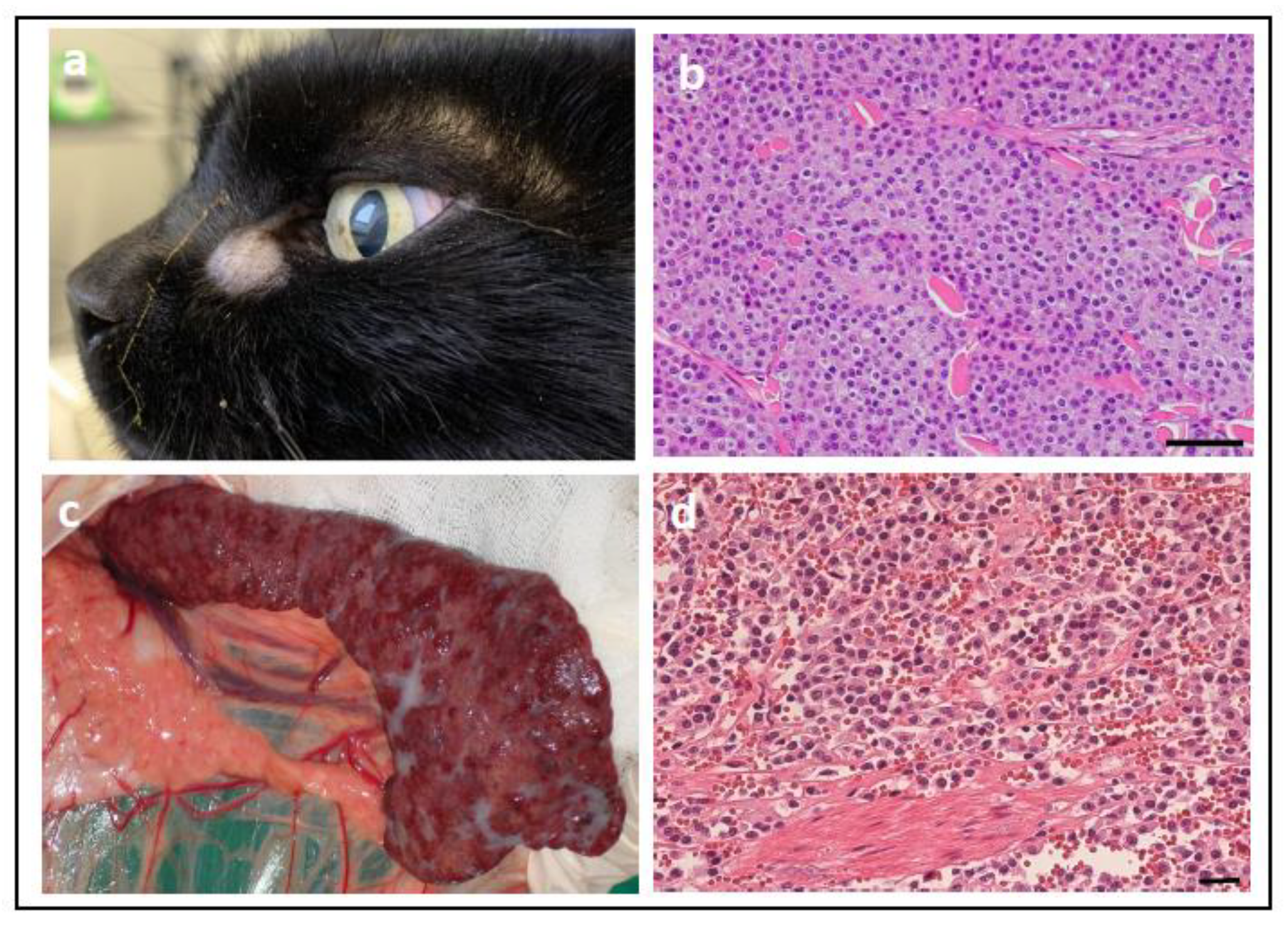
- Ludwig, L.; Dobromylskyj, M.; Wood, G.A.; van der Weyden, L. Feline Oncogenomics: What Do We Know about the Genetics of Cancer in Domestic Cats? Vet. Sci. 2022, 9, 547. [Google Scholar] [CrossRef] [PubMed]
- Rushton, J.G.; Ertl, R.; Klein, D.; Nell, B. Mutation analysis and gene expression profiling of ocular melanomas in cats. Vet. Comp. Oncol. 2017, 15, 1403–1416. [Google Scholar] [CrossRef]
- Rushton, J.G.; Korb, M.; Kummer, S.; Reichart, U.; Fuchs-Baumgartinger, A.; Tichy, A.; Nell, B. Protein expression of KIT, BRAF, GNA11, GNAQ and RASSF1 in feline diffuse iris melanomas. Vet. J. 2019, 249, 33–40. [Google Scholar] [CrossRef]
- Sheikh, E.; Tran, T.; Vranic, S.; Levy, A.; Bonfil, R.D. Role and significance of c-KIT receptor tyrosine kinase in cancer: A review. Bosn. J. Basic. Med. Sci. 2022, 22, 683–698. [Google Scholar] [CrossRef]
- Liang, J.; Wu, Y.L.; Chen, B.J.; Zhang, W.; Tanaka, Y.; Sugiyama, H. The C-kit receptor-mediated signal transduction and tumor-related diseases. Int. J. Biol. Sci. 2013, 9, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Longley, B.J.; Reguera, M.J.; Ma, Y. Classes of c-KIT activating mutations: Proposed mechanisms of action and implications for disease classification and therapy. Leuk. Res. 2001, 25, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Montero, A.C.; Jara-Acevedo, M.; Teodosio, C.; Sanchez, M.L.; Nunez, R.; Prados, A.; Aldanondo, I.; Sanchez, L.; Dominguez, M.; Botana, L.M.; et al. KIT mutation in mast cells and other bone marrow hematopoietic cell lineages in systemic mast cell disorders: A prospective study of the Spanish Network on Mastocytosis (REMA) in a series of 113 patients. Blood 2006, 108, 2366–2372. [Google Scholar] [CrossRef]
- Hirota, S.; Isozaki, K.; Moriyama, Y.; Hashimoto, K.; Nishida, T.; Ishiguro, S.; Kawano, K.; Hanada, M.; Kurata, A.; Takeda, M.; et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 1998, 279, 577–580. [Google Scholar] [CrossRef]
- Abdellateif, M.S.; Bayoumi, A.K.; Mohammed, M.A. c-Kit Receptors as a Therapeutic Target in Cancer: Current Insights. Onco Targets Ther. 2023, 16, 785–799. [Google Scholar] [CrossRef]
- Allen, C.; Hills, R.K.; Lamb, K.; Evans, C.; Tinsley, S.; Sellar, R.; O’Brien, M.; Yin, J.L.; Burnett, A.K.; Linch, D.C.; et al. The importance of relative mutant level for evaluating impact on outcome of KIT, FLT3 and CBL mutations in core-binding factor acute myeloid leukemia. Leukemia 2013, 27, 1891–1901. [Google Scholar] [CrossRef]
- Beadling, C.; Jacobson-Dunlop, E.; Hodi, F.S.; Le, C.; Warrick, A.; Patterson, J.; Town, A.; Harlow, A.; Cruz, F., 3rd; Azar, S.; et al. KIT gene mutations and copy number in melanoma subtypes. Clin. Cancer Res. 2008, 14, 6821–6828. [Google Scholar] [CrossRef] [PubMed]
- Torres-Cabala, C.A.; Wang, W.L.; Trent, J.; Yang, D.; Chen, S.; Galbincea, J.; Kim, K.B.; Woodman, S.; Davies, M.; Plaza, J.A.; et al. Correlation between KIT expression and KIT mutation in melanoma: A study of 173 cases with emphasis on the acral-lentiginous/mucosal type. Mod. Pathol. 2009, 22, 1446–1456. [Google Scholar] [CrossRef]
- London, C.A. Tyrosine kinase inhibitors in veterinary medicine. Top. Companion Anim. Med. 2009, 24, 106–112. [Google Scholar] [CrossRef]
- Flynn, J.P.; Gerriets, V. Imatinib; StatPearls Publishing LLC: St. Petersburg, FL, USA, 2023. [Google Scholar]
- Ma, Y.; Zeng, S.; Metcalfe, D.D.; Akin, C.; Dimitrijevic, S.; Butterfield, J.H.; McMahon, G.; Longley, B.J. The c-KIT mutation causing human mastocytosis is resistant to STI571 and other KIT kinase inhibitors; kinases with enzymatic site mutations show different inhibitor sensitivity profiles than wild-type kinases and those with regulatory-type mutations. Blood 2002, 99, 1741–1744. [Google Scholar] [CrossRef]
- Chen, L.L.; Trent, J.C.; Wu, E.F.; Fuller, G.N.; Ramdas, L.; Zhang, W.; Raymond, A.K.; Prieto, V.G.; Oyedeji, C.O.; Hunt, K.K.; et al. A missense mutation in KIT kinase domain 1 correlates with imatinib resistance in gastrointestinal stromal tumors. Cancer Res. 2004, 64, 5913–5919. [Google Scholar] [CrossRef] [PubMed]
- Nishida, T.; Kanda, T.; Nishitani, A.; Takahashi, T.; Nakajima, K.; Ishikawa, T.; Hirota, S. Secondary mutations in the kinase domain of the KIT gene are predominant in imatinib-resistant gastrointestinal stromal tumor. Cancer Sci. 2008, 99, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Apperley, J.F. Part I: Mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007, 8, 1018–1029. [Google Scholar] [CrossRef]
- Mahadevan, D.; Cooke, L.; Riley, C.; Swart, R.; Simons, B.; Della Croce, K.; Wisner, L.; Iorio, M.; Shakalya, K.; Garewal, H.; et al. A novel tyrosine kinase switch is a mechanism of imatinib resistance in gastrointestinal stromal tumors. Oncogene 2007, 26, 3909–3919. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Longley, B.J.; Wang, X.; Blount, J.L.; Langley, K.; Caughey, G.H. Clustering of activating mutations in c-KIT’s juxtamembrane coding region in canine mast cell neoplasms. J. Investig. Dermatol. 1999, 112, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Zemke, D.; Yamini, B.; Yuzbasiyan-Gurkan, V. Mutations in the juxtamembrane domain of c-KIT are associated with higher grade mast cell tumors in dogs. Vet. Pathol. 2002, 39, 529–535. [Google Scholar] [CrossRef]
- Letard, S.; Yang, Y.; Hanssens, K.; Palmérini, F.; Leventhal, P.S.; Guéry, S.; Moussy, A.; Kinet, J.P.; Hermine, O.; Dubreuil, P. Gain-of-function mutations in the extracellular domain of KIT are common in canine mast cell tumors. Mol. Cancer Res. 2008, 6, 1137–1145. [Google Scholar] [CrossRef]
- Giantin, M.; Vascellari, M.; Morello, E.M.; Capello, K.; Vercelli, A.; Granato, A.; Lopparelli, R.M.; Nassuato, C.; Carminato, A.; Martano, M.; et al. c-KIT messenger RNA and protein expression and mutations in canine cutaneous mast cell tumors: Correlations with post-surgical prognosis. J. Vet. Diagn. Investig. 2012, 24, 116–126. [Google Scholar] [CrossRef]
- Vozdova, M.; Kubickova, S.; Pal, K.; Fröhlich, J.; Fictum, P.; Rubes, J. Recurrent gene mutations detected in canine mast cell tumours by next generation sequencing. Vet. Comp. Oncol. 2020, 18, 509–518. [Google Scholar] [CrossRef]
- London, C.A.; Galli, S.J.; Yuuki, T.; Hu, Z.Q.; Helfand, S.C.; Geissler, E.N. Spontaneous canine mast cell tumors express tandem duplications in the proto-oncogene c-kit. Exp. Hematol. 1999, 27, 689–697. [Google Scholar] [CrossRef]
- Downing, S.; Chien, M.B.; Kass, P.H.; Moore, P.E.; London, C.A. Prevalence and importance of internal tandem duplications in exons 11 and 12 of c-kit in mast cell tumors of dogs. Am. J. Vet. Res. 2002, 63, 1718–1723. [Google Scholar] [CrossRef]
- Webster, J.D.; Yuzbasiyan-Gurkan, V.; Kaneene, J.B.; Miller, R.; Resau, J.H.; Kiupel, M. The role of c-KIT in tumorigenesis: Evaluation in canine cutaneous mast cell tumors. Neoplasia 2006, 8, 104–111. [Google Scholar] [CrossRef]
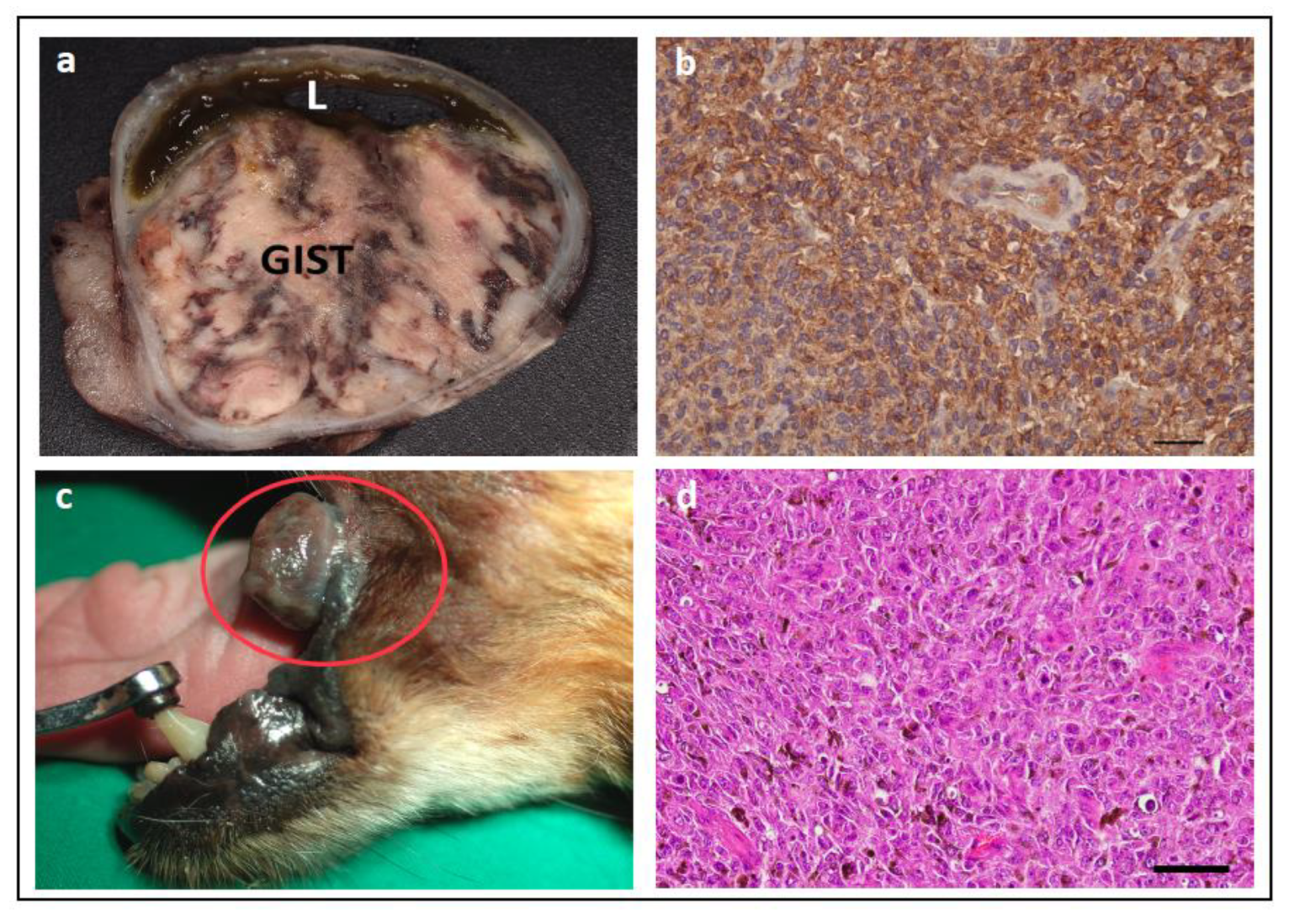
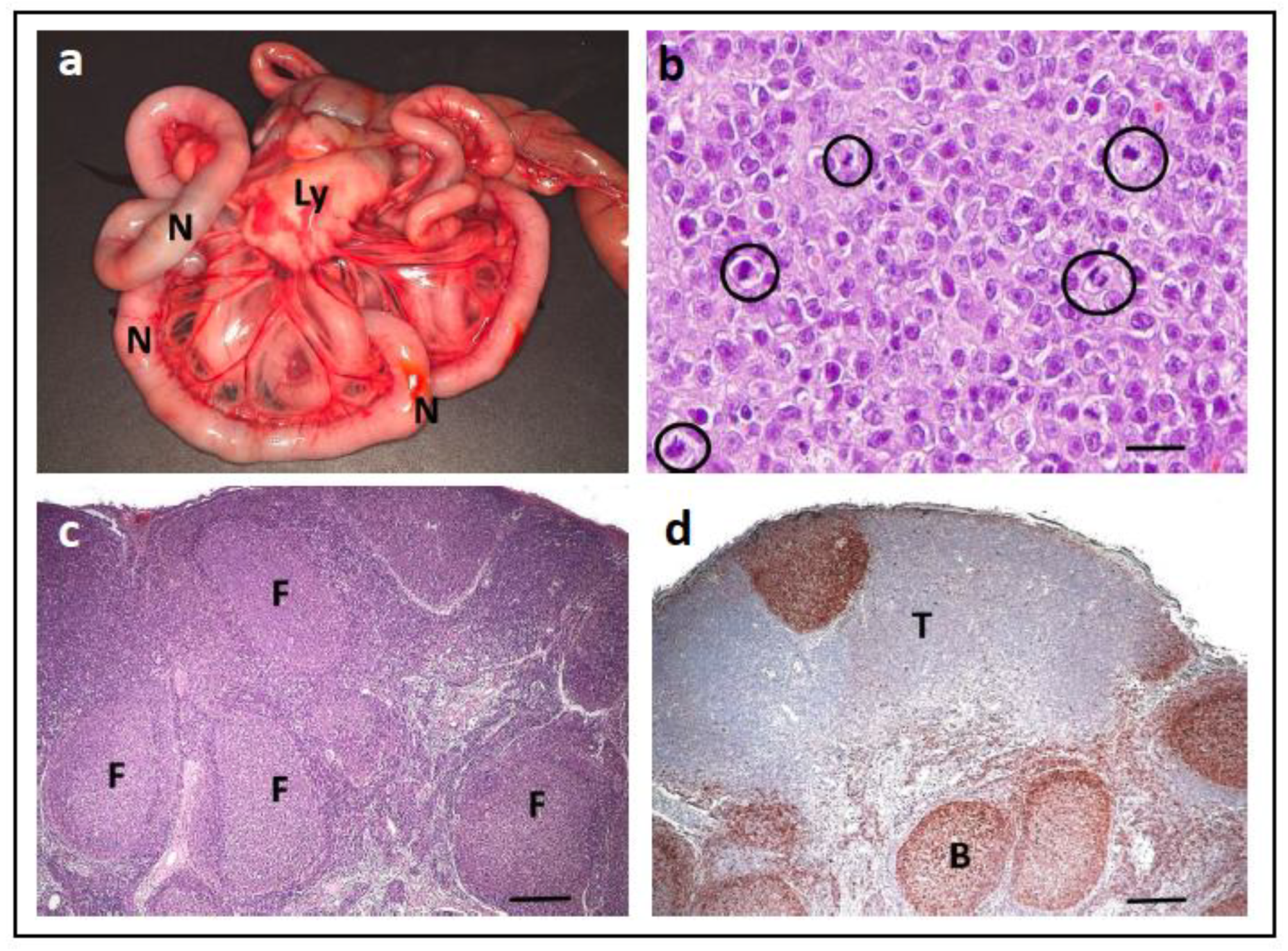
- Frost, D.; Lasota, J.; Miettinen, M. Gastrointestinal stromal tumors and leiomyomas in the dog: A histopathologic, immunohistochemical, and molecular genetic study of 50 cases. Vet. Pathol. 2003, 40, 42–54. [Google Scholar] [CrossRef]
- Gregory-Bryson, E.; Bartlett, E.; Kiupel, M.; Hayes, S.; Yuzbasiyan-Gurkan, V. Canine and human gastrointestinal stromal tumors display similar mutations in c-KIT exon 11. BMC Cancer 2010, 10, 559. [Google Scholar] [CrossRef]
