Octoploids Show Enhanced Salt Tolerance through Chromosome Doubling in Switchgrass (Panicum virgatum L.)
Abstract
:1. Introduction
2. Results
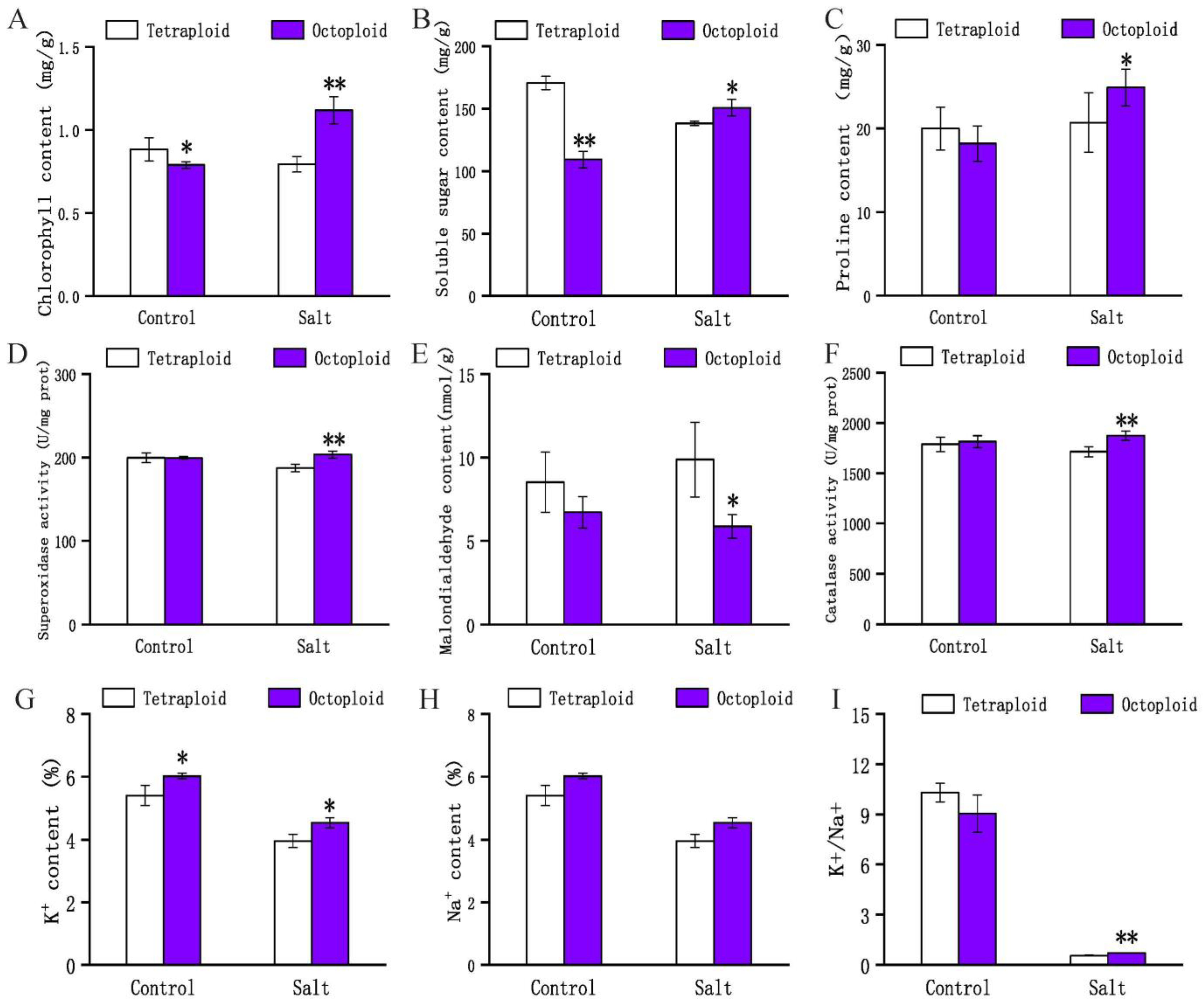
2.1. Phenotypic and Physiological Changes in Tetraploid and Octoploid Switchgrass in Response to Salt Stress
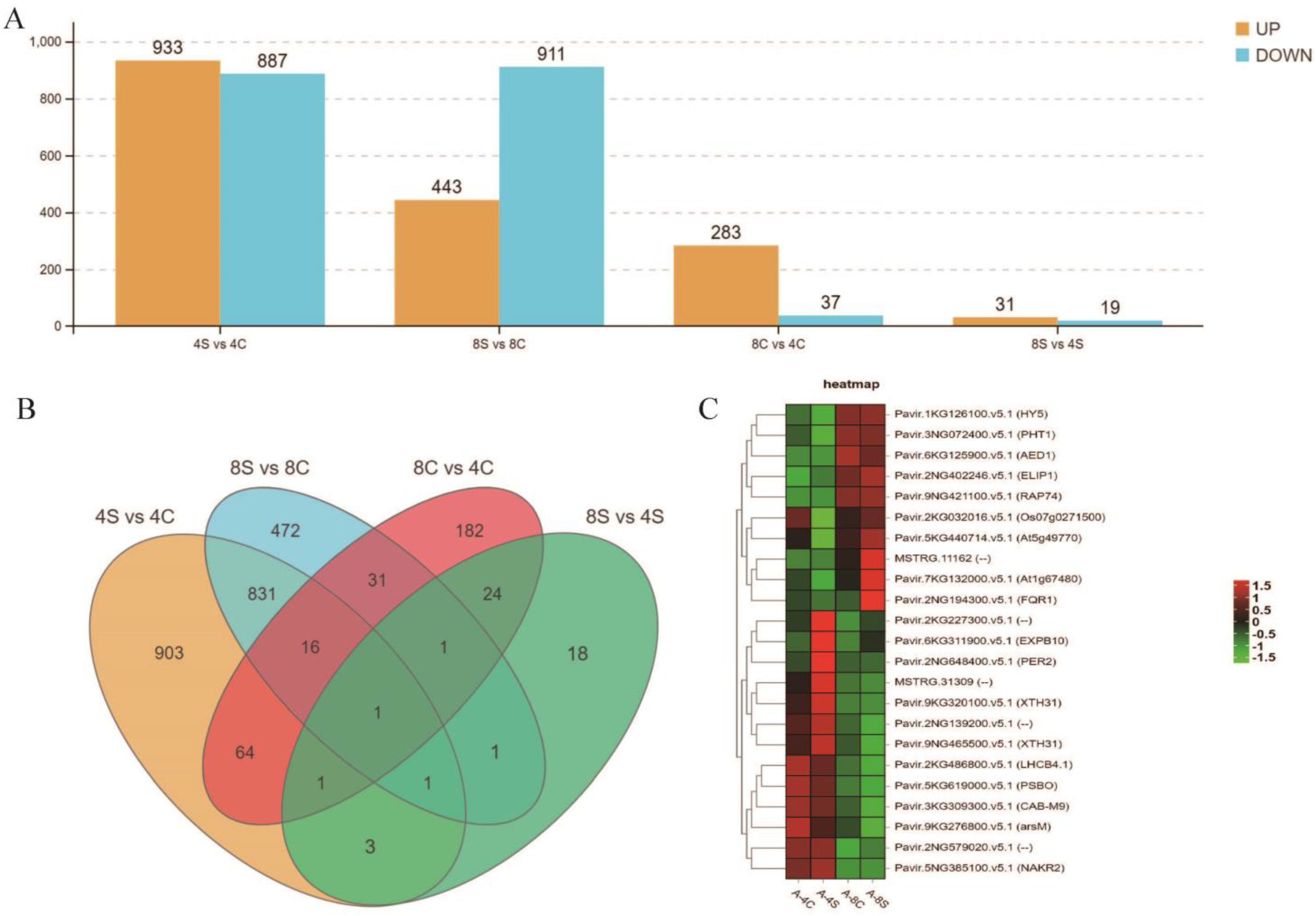
2.2. Transcriptome Sequencing
2.3. Identification of Differentially Expressed Genes
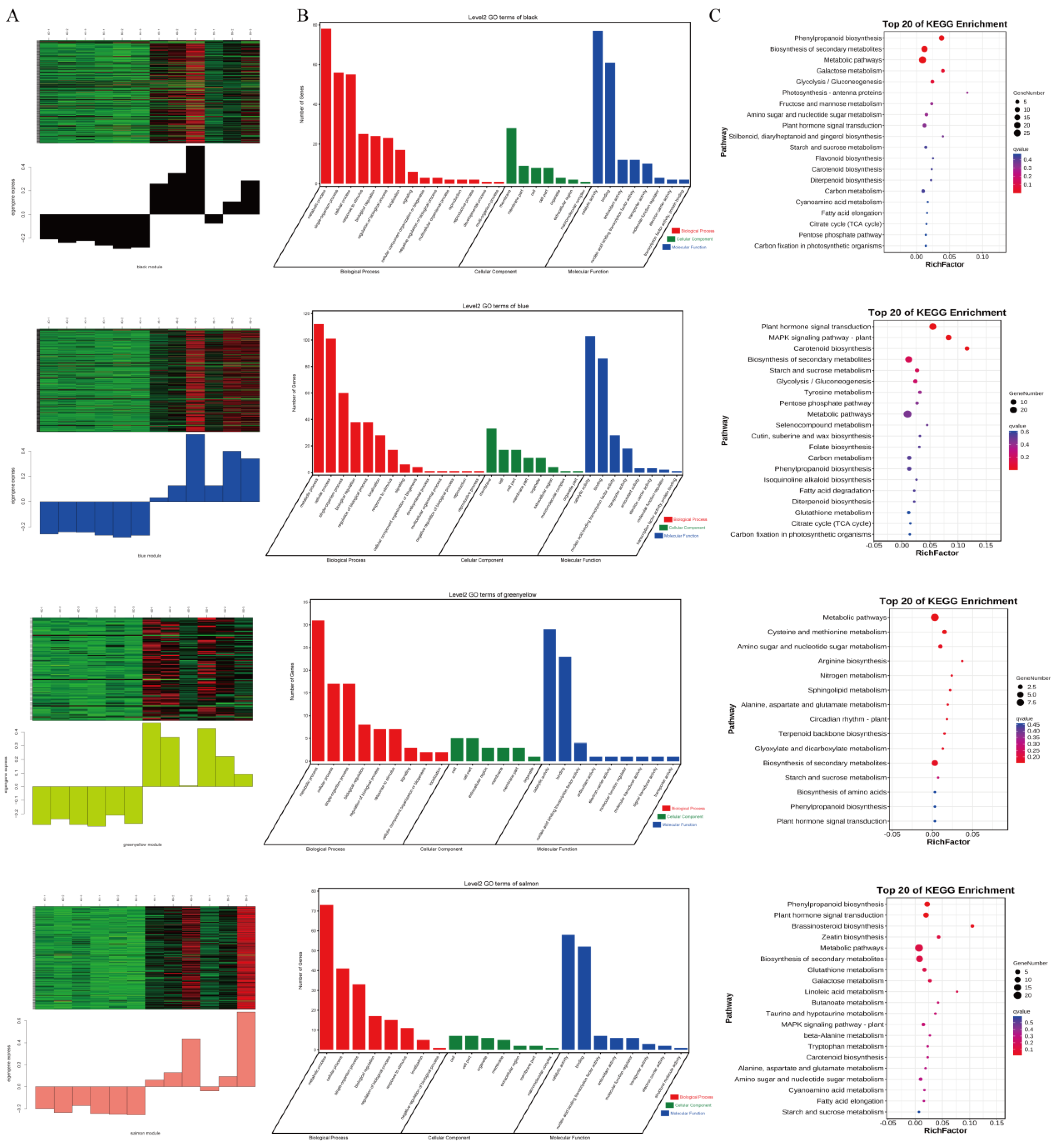
2.4. Go and KEGG Pathway Analysis
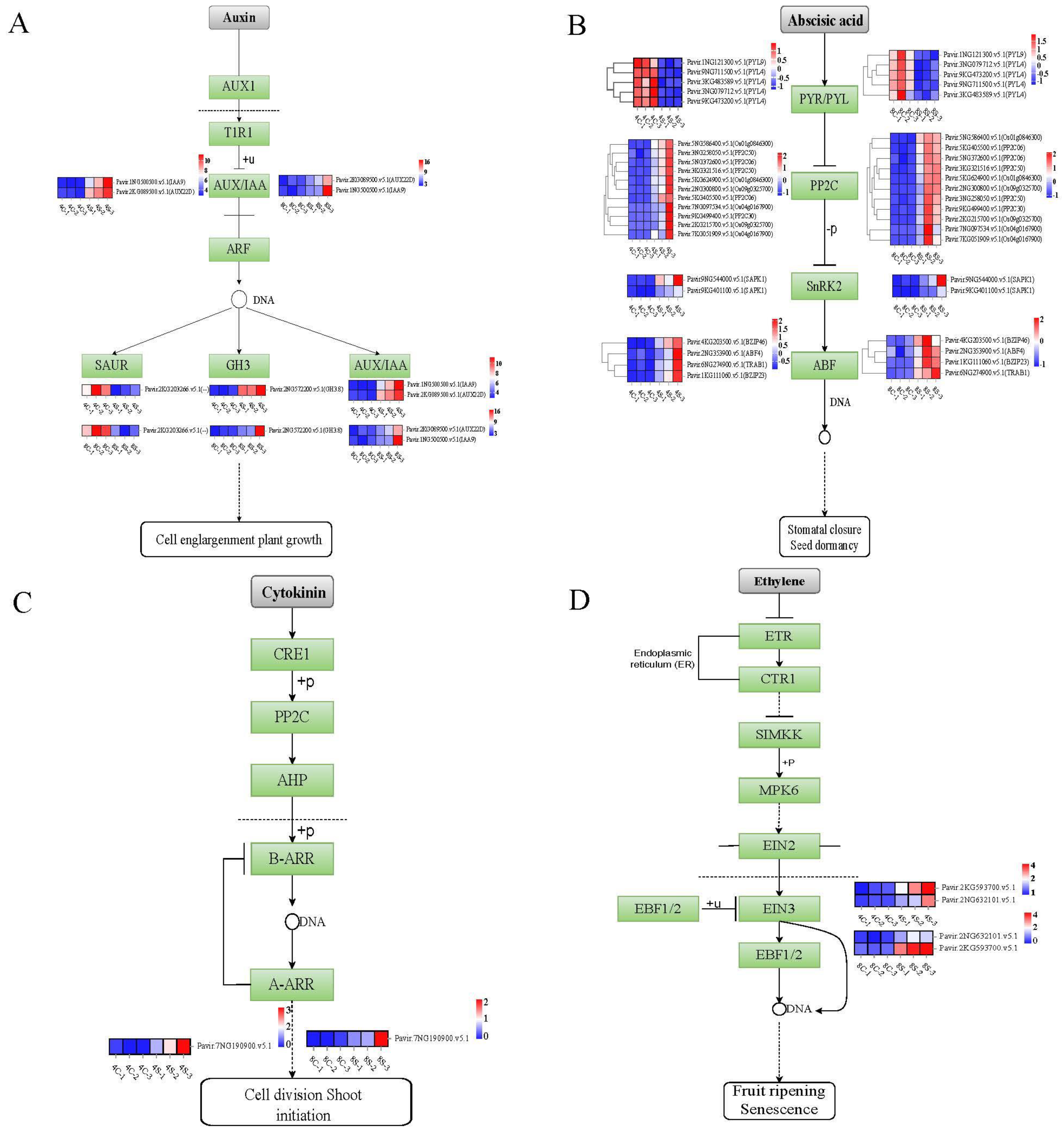
2.5. Hormonal Signaling Is Altered in Octoploids under Salt Stress
2.6. WGCNA Module Generation and Functional Enrichment Analysis
2.7. Gene Expression Validation by qRT-PCR Analysis
3. Discussion
3.1. Morphological and Physiological Responses of 4× and 8× Switchgrass under Salt Stress