- Hayes, S.; Yuzbasiyan-Gurkan, V.; Gregory-Bryson, E.; Kiupel, M. Classification of canine nonangiogenic, nonlymphogenic, gastrointestinal sarcomas based on microscopic, immunohistochemical, and molecular characteristics. Vet. Pathol. 2013, 50, 779–788. [Google Scholar] [CrossRef]
- Kobayashi, M.; Kuroki, S.; Ito, K.; Yasuda, A.; Sawada, H.; Ono, K.; Washizu, T.; Bonkobara, M. Imatinib-associated tumour response in a dog with a non-resectable gastrointestinal stromal tumour harbouring a c-kit exon 11 deletion mutation. Vet. J. 2013, 198, 271–274. [Google Scholar] [CrossRef]
- Takanosu, M.; Amano, S.; Kagawa, Y. Analysis of c-KIT exon 11 mutations in canine gastrointestinal stromal tumours. Vet. J. 2016, 207, 118–123. [Google Scholar] [CrossRef]
- Morini, M.; Gentilini, F.; Turba, M.E.; Gobbo, F.; Mandrioli, L.; Bettini, G. Mutational Analysis of c-KIT and PDGFRA in Canine Gastrointestinal Stromal Tumors (GISTs). Vet. Sci. 2022, 9, 376. [Google Scholar] [CrossRef]
- Irie, M.; Takeuchi, Y.; Ohtake, Y.; Suzuki, H.; Nagata, N.; Miyoshi, T.; Kagawa, Y.; Yamagami, T. Imatinib mesylate treatment in a dog with gastrointestinal stromal tumors with a c-kit mutation. J. Vet. Med. Sci. 2015, 77, 1535–1539. [Google Scholar] [CrossRef]
- Murakami, A.; Mori, T.; Sakai, H.; Murakami, M.; Yanai, T.; Hoshino, Y.; Maruo, K. Analysis of KIT expression and KIT exon 11 mutations in canine oral malignant melanomas. Vet. Comp. Oncol. 2011, 9, 219–224. [Google Scholar] [CrossRef]
- Chu, P.Y.; Pan, S.L.; Liu, C.H.; Lee, J.; Yeh, L.S.; Liao, A.T. KIT gene exon 11 mutations in canine malignant melanoma. Vet. J. 2013, 196, 226–230. [Google Scholar] [CrossRef]
- Tani, H.; Miyamoto, R.; Noguchi, S.; Kurita, S.; Nagashima, T.; Michishita, M.; Yayoshi, N.; Tamura, K.; Bonkobara, M. A canine case of malignant melanoma carrying a KIT c.1725_1733del mutation treated with toceranib: A case report and in vitro analysis. BMC Vet. Res. 2021, 17, 147. [Google Scholar] [CrossRef]
- Brocca, G.; Poncina, B.; Sammarco, A.; Cavicchioli, L.; Castagnaro, M. KIT Somatic Mutations and Immunohistochemical Expression in Canine Oral Melanoma. Animals 2020, 10, 2370. [Google Scholar] [CrossRef]
- Wong, K.; van der Weyden, L.; Schott, C.R.; Foote, A.; Constantino-Casas, F.; Smith, S.; Dobson, J.M.; Murchison, E.P.; Wu, H.; Yeh, I.; et al. Cross-species genomic landscape comparison of human mucosal melanoma with canine oral and equine melanoma. Nat. Commun. 2019, 10, 353. [Google Scholar] [CrossRef]
- Smedley, R.C.; Thaiwong, T.; Deeth, L.E.; Kiupel, M. Correlation Between KIT Expression and c-Kit Mutations in 2 Subtypes of Canine Oral Melanocytic Neoplasms. Vet. Pathol. 2021, 58, 683–691. [Google Scholar] [CrossRef]
- Conrad, D.; Kehl, A.; Beitzinger, C.; Metzler, T.; Steiger, K.; Pfarr, N.; Fischer, K.; Klopfleisch, R.; Aupperle-Lellbach, H. Molecular Genetic Investigation of Digital Melanoma in Dogs. Vet. Sci. 2022, 9, 56. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Fujino, Y.; Watanabe, M.; Takahashi, M.; Nakagawa, T.; Takeuchi, A.; Bonkobara, M.; Kobayashi, T.; Ohno, K.; Uchida, K.; et al. Validation of the prognostic value of histopathological grading or c-kit mutation in canine cutaneous mast cell tumours: A retrospective cohort study. Vet. J. 2013, 196, 492–498. [Google Scholar] [CrossRef]
- Tamlin, V.S.; Kessell, A.E.; McCoy, R.J.; Dobson, E.C.; Smith, T.S.; Hebart, M.; Brown, L.; Mitrovic, D.; Peaston, A.E. Prevalence of exon 11 internal tandem duplications in the C-KIT proto-oncogene in Australian canine mast cell tumours. Aust. Vet. J. 2017, 95, 386–391. [Google Scholar] [CrossRef]
- Horta, R.S.; Lavalle, G.E.; Monteiro, L.N.; Souza, M.C.C.; Cassali, G.D.; Araújo, R.B. Assessment of Canine Mast Cell Tumor Mortality Risk Based on Clinical, Histologic, Immunohistochemical, and Molecular Features. Vet. Pathol. 2018, 55, 212–223. [Google Scholar] [CrossRef]
- Brocks, B.A.W.; Bertram, C.A.; Bartel, A.; Kirpensteijn, J.; Collins-Webb, A.; Catlin, C.; Thaiwong, T.; Kiupel, M. Internal Tandem Duplication of Exon 8 of c-kit Is Associated with Longer Total Survival in Canine Cutaneous Mast Cell Tumors. Vet. Pathol. 2021, 58, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Kiupel, M.; Camus, M. Diagnosis and Prognosis of Canine Cutaneous Mast Cell Tumors. Vet. Clin. N. Am. Small Anim. Pract. 2019, 49, 819–836. [Google Scholar] [CrossRef]
- de Nardi, A.B.; Dos Santos Horta, R.; Fonseca-Alves, C.E.; de Paiva, F.N.; Linhares, L.C.M.; Firmo, B.F.; Ruiz Sueiro, F.A.; de Oliveira, K.D.; Lourenço, S.V.; De Francisco Strefezzi, R.; et al. Diagnosis, Prognosis and Treatment of Canine Cutaneous and Subcutaneous Mast Cell Tumors. Cells 2022, 11, 618. [Google Scholar] [CrossRef] [PubMed]
- Bellamy, E.; Berlato, D. Canine cutaneous and subcutaneous mast cell tumours: A narrative review. J. Small Anim. Pract. 2022, 63, 497–511. [Google Scholar] [CrossRef] [PubMed]
- London, C.A.; Malpas, P.B.; Wood-Follis, S.L.; Boucher, J.F.; Rusk, A.W.; Rosenberg, M.P.; Henry, C.J.; Mitchener, K.L.; Klein, M.K.; Hintermeister, J.G.; et al. Multi-center, placebo-controlled, double-blind, randomized study of oral toceranib phosphate (SU11654), a receptor tyrosine kinase inhibitor, for the treatment of dogs with recurrent (either local or distant) mast cell tumor following surgical excision. Clin. Cancer Res. 2009, 15, 3856–3865. [Google Scholar] [CrossRef] [PubMed]
- Blackwood, L.; Murphy, S.; Buracco, P.; De Vos, J.P.; De Fornel-Thibaud, P.; Hirschberger, J.; Kessler, M.; Pastor, J.; Ponce, F.; Savary-Bataille, K.; et al. European consensus document on mast cell tumours in dogs and cats. Vet. Comp. Oncol. 2012, 10, e1–e29. [Google Scholar] [CrossRef]
- Pryer, N.K.; Lee, L.B.; Zadovaskaya, R.; Yu, X.; Sukbuntherng, J.; Cherrington, J.M.; London, C.A. Proof of target for SU11654: Inhibition of KIT phosphorylation in canine mast cell tumors. Clin. Cancer Res. 2003, 9, 5729–5734. [Google Scholar]
- Dubreuil, P.; Letard, S.; Ciufolini, M.; Gros, L.; Humbert, M.; Castéran, N.; Borge, L.; Hajem, B.; Lermet, A.; Sippl, W.; et al. Masitinib (AB1010), a potent and selective tyrosine kinase inhibitor targeting KIT. PLoS ONE 2009, 4, e7258. [Google Scholar] [CrossRef] [PubMed]
- London, C.A.; Hannah, A.L.; Zadovoskaya, R.; Chien, M.B.; Kollias-Baker, C.; Rosenberg, M.; Downing, S.; Post, G.; Boucher, J.; Shenoy, N.; et al. Phase I dose-escalating study of SU11654, a small molecule receptor tyrosine kinase inhibitor, in dogs with spontaneous malignancies. Clin. Cancer Res. 2003, 9, 2755–2768. [Google Scholar] [PubMed]
- Hahn, K.A.; Ogilvie, G.; Rusk, T.; Devauchelle, P.; Leblanc, A.; Legendre, A.; Powers, B.; Leventhal, P.S.; Kinet, J.P.; Palmerini, F.; et al. Masitinib is safe and effective for the treatment of canine mast cell tumors. J. Vet. Intern. Med. 2008, 22, 1301–1309. [Google Scholar] [CrossRef]
- Hahn, K.A.; Legendre, A.M.; Shaw, N.G.; Phillips, B.; Ogilvie, G.K.; Prescott, D.M.; Atwater, S.W.; Carreras, J.K.; Lana, S.E.; Ladue, T.; et al. Evaluation of 12- and 24-month survival rates after treatment with masitinib in dogs with nonresectable mast cell tumors. Am. J. Vet. Res. 2010, 71, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Kobayashi, T.; Oshima, F.; Fukazawa, E.; Yamagami, T.; Shiraishi, Y.; Takanosu, M. Imatinib responsiveness in canine mast cell tumors carrying novel mutations of c-KIT exon 11. J. Vet. Med. Sci. 2014, 76, 545–548. [Google Scholar] [CrossRef] [PubMed]
- Horta, R.D.S.; Giuliano, A.; Lavalle, G.E.; Costa, M.P.; de Araújo, R.B.; Constantino-Casas, F.; Dobson, J.M. Clinical, histological, immunohistochemical and genetic factors associated with measurable response of high-risk canine mast cell tumours to tyrosine kinase inhibitors. Oncol. Lett. 2018, 15, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Isotani, M.; Ishida, N.; Tominaga, M.; Tamura, K.; Yagihara, H.; Ochi, S.; Kato, R.; Kobayashi, T.; Fujita, M.; Fujino, Y.; et al. Effect of tyrosine kinase inhibition by imatinib mesylate on mast cell tumors in dogs. J. Vet. Intern. Med. 2008, 22, 985–988. [Google Scholar] [CrossRef] [PubMed]
- Weishaar, K.M.; Ehrhart, E.J.; Avery, A.C.; Charles, J.B.; Elmslie, R.E.; Vail, D.M.; London, C.A.; Clifford, C.A.; Eickhoff, J.C.; Thamm, D.H. c-Kit Mutation and Localization Status as Response Predictors in Mast Cell Tumors in Dogs Treated with Prednisone and Toceranib or Vinblastine. J. Vet. Intern. Med. 2018, 32, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Sugisaki, O.; Ishii, N.; Yamada, O.; Ito, K.; Kuroki, S.; Sasaki, Y.; Ono, K.; Washizu, T.; Bonkobara, M. Canine intestinal mast cell tumor with c-kit exon 8 mutation responsive to imatinib therapy. Vet. J. 2012, 193, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Verstovsek, S.; Akin, C.; Manshouri, T.; Quintás-Cardama, A.; Huynh, L.; Manley, P.; Tefferi, A.; Cortes, J.; Giles, F.J.; Kantarjian, H. Effects of AMN107, a novel aminopyrimidine tyrosine kinase inhibitor, on human mast cells bearing wild-type or mutated codon 816 c-kit. Leuk. Res. 2006, 30, 1365–1370. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Kobayashi, M.; Bonkobara, M.; Takanosu, M. Identification of a secondary mutation in the KIT kinase domain correlated with imatinib-resistance in a canine mast cell tumor. Vet. Immunol. Immunopathol. 2017, 188, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Kuroki, S.; Tanaka, Y.; Moriya, Y.; Kozutumi, Y.; Uehara, Y.; Ono, K.; Tamura, K.; Washizu, T.; Bonkobara, M. Molecular changes associated with the development of resistance to imatinib in an imatinib-sensitive canine neoplastic mast cell line carrying a KIT c.1523A>T mutation. Eur. J. Haematol. 2015, 95, 524–531. [Google Scholar] [CrossRef]
- Gentilini, F.; Turba, M.E.; Dally, C.; Takanosu, M.; Kurita, S.; Bonkobara, M. The secondary KIT mutation p.Ala510Val in a cutaneous mast cell tumour carrying the activating mutation p.Asn508Ile confers resistance to masitinib in dogs. BMC Vet. Res. 2020, 16, 64. [Google Scholar] [CrossRef]
- Tamlin, V.S.; Bottema, C.D.K.; Peaston, A.E. Comparative aspects of mast cell neoplasia in animals and the role of KIT in prognosis and treatment. Vet. Med. Sci. 2020, 6, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.W.; Swinbourne, F.; Parry, A.; Baines, L. Successful treatment of a metastatic, gastrointestinal stromal tumour in a dog with toceranib phosphate (Palladia). J. Small Anim. Pract. 2017, 58, 416–418. [Google Scholar] [CrossRef]
- Berger, E.P.; Johannes, C.M.; Jergens, A.E.; Allenspach, K.; Powers, B.E.; Du, Y.; Mochel, J.P.; Fox, L.E.; Musser, M.L. Retrospective evaluation of toceranib phosphate (Palladia®) use in the treatment of gastrointestinal stromal tumors of dogs. J. Vet. Intern. Med. 2018, 32, 2045–2053. [Google Scholar] [CrossRef] [PubMed]
- Treggiari, E.; Giantin, M.; Ferro, S.; Romanelli, G. Canine gastrointestinal stromal tumours treated with surgery and imatinib mesylate: Three cases (2018–2020). J. Small Anim. Pract. 2023, 64, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Isotani, M.; Yamada, O.; Lachowicz, J.L.; Tamura, K.; Yagihara, H.; Fujino, Y.; Ono, K.; Washizu, T.; Bonkobara, M. Mutations in the fifth immunoglobulin-like domain of kit are common and potentially sensitive to imatinib mesylate in feline mast cell tumours. Br. J. Haematol. 2010, 148, 144–153. [Google Scholar] [CrossRef]
- Sabattini, S.; Guadagni Frizzon, M.; Gentilini, F.; Turba, M.E.; Capitani, O.; Bettini, G. Prognostic significance of Kit receptor tyrosine kinase dysregulations in feline cutaneous mast cell tumors. Vet. Pathol. 2013, 50, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Dank, G.; Chien, M.B.; London, C.A. Activating mutations in the catalytic or juxtamembrane domain of c-kit in splenic mast cell tumors of cats. Am. J. Vet. Res. 2002, 63, 1129–1133. [Google Scholar] [CrossRef]
- Sabattini, S.; Barzon, G.; Giantin, M.; Lopparelli, R.M.; Dacasto, M.; Prata, D.; Bettini, G. Kit receptor tyrosine kinase dysregulations in feline splenic mast cell tumours. Vet. Comp. Oncol. 2017, 15, 1051–1061. [Google Scholar] [CrossRef]
- Isotani, M.; Tamura, K.; Yagihara, H.; Hikosaka, M.; Ono, K.; Washizu, T.; Bonkobara, M. Identification of a c-kit exon 8 internal tandem duplication in a feline mast cell tumor case and its favorable response to the tyrosine kinase inhibitor imatinib mesylate. Vet. Immunol. Immunopathol. 2006, 114, 168–172. [Google Scholar] [CrossRef]
- Sabattini, S.; Giantin, M.; Barbanera, A.; Zorro Shahidian, L.; Dacasto, M.; Zancanella, V.; Prata, D.; Trivigno, E.; Bettini, G. Feline intestinal mast cell tumours: Clinicopathological characterisation and KIT mutation analysis. J. Feline Med. Surg. 2016, 18, 280–289. [Google Scholar] [CrossRef]
- Tamlin, V.S.; Bottema, C.D.K.; Campbell-Ward, M.L.; Hanshaw, D.; Peaston, A.E. KIT mutations in mast cell tumours from cheetahs (Acinonyx jubatus) and domestic cats (Felis catus). Vet. Comp. Oncol. 2021, 19, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Žagar, Ž.; Schmidt, J.M. A Scoping Review on Tyrosine Kinase Inhibitors in Cats: Current Evidence and Future Directions. Animals 2023, 13, 3059. [Google Scholar] [CrossRef] [PubMed]
- Berger, E.P.; Johannes, C.M.; Post, G.S.; Rothchild, G.; Shiu, K.B.; Wetzel, S.; Fox, L.E. Retrospective evaluation of toceranib phosphate (Palladia) use in cats with mast cell neoplasia. J. Feline Med. Surg. 2018, 20, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Hadzijusufovic, E.; Peter, B.; Rebuzzi, L.; Baumgartner, C.; Gleixner, K.V.; Gruze, A.; Thaiwong, T.; Pickl, W.F.; Yuzbasiyan-Gurkan, V.; Willmann, M.; et al. Growth-inhibitory effects of four tyrosine kinase inhibitors on neoplastic feline mast cells exhibiting a Kit exon 8 ITD mutation. Vet. Immunol. Immunopathol. 2009, 132, 243–250. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Shosu, K.; Tsuji, K.; Shimoyama, Y.; Miyama, T.S.; Baba, K.; Okuda, M.; Itamoto, K.; Igase, M.; Mizuno, T. Intratumoral heterogeneity of c-KIT mutations in a feline splenic mast cell tumor and their functional effects on cell proliferation. Sci. Rep. 2022, 12, 15791. [Google Scholar] [CrossRef]
- Fujii, Y.; Iwasaki, R.; Ikeda, S.; Chimura, S.; Goto, M.; Yoshizaki, K.; Sakai, H.; Ito, N.; Mori, T. Gastrointestinal stromal tumour lacking mutations in the KIT and PDGFRA genes in a cat. J. Small Anim. Pract. 2022, 63, 239–243. [Google Scholar] [CrossRef]
- Vernau, W.; Moore, P.F. An immunophenotypic study of canine leukemias and preliminary assessment of clonality by polymerase chain reaction. Vet. Immunol. Immunopathol. 1999, 69, 145–164. [Google Scholar] [CrossRef]
- Burnett, R.C.; Vernau, W.; Modiano, J.F.; Olver, C.S.; Moore, P.F.; Avery, A.C. Diagnosis of canine lymphoid neoplasia using clonal rearrangements of antigen receptor genes. Vet. Pathol. 2003, 40, 32–41. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, J.J.; Langerak, A.W.; Brüggemann, M.; Evans, P.A.; Hummel, M.; Lavender, F.L.; Delabesse, E.; Davi, F.; Schuuring, E.; García-Sanz, R.; et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: Report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003, 17, 2257–2317. [Google Scholar] [CrossRef]
- Gentilini, F.; Calzolari, C.; Turba, M.E.; Bettini, G.; Famigli-Bergamini, P. GeneScanning analysis of Ig/TCR gene rearrangements to detect clonality in canine lymphomas. Vet. Immunol. Immunopathol. 2009, 127, 47–56. [Google Scholar] [CrossRef]
- Langerak, A.W.; Groenen, P.J.; Brüggemann, M.; Beldjord, K.; Bellan, C.; Bonello, L.; Boone, E.; Carter, G.I.; Catherwood, M.; Davi, F.; et al. EuroClonality/BIOMED-2 guidelines for interpretation and reporting of Ig/TCR clonality testing in suspected lymphoproliferations. Leukemia 2012, 26, 2159–2171. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.M.; Moore, P.F. A novel clonality assay for the assessment of canine T cell proliferations. Vet. Immunol. Immunopathol. 2012, 145, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Hammer, S.E.; Groiss, S.; Fuchs-Baumgartinger, A.; Nedorost, N.; Gress, V.; Luckschander-Zeller, N.; Saalmüller, A.; Schwendenwein, I.; Rütgen, B.C. Characterization of a PCR-based lymphocyte clonality assay as a complementary tool for the diagnosis of feline lymphoma. Vet. Comp. Oncol. 2017, 15, 1354–1369. [Google Scholar] [CrossRef] [PubMed]
- Radtanakatikanon, A.; Moore, P.F.; Keller, S.M.; Vernau, W. Novel clonality assays for T cell lymphoma in cats targeting the T cell receptor beta, T cell receptor delta, and T cell receptor gamma loci. J. Vet. Intern. Med. 2021, 35, 2865–2875. [Google Scholar] [CrossRef] [PubMed]
- Melendez-Lazo, A.; Jasensky, A.K.; Jolly-Frahija, I.T.; Kehl, A.; Müller, E.; Mesa-Sánchez, I. Clonality testing in the lymph nodes from dogs with lymphadenomegaly due to Leishmania infantum infection. PLoS ONE 2019, 14, e0226336. [Google Scholar] [CrossRef]
- Wallden, B.; Storhoff, J.; Nielsen, T.; Dowidar, N.; Schaper, C.; Ferree, S.; Liu, S.; Leung, S.; Geiss, G.; Snider, J.; et al. Development and verification of the PAM50-based Prosigna breast cancer gene signature assay. BMC Med. Genom. 2015, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Haan, J.C.; Bhaskaran, R.; Ellappalayam, A.; Bijl, Y.; Griffioen, C.J.; Lujinovic, E.; Audeh, W.M.; Penault-Llorca, F.; Mittempergher, L.; Glas, A.M. MammaPrint and BluePrint comprehensively capture the cancer hallmarks in early-stage breast cancer patients. Genes. Chromosomes Cancer 2022, 61, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Constantinidou, A.; Marcou, Y.; Toss, M.S.; Simmons, T.; Bernhisel, R.; Hughes, E.; Probst, B.; Meek, S.; Kakouri, E.; Georgiou, G.; et al. Clinical Validation of EndoPredict in Pre-Menopausal Women with ER-Positive, HER2-Negative Primary Breast Cancer. Clin. Cancer Res. 2022, 28, 4435–4443. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Tang, G.; Shak, S.; Kim, C.; Baker, J.; Kim, W.; Cronin, M.; Baehner, F.L.; Watson, D.; Bryant, J.; et al. Gene Expression and Benefit of Chemotherapy in Women with Node-Negative, Estrogen Receptor-Positive Breast Cancer. J. Clin. Oncol. 2023, 41, 3565–3575. [Google Scholar] [CrossRef]
- Johnson, A.; Khotskaya, Y.B.; Brusco, L.; Zeng, J.; Holla, V.; Bailey, A.M.; Litzenburger, B.C.; Sanchez, N.; Shufean, M.A.; Piha-Paul, S.; et al. Clinical Use of Precision Oncology Decision Support. JCO Precis. Oncol. 2017, 2017, 12. [Google Scholar] [CrossRef]
- Johnson, A.; Ng, P.K.; Kahle, M.; Castillo, J.; Amador, B.; Wang, Y.; Zeng, J.; Holla, V.; Vu, T.; Su, F.; et al. Actionability classification of variants of unknown significance correlates with functional effect. NPJ Precis. Oncol. 2023, 7, 67. [Google Scholar] [CrossRef]
- Wong, K.; Ludwig, L.; Krijgsman, O.; Adams, D.J.; Wood, G.A.; van der Weyden, L. Comparison of the oncogenomic landscape of canine and feline hemangiosarcoma shows novel parallels with human angiosarcoma. Dis. Model. Mech. 2021, 14, dmm.049044. [Google Scholar] [CrossRef]
- Rodney, A.R.; Skidmore, Z.L.; Grenier, J.K.; Griffith, O.L.; Miller, A.D.; Chu, S.; Ahmed, F.; Bryan, J.N.; Peralta, S.; Warren, W.C. Genomic landscape and gene expression profiles of feline oral squamous cell carcinoma. Front. Vet. Sci. 2023, 10, 1079019. [Google Scholar] [CrossRef]
- Chon, E.; Hendricks, W.; White, M.; Rodrigues, L.; Haworth, D.; Post, G. Precision Medicine in Veterinary Science. Vet. Clin. N. Am. Small Anim. Pract. 2024, 54, 501–521. [Google Scholar] [CrossRef]
- Sakthikumar, S.; Warrier, M.; Whitley, D.; Facista, S.; Adkins, J.; Aman, S.; Tsinajinnie, D.; Duran, N.; Siravegna, G.; Ahmed, Z.; et al. Genomic analysis across 53 canine cancer types reveals novel mutations and high clinical actionability potential. Vet. Comp. Oncol. 2024, 22, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Rodrigues, L.; Post, G.; Harvey, G.; White, M.; Miller, A.; Lambert, L.; Lewis, B.; Lopes, C.; Zou, J. Analyses of canine cancer mutations and treatment outcomes using real-world clinico-genomics data of 2119 dogs. NPJ Precis. Oncol. 2023, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Chon, E.; Wang, G.; Whitley, D.; Sakthikumar, S.; Warrier, M.; Wong, S.; Duran, N.; Adkins, J.; Boateng, M.; Zhu, Z.; et al. Genomic tumor analysis provides clinical guidance for the management of diagnostically challenging cancers in dogs. J. Am. Vet. Med. Assoc. 2023, 261, 668–677. [Google Scholar] [CrossRef]
- Chon, E.; Sakthikumar, S.; Tang, M.; Hamilton, M.J.; Vaughan, A.; Smith, A.; Sommer, B.; Robat, C.; Manley, C.; Mullin, C.; et al. Novel genomic prognostic biomarkers for dogs with cancer. J. Vet. Intern. Med. 2023, 37, 2410–2421. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Andrews, K.S.; Brooks, D.; Fedewa, S.A.; Manassaram-Baptiste, D.; Saslow, D.; Wender, R.C. Cancer screening in the United States, 2019: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J. Clin. 2019, 69, 184–210. [Google Scholar] [CrossRef]
- Rafalko, J.M.; Kruglyak, K.M.; McCleary-Wheeler, A.L.; Goyal, V.; Phelps-Dunn, A.; Wong, L.K.; Warren, C.D.; Brandstetter, G.; Rosentel, M.C.; DiMarzio, L.; et al. Age at cancer diagnosis by breed, weight, sex, and cancer type in a cohort of more than 3000 dogs: Determining the optimal age to initiate cancer screening in canine patients. PLoS ONE 2023, 18, e0280795. [Google Scholar] [CrossRef]
- Flory, A.; Kruglyak, K.M.; Tynan, J.A.; McLennan, L.M.; Rafalko, J.M.; Fiaux, P.C.; Hernandez, G.E.; Marass, F.; Nakashe, P.; Ruiz-Perez, C.A.; et al. Clinical validation of a next-generation sequencing-based multi-cancer early detection “liquid biopsy” blood test in over 1000 dogs using an independent testing set: The CANcer Detection in Dogs (CANDiD) study. PLoS ONE 2022, 17, e0266623. [Google Scholar] [CrossRef]
- O’Kell, A.L.; Lytle, K.M.; Cohen, T.A.; Wong, L.K.; Sandford, E.; Rafalko, J.M.; Brandstetter, G.; DiMarzio, L.R.; Phelps-Dunn, A.; Rosentel, M.C.; et al. Clinical experience with next-generation sequencing-based liquid biopsy testing for cancer detection in dogs: A review of 1500 consecutive clinical cases. J. Am. Vet. Med. Assoc. 2023, 261, 827–836. [Google Scholar] [CrossRef]
- Moe, L.; Lium, B. Hereditary multifocal renal cystadenocarcinomas and nodular dermatofibrosis in 51 German shepherd dogs. J. Small Anim. Pract. 1997, 38, 498–505. [Google Scholar] [CrossRef]
- Lingaas, F.; Comstock, K.E.; Kirkness, E.F.; Sørensen, A.; Aarskaug, T.; Hitte, C.; Nickerson, M.L.; Moe, L.; Schmidt, L.S.; Thomas, R.; et al. A mutation in the canine BHD gene is associated with hereditary multifocal renal cystadenocarcinoma and nodular dermatofibrosis in the German Shepherd dog. Hum. Mol. Genet. 2003, 12, 3043–3053. [Google Scholar] [CrossRef]
- Yu, Y.; Krupa, A.; Keesler, R.I.; Grinwis, G.C.M.; de Ruijsscher, M.; de Vos, J.; Groenen, M.A.M.; Crooijmans, R. Familial follicular cell thyroid carcinomas in a large number of Dutch German longhaired pointers. Vet. Comp. Oncol. 2022, 20, 227–234. [Google Scholar] [CrossRef]
- Yu, Y.; Bovenhuis, H.; Wu, Z.; Laport, K.; Groenen, M.A.M.; Crooijmans, R. Deleterious Mutations in the TPO Gene Associated with Familial Thyroid Follicular Cell Carcinoma in Dutch German Longhaired Pointers. Genes 2021, 12, 997. [Google Scholar] [CrossRef]
- Fulton, A.J.; Nemec, A.; Murphy, B.G.; Kass, P.H.; Verstraete, F.J. Risk factors associated with survival in dogs with nontonsillar oral squamous cell carcinoma 31 cases (1990–2010). J. Am. Vet. Med. Assoc. 2013, 243, 696–702. [Google Scholar] [CrossRef]
- Marconato, L.; Murgia, D.; Finotello, R.; Meier, V.; Morello, E.M.; Pisoni, L.; Foglia, A.; Guerra, D.; Chalfon, C.; Aralla, M.; et al. Clinical Features and Outcome of 79 Dogs with Digital Squamous Cell Carcinoma Undergoing Treatment: A SIONCOV Observational Study. Front. Vet. Sci. 2021, 8, 645982. [Google Scholar] [CrossRef]
- Wobeser, B.K.; Kidney, B.A.; Powers, B.E.; Withrow, S.J.; Mayer, M.N.; Spinato, M.T.; Allen, A.L. Diagnoses and clinical outcomes associated with surgically amputated canine digits submitted to multiple veterinary diagnostic laboratories. Vet. Pathol. 2007, 44, 355–361. [Google Scholar] [CrossRef]
- Grassinger, J.M.; Floren, A.; Müller, T.; Cerezo-Echevarria, A.; Beitzinger, C.; Conrad, D.; Törner, K.; Staudacher, M.; Aupperle-Lellbach, H. Digital Lesions in Dogs: A Statistical Breed Analysis of 2912 Cases. Vet. Sci. 2021, 8, 136. [Google Scholar] [CrossRef]
- Marino, D.J.; Matthiesen, D.T.; Stefanacci, J.D.; Moroff, S.D. Evaluation of dogs with digit masses: 117 cases (1981–1991). J. Am. Vet. Med. Assoc. 1995, 207, 726–728. [Google Scholar] [CrossRef]
- Henry, C.J.; Brewer, W.G., Jr.; Whitley, E.M.; Tyler, J.W.; Ogilvie, G.K.; Norris, A.; Fox, L.E.; Morrison, W.B.; Hammer, A.; Vail, D.M.; et al. Canine digital tumors: A veterinary cooperative oncology group retrospective study of 64 dogs. J. Vet. Intern. Med. 2005, 19, 720–724. [Google Scholar] [CrossRef]
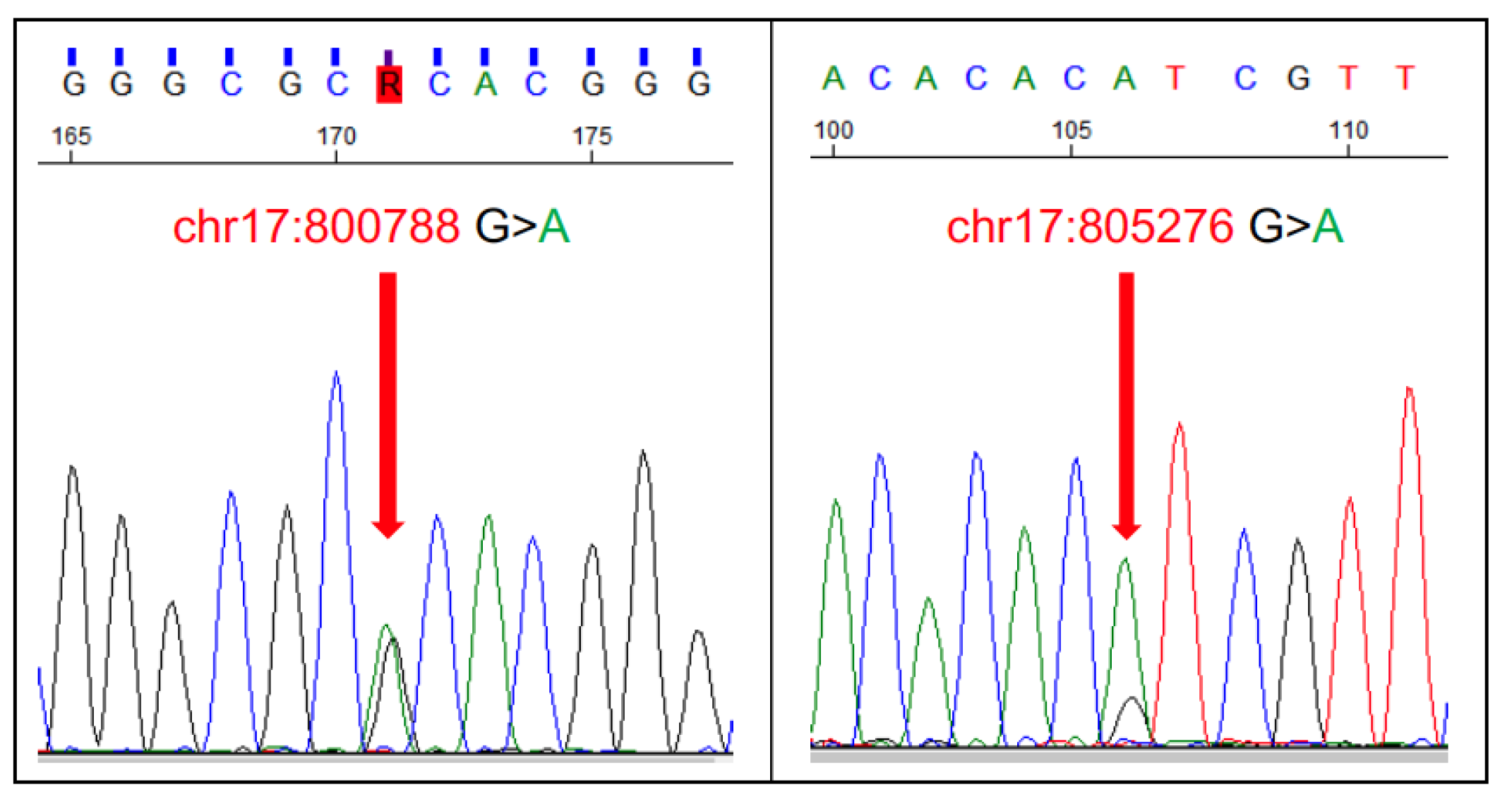
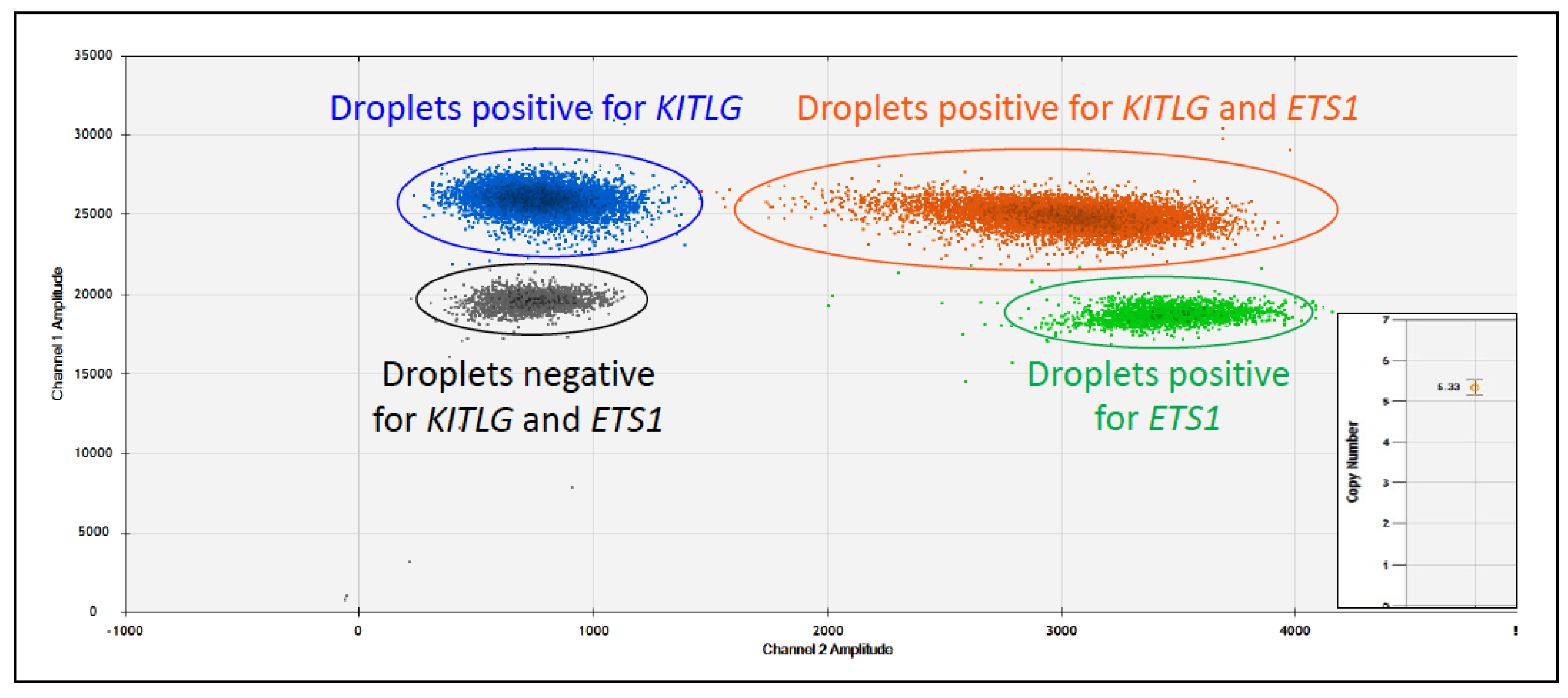
- Aupperle-Lellbach, H.; Heidrich, D.; Kehl, A.; Conrad, D.; Brockmann, M.; Törner, K.; Beitzinger, C.; Müller, T. KITLG Copy Number Germline Variations in Schnauzer Breeds and Their Relevance in Digital Squamous Cell Carcinoma in Black Giant Schnauzers. Vet. Sci. 2023, 10, 147. [Google Scholar] [CrossRef]
- Karyadi, D.M.; Karlins, E.; Decker, B.; vonHoldt, B.M.; Carpintero-Ramirez, G.; Parker, H.G.; Wayne, R.K.; Ostrander, E.A. A copy number variant at the KITLG locus likely confers risk for canine squamous cell carcinoma of the digit. PLoS Genet. 2013, 9, e1003409. [Google Scholar] [CrossRef]
- Weich, K.; Affolter, V.; York, D.; Rebhun, R.; Grahn, R.; Kallenberg, A.; Bannasch, D. Pigment Intensity in Dogs is Associated with a Copy Number Variant Upstream of KITLG. Genes 2020, 11, 75. [Google Scholar] [CrossRef]
- Bannasch, D.L.; Affolter, V.K.; York, D.; Rebhun, R.B.; Grahn, R.A.; Weich, K.M.; Kallenberg, A.; Weich, K.; Kallenberg, A. Correction: Pigment Intensity in Dogs Is Associated with a Copy Number Variant Upstream of KITLG. Genes 2020, 11, 75. Genes 2021, 12, 357. [Google Scholar] [CrossRef] [PubMed]
- Cerezo-Echevarria, A.; Kehl, A.; Beitzinger, C.; Müller, T.; Klopfleisch, R.; Aupperle-Lellbach, H. Evaluating the Histologic Grade of Digital Squamous Cell Carcinomas in Dogs and Copy Number Variation of KIT Ligand—A Correlation Study. Vet. Sci. 2023, 10, 88. [Google Scholar] [CrossRef]
- Fulmer, A.K.; Mauldin, G.E. Canine histiocytic neoplasia: An overview. Can. Vet. J. 2007, 48, 1041–1043, 1046–1050. [Google Scholar]
- Affolter, V.K.; Moore, P.F. Localized and disseminated histiocytic sarcoma of dendritic cell origin in dogs. Vet. Pathol. 2002, 39, 74–83. [Google Scholar] [CrossRef]
- Abadie, J.; Hédan, B.; Cadieu, E.; De Brito, C.; Devauchelle, P.; Bourgain, C.; Parker, H.G.; Vaysse, A.; Margaritte-Jeannin, P.; Galibert, F.; et al. Epidemiology, pathology, and genetics of histiocytic sarcoma in the Bernese mountain dog breed. J. Hered. 2009, 100 (Suppl. S1), S19–S27. [Google Scholar] [CrossRef] [PubMed]
- Shearin, A.L.; Hedan, B.; Cadieu, E.; Erich, S.A.; Schmidt, E.V.; Faden, D.L.; Cullen, J.; Abadie, J.; Kwon, E.M.; Gröne, A.; et al. The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidemiol. Biomarkers Prev. 2012, 21, 1019–1027. [Google Scholar] [CrossRef]
- Arendt, M.L.; Melin, M.; Tonomura, N.; Koltookian, M.; Courtay-Cahen, C.; Flindall, N.; Bass, J.; Boerkamp, K.; Megquir, K.; Youell, L.; et al. Genome-Wide Association Study of Golden Retrievers Identifies Germ-Line Risk Factors Predisposing to Mast Cell Tumours. PLoS Genet. 2015, 11, e1005647. [Google Scholar] [CrossRef]
- Hédan, B.; Cadieu, É.; Rimbault, M.; Vaysse, A.; Dufaure de Citres, C.; Devauchelle, P.; Botherel, N.; Abadie, J.; Quignon, P.; Derrien, T.; et al. Identification of common predisposing loci to hematopoietic cancers in four dog breeds. PLoS Genet. 2021, 17, e1009395. [Google Scholar] [CrossRef]
- Yarden, Y.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Allarakha, A.; Gao, Y.; Jiang, H.; Wang, P.J. Prediction and prognosis of biologically aggressive breast cancers by the combination of DWI/DCE-MRI and immunohistochemical tumor markers. Discov. Med. 2019, 27, 7–15. [Google Scholar]
- Bose, R.; Kavuri, S.M.; Searleman, A.C.; Shen, W.; Shen, D.; Koboldt, D.C.; Monsey, J.; Goel, N.; Aronson, A.B.; Li, S.; et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013, 3, 224–237. [Google Scholar] [CrossRef]
- Ahn, S.; Woo, J.W.; Lee, K.; Park, S.Y. HER2 status in breast cancer: Changes in guidelines and complicating factors for interpretation. J. Pathol. Transl. Med. 2020, 54, 34–44. [Google Scholar] [CrossRef]
- Molinelli, C.; Jacobs, F.; Marchiò, C.; Pitto, F.; Cosso, M.; Spinaci, S.; de Azambuja, E.; Schettini, F.; Agostinetto, E.; Lambertini, M. HER2-Low Breast Cancer: Where Are We? Breast Care 2022, 17, 533–545. [Google Scholar] [CrossRef]
- Molinelli, C.; Jacobs, F.; Agostinetto, E.; Nader-Marta, G.; Ceppi, M.; Bruzzone, M.; Blondeaux, E.; Schettini, F.; Prat, A.; Viale, G.; et al. Prognostic value of HER2-low status in breast cancer: A systematic review and meta-analysis. ESMO Open 2023, 8, 101592. [Google Scholar] [CrossRef] [PubMed]
- Rassy, E.; Rached, L.; Pistilli, B. Antibody drug conjugates targeting HER2: Clinical development in metastatic breast cancer. Breast 2022, 66, 217–226. [Google Scholar] [CrossRef]
- Riudavets, M.; Sullivan, I.; Abdayem, P.; Planchard, D. Targeting HER2 in non-small-cell lung cancer (NSCLC): A glimpse of hope? An updated review on therapeutic strategies in NSCLC harbouring HER2 alterations. ESMO Open 2021, 6, 100260. [Google Scholar] [CrossRef]
- Singer, J.; Weichselbaumer, M.; Stockner, T.; Mechtcheriakova, D.; Sobanov, Y.; Bajna, E.; Wrba, F.; Horvat, R.; Thalhammer, J.G.; Willmann, M.; et al. Comparative oncology: ErbB-1 and ErbB-2 homologues in canine cancer are susceptible to cetuximab and trastuzumab targeting. Mol. Immunol. 2012, 50, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Ressel, L.; Puleio, R.; Loria, G.R.; Vannozzi, I.; Millanta, F.; Caracappa, S.; Poli, A. HER-2 expression in canine morphologically normal, hyperplastic and neoplastic mammary tissues and its correlation with the clinical outcome. Res. Vet. Sci. 2013, 94, 299–305. [Google Scholar] [CrossRef]
- Brunetti, B.; Bacci, B.; Sarli, G.; Pancioni, E.; Muscatello, L.V. Immunohistochemical Screening of HER2 in Canine Carcinomas: A Preliminary Study. Animals 2021, 11, 1006. [Google Scholar] [CrossRef]
- Yoshimoto, S.; Kato, D.; Kamoto, S.; Yamamoto, K.; Tsuboi, M.; Shinada, M.; Ikeda, N.; Tanaka, Y.; Yoshitake, R.; Eto, S.; et al. Detection of human epidermal growth factor receptor 2 overexpression in canine anal sac gland carcinoma. J. Vet. Med. Sci. 2019, 81, 1034–1039. [Google Scholar] [CrossRef]
- Sakai, K.; Maeda, S.; Saeki, K.; Yoshitake, R.; Goto-Koshino, Y.; Nakagawa, T.; Nishimura, R.; Yonezawa, T.; Matsuki, N. ErbB2 Copy Number Aberration in Canine Urothelial Carcinoma Detected by a Digital Polymerase Chain Reaction Assay. Vet. Pathol. 2020, 57, 56–65. [Google Scholar] [CrossRef]
- Sakai, K.; Chambers, J.K.; Uchida, K.; Nakagawa, T.; Nishimura, R.; Yonezawa, T.; Maeda, S. ErbB2 copy number gain is associated with adverse outcome in canine mammary carcinoma. J. Vet. Med. Sci. 2021, 83, 370–377. [Google Scholar] [CrossRef]
- Muscatello, L.V.; Gobbo, F.; Di Oto, E.; Sarli, G.; De Maria, R.; De Leo, A.; Tallini, G.; Brunetti, B. HER2 Overexpression and Cytogenetical Patterns in Canine Mammary Carcinomas. Vet. Sci. 2022, 9, 583. [Google Scholar] [CrossRef] [PubMed]
- Lorch, G.; Sivaprakasam, K.; Zismann, V.; Perdigones, N.; Contente-Cuomo, T.; Nazareno, A.; Facista, S.; Wong, S.; Drenner, K.; Liang, W.S.; et al. Identification of Recurrent Activating HER2 Mutations in Primary Canine Pulmonary Adenocarcinoma. Clin. Cancer Res. 2019, 25, 5866–5877. [Google Scholar] [CrossRef]
- Yu, T.W.; Yamamoto, H.; Morita, S.; Fukushima, R.; Elbadawy, M.; Usui, T.; Sasaki, K. Comparative pharmacokinetics of tyrosine kinase inhibitor, lapatinib, in dogs and cats following single oral administration. J. Vet. Med. Sci. 2024, 86, 317–321. [Google Scholar] [CrossRef]
- Millanta, F.; Calandrella, M.; Citi, S.; Della Santa, D.; Poli, A. Overexpression of HER-2 in feline invasive mammary carcinomas: An immunohistochemical survey and evaluation of its prognostic potential. Vet. Pathol. 2005, 42, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Soares, M.; Correia, J.; Rodrigues, P.; Simões, M.; de Matos, A.; Ferreira, F. Feline HER2 protein expression levels and gene status in feline mammary carcinoma: Optimization of immunohistochemistry (IHC) and in situ hybridization (ISH) techniques. Microsc. Microanal. 2013, 19, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Muscatello, L.V.; Di Oto, E.; Sarli, G.; Monti, V.; Foschini, M.P.; Benazzi, C.; Brunetti, B. HER2 Amplification Status in Feline Mammary Carcinoma: A Tissue Microarray-Fluorescence In Situ Hydridization-Based Study. Vet. Pathol. 2019, 56, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Maniscalco, L.; Iussich, S.; de Las Mulas, J.M.; Millán, Y.; Biolatti, B.; Sasaki, N.; Nakagawa, T.; De Maria, R. Activation of AKT in feline mammary carcinoma: A new prognostic factor for feline mammary tumours. Vet. J. 2012, 191, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Muscatello, L.V.; Oto, E.D.; Dignazzi, M.; Murphy, W.J.; Porcellato, I.; De Maria, R.; Raudsepp, T.; Foschini, M.P.; Sforna, M.; Benazzi, C.; et al. HER2 Overexpression and Amplification in Feline Pulmonary Carcinoma. Vet. Pathol. 2021, 58, 527–530. [Google Scholar] [CrossRef] [PubMed]
- Gameiro, A.; Almeida, F.; Nascimento, C.; Correia, J.; Ferreira, F. Tyrosine Kinase Inhibitors Are Promising Therapeutic Tools for Cats with HER2-Positive Mammary Carcinoma. Pharmaceutics 2021, 13, 346. [Google Scholar] [CrossRef] [PubMed]
- Gameiro, A.; Nascimento, C.; Correia, J.; Ferreira, F. HER2-Targeted Immunotherapy and Combined Protocols Showed Promising Antiproliferative Effects in Feline Mammary Carcinoma Cell-Based Models. Cancers 2021, 13, 2007. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef]
- VanLandingham, N.K.; Nazarenko, A.; Grandis, J.R.; Johnson, D.E. The mutational profiles and corresponding therapeutic implications of PI3K mutations in cancer. Adv. Biol. Regul. 2023, 87, 100934. [Google Scholar] [CrossRef] [PubMed]
- Alsaihati, B.A.; Ho, K.L.; Watson, J.; Feng, Y.; Wang, T.; Dobbin, K.K.; Zhao, S. Canine tumor mutational burden is correlated with TP53 mutation across tumor types and breeds. Nat. Commun. 2021, 12, 4670. [Google Scholar] [CrossRef]
- Wang, G.; Wu, M.; Maloneyhuss, M.A.; Wojcik, J.; Durham, A.C.; Mason, N.J.; Roth, D.B. Actionable mutations in canine hemangiosarcoma. PLoS ONE 2017, 12, e0188667. [Google Scholar] [CrossRef] [PubMed]
- Megquier, K.; Turner-Maier, J.; Swofford, R.; Kim, J.-H.; Sarver, A.L.; Wang, C.; Sakthikumar, S.; Johnson, J.; Koltookian, M.; Lewellen, M.; et al. Comparative Genomics Reveals Shared Mutational Landscape in Canine Hemangiosarcoma and Human Angiosarcoma. Mol. Cancer Res. 2019, 17, 2410–2421. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wu, M.; Durham, A.C.; Radaelli, E.; Mason, N.J.; Xu, X.; Roth, D.B. Molecular subtypes in canine hemangiosarcoma reveal similarities with human angiosarcoma. PLoS ONE 2020, 15, e0229728. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Megquier, K.; Thomas, R.; Sarver, A.L.; Song, J.M.; Kim, Y.T.; Cheng, N.; Schulte, A.J.; Linden, M.A.; Murugan, P.; et al. Genomically Complex Human Angiosarcoma and Canine Hemangiosarcoma Establish Convergent Angiogenic Transcriptional Programs Driven by Novel Gene Fusions. Mol. Cancer Res. 2021, 19, 847–861. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Hwang, H.J.; Noh, H.J.; Shin, T.J.; Cho, J.Y. Somatic Mutation of PIK3CA (H1047R) Is a Common Driver Mutation Hotspot in Canine Mammary Tumors as Well as Human Breast Cancers. Cancers 2019, 11, 2006. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.M.; Yang, I.S.; Seung, B.J.; Lee, S.; Kim, D.; Ha, Y.J.; Seo, M.K.; Kim, K.K.; Kim, H.S.; Cheong, J.H.; et al. Cross-species oncogenic signatures of breast cancer in canine mammary tumors. Nat. Commun. 2020, 11, 3616. [Google Scholar] [CrossRef] [PubMed]
- Arendt, M.L.; Sakthikumar, S.; Melin, M.; Elvers, I.; Rivera, P.; Larsen, M.; Saellström, S.; Lingaas, F.; Rönnberg, H.; Lindblad-Toh, K. PIK3CA is recurrently mutated in canine mammary tumors, similarly to in human mammary neoplasia. Sci. Rep. 2023, 13, 632. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.B.; Anderson, K.J.; Boudreau, C.E.; Martinez-Ledesma, E.; Kocakavuk, E.; Johnson, K.C.; Barthel, F.P.; Varn, F.S.; Kassab, C.; Ling, X.; et al. Comparative Molecular Life History of Spontaneous Canine and Human Gliomas. Cancer Cell 2020, 37, 243–257.e7. [Google Scholar] [CrossRef] [PubMed]
- Sakthikumar, S.; Elvers, I.; Kim, J.; Arendt, M.L.; Thomas, R.; Turner-Maier, J.; Swofford, R.; Johnson, J.; Schumacher, S.E.; Alföldi, J.; et al. SETD2 Is Recurrently Mutated in Whole-Exome Sequenced Canine Osteosarcoma. Cancer Res. 2018, 78, 3421–3431. [Google Scholar] [CrossRef]
- Gardner, H.L.; Sivaprakasam, K.; Briones, N.; Zismann, V.; Perdigones, N.; Drenner, K.; Facista, S.; Richholt, R.; Liang, W.; Aldrich, J.; et al. Canine osteosarcoma genome sequencing identifies recurrent mutations in DMD and the histone methyltransferase gene SETD2. Commun. Biol. 2019, 2, 266. [Google Scholar] [CrossRef]
- Maeda, M.; Ochiai, K.; Michishita, M.; Morimatsu, M.; Sakai, H.; Kinoshita, N.; Sakaue, M.; Onozawa, E.; Azakami, D.; Yamamoto, M.; et al. In vitro anticancer effects of alpelisib against PIK3CA-mutated canine hemangiosarcoma cell lines. Oncol. Rep. 2022, 47, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yeom, J.; Cho, Y.; Ahn, S.; Jeung, S. Anticancer effects of alpelisib on PIK3CA-mutated canine mammary tumor cell lines. Front. Vet. Sci. 2023, 10, 1279535. [Google Scholar] [CrossRef]
- Inglebert, M.; Dettwiler, M.; Hahn, K.; Letko, A.; Drogemuller, C.; Doench, J.; Brown, A.; Memari, Y.; Davies, H.R.; Degasperi, A.; et al. A living biobank of canine mammary tumor organoids as a comparative model for human breast cancer. Sci. Rep. 2022, 12, 18051. [Google Scholar] [CrossRef]
- Murase, Y.; Hosoya, K.; Sato, T.; Kim, S.; Okumura, M. Antitumor activity of the dual PI3K/mTOR inhibitor gedatolisib and the involvement of ABCB1 in gedatolisib resistance in canine tumor cells. Oncol. Rep. 2022, 47, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, B.; Downward, J. Genomic Determinants of PI3K Pathway Inhibitor Response in Cancer. Front. Oncol. 2012, 2, 109. [Google Scholar] [CrossRef] [PubMed]
- Tonomura, N.; Elvers, I.; Thomas, R.; Megquier, K.; Turner-Maier, J.; Howald, C.; Sarver, A.L.; Swofford, R.; Frantz, A.M.; Ito, D.; et al. Genome-wide association study identifies shared risk loci common to two malignancies in golden retrievers. PLoS Genet. 2015, 11, e1004922. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.M.; Parker, H.G.; Rutteman, G.R.; Plassais, J.; Grinwis, G.C.M.; Harris, A.C.; Lana, S.E.; Ostrander, E.A. Multi-omics approach identifies germline regulatory variants associated with hematopoietic malignancies in retriever dog breeds. PLoS Genet. 2021, 17, e1009543. [Google Scholar] [CrossRef] [PubMed]
- Yoshizaki, K.; Hirata, A.; Nishii, N.; Kawabe, M.; Goto, M.; Mori, T.; Sakai, H. Familial adenomatous polyposis in dogs: Hereditary gastrointestinal polyposis in Jack Russell Terriers with germline APC mutations. Carcinogenesis 2021, 42, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Yoneji, W.; Yoshizaki, K.; Hirota, T.; Yoneji, K.; Yoshikawa, R.; Mori, T.; Sakai, H.; Hirata, A. First Evidence of Familial Transmission of Hereditary Gastrointestinal Polyposis Associated with Germline APC Variant in Jack Russell Terriers. Vet. Sci. 2023, 10, 439. [Google Scholar] [CrossRef]
- Rivera, P.; Melin, M.; Biagi, T.; Fall, T.; Häggström, J.; Lindblad-Toh, K.; von Euler, H. Mammary tumor development in dogs is associated with BRCA1 and BRCA2. Cancer Res. 2009, 69, 8770–8774. [Google Scholar] [CrossRef]
- Enginler, S.O.; Akış, I.; Toydemir, T.S.; Oztabak, K.; Haktanir, D.; Gündüz, M.C.; Kırşan, I.; Fırat, I. Genetic variations of BRCA1 and BRCA2 genes in dogs with mammary tumours. Vet. Res. Commun. 2014, 38, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Maués, T.; El-Jaick, K.B.; Costa, F.B.; Araujo, G.E.F.; Soares, M.V.G.; Moreira, A.S.; Ferreira, M.L.G.; Ferreira, A.M.R. Common germline haplotypes and genotypes identified in BRCA2 exon 11 of dogs with mammary tumours and histopathological analyses. Vet. Comp. Oncol. 2018, 16, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, D.; Di Domenico, M.; Defourny, S.V.P.; Malatesta, D.; Di Teodoro, G.; Martino, M.; Viola, A.; D’Alterio, N.; Cammà, C.; Modesto, P.; et al. Validation of AmpliSeq NGS Panel for BRCA1 and BRCA2 Variant Detection in Canine Formalin-Fixed Paraffin-Embedded Mammary Tumors. Life 2022, 12, 851. [Google Scholar] [CrossRef] [PubMed]
- Melin, M.; Rivera, P.; Arendt, M.; Elvers, I.; Murén, E.; Gustafson, U.; Starkey, M.; Borge, K.S.; Lingaas, F.; Häggström, J.; et al. Genome-Wide Analysis Identifies Germ-Line Risk Factors Associated with Canine Mammary Tumours. PLoS Genet. 2016, 12, e1006029. [Google Scholar] [CrossRef] [PubMed]
- Biasoli, D.; Compston-Garnett, L.; Ricketts, S.L.; Birand, Z.; Courtay-Cahen, C.; Fineberg, E.; Arendt, M.; Boerkamp, K.; Melin, M.; Koltookian, M.; et al. A synonymous germline variant in a gene encoding a cell adhesion molecule is associated with cutaneous mast cell tumour development in Labrador and Golden Retrievers. PLoS Genet. 2019, 15, e1007967. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Stephenson, B.; Hauck, M.; Dillberger, J. Heritability and segregation analysis of osteosarcoma in the Scottish deerhound. Genomics 2007, 90, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, E.K.; Sigurdsson, S.; Ivansson, E.; Thomas, R.; Elvers, I.; Wright, J.; Howald, C.; Tonomura, N.; Perloski, M.; Swofford, R.; et al. Genome-wide analyses implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near CDKN2A/B. Genome Biol. 2013, 14, R132. [Google Scholar] [CrossRef] [PubMed]
- Letko, A.; Minor, K.M.; Norton, E.M.; Marinescu, V.D.; Drögemüller, M.; Ivansson, E.; Megquier, K.; Noh, H.J.; Starkey, M.; Friedenberg, S.G.; et al. Genome-Wide Analyses for Osteosarcoma in Leonberger Dogs Reveal the CDKN2A/B Gene Locus as a Major Risk Locus. Genes 2021, 12, 1964. [Google Scholar] [CrossRef] [PubMed]
- Hendrick, M.J.; Shofer, F.S.; Goldschmidt, M.H.; Haviland, J.C.; Schelling, S.H.; Engler, S.J.; Gliatto, J.M. Comparison of fibrosarcomas that developed at vaccination sites and at nonvaccination sites in cats: 239 cases (1991–1992). J. Am. Vet. Med. Assoc. 1994, 205, 1425–1429. [Google Scholar] [CrossRef]
- Banerji, N.; Kapur, V.; Kanjilal, S. Association of germ-line polymorphisms in the feline p53 gene with genetic predisposition to vaccine-associated feline sarcoma. J. Hered. 2007, 98, 421–427. [Google Scholar] [CrossRef]
- Mucha, D.; Laberke, S.; Meyer, S.; Hirschberger, J. Lack of association between p53 SNP and FISS in a cat population from Germany. Vet. Comp. Oncol. 2014, 12, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Baptista, C.S.; Santos, S.; Laso, A.; Bastos, E.; Avila, S.; Guedes-Pinto, H.; Gärtner, F.; Gut, I.G.; Castrillo, J.L.; Chaves, R. Sequence variation and mRNA expression of the TWIST1 gene in cats with mammary hyperplasia and neoplasia. Vet. J. 2012, 191, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Govoni, V.M.; Da Silva, T.C.; Guerra, J.M.; Pereira, I.V.A.; Queiroga, F.L.; Cogliati, B. Genetic variants of BRCA1 and BRCA2 genes in cats with mammary gland carcinoma. Vet. Comp. Oncol. 2021, 19, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Batis, N.; Brooks, J.M.; Payne, K.; Sharma, N.; Nankivell, P.; Mehanna, H. Lack of predictive tools for conventional and targeted cancer therapy: Barriers to biomarker development and clinical translation. Adv. Drug Deliv. Rev. 2021, 176, 113854. [Google Scholar] [CrossRef] [PubMed]
- Vaught, J.B.; Caboux, E.; Hainaut, P. International efforts to develop biospecimen best practices. Cancer Epidemiol. Biomark. Prev. 2010, 19, 912–915. [Google Scholar] [CrossRef]
- Pascual, J.; Attard, G.; Bidard, F.C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef]
- Mandelker, D.; Donoghue, M.; Talukdar, S.; Bandlamudi, C.; Srinivasan, P.; Vivek, M.; Jezdic, S.; Hanson, H.; Snape, K.; Kulkarni, A.; et al. Germline-focussed analysis of tumour-only sequencing: Recommendations from the ESMO Precision Medicine Working Group. Ann. Oncol. 2019, 30, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Fürdös, I.; Fazekas, J.; Singer, J.; Jensen-Jarolim, E. Translating clinical trials from human to veterinary oncology and back. J. Transl. Med. 2015, 13, 265. [Google Scholar] [CrossRef] [PubMed]
- Adami, C.; Murrell, J.; Fordyce, P. Ethical considerations in clinical veterinary research. Vet. J. 2023, 300–302, 106026. [Google Scholar] [CrossRef]
- Hampshire, V.A. Regulatory Issues Surrounding the Use of Companion Animals in Clinical Investigations, Trials, and Studies. ILAR J. 2003, 44, 191–196. [Google Scholar] [CrossRef]
- Pestrin, M.; Bessi, S.; Galardi, F.; Truglia, M.; Biggeri, A.; Biagioni, C.; Cappadona, S.; Biganzoli, L.; Giannini, A.; Di Leo, A. Correlation of HER2 status between primary tumors and corresponding circulating tumor cells in advanced breast cancer patients. Breast Cancer Res. Treat. 2009, 118, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Stanta, G.; Bonin, S. Overview on Clinical Relevance of Intra-Tumor Heterogeneity. Front. Med. 2018, 5, 85. [Google Scholar] [CrossRef] [PubMed]








| Tumour Type | Affected Breed(s) | Molecular Alteration | Ref |
|---|---|---|---|
| B-cell lymphoma | BMD, GR, FCR | Two loci on CFA5 | [186,229,230] |
| Gastro-intestinal polyposis | Jack Russell Terrier (in Japan) | 2-bp substitution in APC. The second base pair substitution is a missense mutation (APC p.K155X), resulting in a truncated protein. Transgenerational transmission of hereditary gastrointestinal polyposis associated with the germline APC variant has been observed in Jack Russell Terrier lineages in Japan | [231,232] |
| HS | GR, BMD | Haplotypes harbouring the hyaluronidase genes HYAL1, HYAL2, and HYAL3 on CFA20 | [186] |
| HSA | GR | Two loci on CFA5 (same loci as B-cell lymphoma) | [229] |
| Mammary tumour | ESS and a variety of breeds | SNPs in BRCA1 and BRCA2 | [233,234,235,236] |
| Mammary tumour | ESS | Haplotype on CFA11 spanning a 446 kb region, overlapping CDK5RAP2 and two loci on CFA27 | [237] |
| MCT | GR | SNP in GNAI2, introducing an alternative splice form of the gene and resulting in a truncated protein in European and US populations. Haplotypes harbouring the hyaluronidase genes HYAL1, HYAL2, and HYAL3 on CFA20 in European populations. Haplotypes harbouring the hyaluronidase genes HYAL4, SPAM1, and HYALP1 on CFA14 in US populations | [185] |
| MCT | BMD | Haplotypes harbouring the hyaluronidase genes HYAL1, HYAL2 and HYAL3, on CFA20 | [186] |
| MCT (skin) | GR, Labrador Retriever | Synonymous variant in DSCAM on CFA31 | [238] |
| OSA | Scottish Deerhound | Haplotype on CFA34 | [239] |
| OSA | Greyhound, Leonberger | A rearranged locus 150 kilobases upstream of CDKN2A/B on CFA11 alters the regulation of downstream genes | [240,241] |
| Comparator | Human Medicine | Veterinary Medicine | Veterinary Medicine Hurdles |
|---|---|---|---|
| Techniques used in routine diagnostics | A broad spectrum of techniques are available. | Mainly PCR/ddPCR. Sequencing is starting to be used, but there has been no analysis of methylation or translocation status. | The stage of research is about 20 years behind that of human medicine. |
| Use of liquid biopsies | There is a lot of research on ctDNA monitoring of patients with specific tumour types and liquid biopsies routinely used in the clinic. | Very few ctDNA monitoring studies in dogs and none in cats. Liquid biopsies are not routinely used in the clinic. | Limited knowledge about genetic alterations of the tumours exists (the use of liquid biopsies is in an early stage of research). |
| Use of multi-gene sequencing panels in routine diagnostics | Multiple panels are available and routinely used in the clinic. | A few panels are currently available, but they are not part of routine diagnostics. | Financial limitations because owners have to pay and the tests are expensive. |
| Use of somatic alterations found in tumours as molecular biomarkers in the clinic | Numerous diagnostic, prognostic, predictive, and screening molecular biomarkers are clinically available for specific tumour types (eg., lung, breast, colorectal, and thyroid cancer). | Only two are available in the clinic: BRAF (used as a diagnostic and predictive biomarker, also used in a monitoring setting) and KIT (used as a prognostic and predictive biomarker). | Limited knowledge about genetic alterations of the tumours, and limitations in study design and availability of clinical trials to allow translation of research findings to the clinic. The cost to the owner is also an issue. |
| Use of germline alterations as molecular biomarkers in the clinic | Numerous molecular biomarkers are routinely used in the clinic for screening and monitoring patients (e.g., BRCA1/2). | Some germline variants have been identified as risk factors and may be used when considering breeding aspects. | Pedigrees must be available. The cost to the owner is also an issue. |
| Treatment opportunities available | Tumour boards, standardized therapeutic concepts, and intense monitoring of the patients with first, second, and third-line therapies in case of recurrence or resistance in specific tumour diseases. | Rarely do tumour boards exist, lack of uniform protocols for the management of cancer, and limited knowledge on dosage/effectivity/side effects of new drugs (or when using human drugs off-label). | Very high costs for the owners, limited availability of new drugs (even limited availability of access to chemotherapy and radiation therapy). |
| State of clinical studies/research | Existence of cancer centres and cancer registries. Numerous medical universities are carrying out clinical research. Clinical studies conducted by the pharmaceutical industry. | There are no cancer centers or cancer registries. There are few veterinary universities and no sponsorship of clinical studies by the pharmaceutical industry. | There are few studies involving canine tumours, even less feline tumours. Poor compliance by the owners. Follow-up data are very difficult to collect. |
| Financial aspects | Sponsoring of research/clinical trials by pharmaceutical industry. Government subsidisation (and/or private insurance coverage) of some diagnostic and treatment costs after the establishment of the method. | There is a very small investment by pharmaceutical industry into sponsoring research/clinical trials. There is no government subsidisation of any costs, and pet insurance only offers limited coverage of diagnostic/treatment costs (and often does not cover precision medicine-related costs as not yet considered ‘routine’). | The owner may not be able to afford the expensive diagnostic tests/treatments and thus decide to euthanise the pet instead of using precision medicine. |
| Ethics | Rules exist regarding patient counselling, patient data confidentiality, ensuring the patient’s best interests, etc. The patient decides on the use of precision medicine after discussion with the clinician. | No defined rules on patient conselling and data confidentiality. The owner decides on the use of precision medicine based on financial and personal reasons, which may not necessarily be in the animal’s best interests. | Technical and financial limitations. There are also limitations in terms of the education of veterinarians about the field of precision medicine (due to its new/emerging/nature). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aupperle-Lellbach, H.; Kehl, A.; de Brot, S.; van der Weyden, L. Clinical Use of Molecular Biomarkers in Canine and Feline Oncology: Current and Future. Vet. Sci. 2024, 11, 199. https://doi.org/10.3390/vetsci11050199
Aupperle-Lellbach H, Kehl A, de Brot S, van der Weyden L. Clinical Use of Molecular Biomarkers in Canine and Feline Oncology: Current and Future. Veterinary Sciences. 2024; 11(5):199. https://doi.org/10.3390/vetsci11050199
Chicago/Turabian StyleAupperle-Lellbach, Heike, Alexandra Kehl, Simone de Brot, and Louise van der Weyden. 2024. "Clinical Use of Molecular Biomarkers in Canine and Feline Oncology: Current and Future" Veterinary Sciences 11, no. 5: 199. https://doi.org/10.3390/vetsci11050199