3.2. Comparative Transcriptomes of 4× and 8× Switchgrass in Response to Salt Stress
3.3. Expression of Genes Associated with Plant Hormone Signal Transduction
4. Material and Methods
4.1. Plant Material, Growth Conditions, and Treatments
4.2. Measurement of Leaf Chlorophyll Content
4.3. Measurement of Leaf Soluble Sugar and Proline Contents
4.4. Measurement of Superoxide Dismutase (SOD) and Catalase (CAT) Activities and Malondialdehyde (MDA) Content in Leaves
4.5. Potassium (K+) and Sodium (Na+) Content in Leaves
4.6. Illumina Transcriptome Library Preparation and Sequencing
4.7. Transcriptome Sequencing Data Analysis
4.8. Gene Ontology (GO) Function and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analyses
4.9. Weighted Gene Co-Expression Network Analysis (WGCNA)
4.10. Quantitative RT-PCR (qRT-PCR) Analysis
4.11. Statistical Analysis of Physiological Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, K.Q.; Sun, G.L.; Stewart, C.N.; Xiao, P.; Zhang, B.H. MicroRNA expression analysis in the cellulosic biofuel crop switchgrass (Panicum virgatum) under abiotic stress. PLoS ONE 2012, 7, e32017. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.K.; Voigt, T.B.; Kim, S.; Lee, D.K. Determining effects of sodicity and salinity on switchgrass and prairie cordgrass germination and plant growth. Ind. Crops Prod. 2015, 64, 79–87. [Google Scholar] [CrossRef]
- Bouton, J.H. Molecular breeding of switchgrass for use as a biofuel crop. Curr. Opin. Genet. Dev. 2007, 17, 553–558. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, S.B.; Adams Kszos, L. Development of switchgrass (Panicum virgatum) as a bioenergy feedstock in the United States. Biomass Bioenergy 2005, 28, 515–535. [Google Scholar] [CrossRef]
- McLaughlin, S.B.; Kiniry, J.R.; Taliaferro, C.M.; De La Torre Ugarte, D. Projecting yield and utilization potential of switchgrass as an energy crop. Adv. Agron. 2006, 90, 267–297. [Google Scholar] [CrossRef]
- Madhugiri, N.-R.; Jaya, R.S.; Charles, K.; C Neal, S.J. Advances in biotechnology and genomics of switchgrass. Biotechnol. Biofuels 2013, 6, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Duo, T.Q.; Wang, F.D.; Zhang, X.Z.; Yang, Z.Z.; Hu, G.F. De novo transcriptome in roots of switchgrass (Panicum virgatum L.) reveals gene expression dynamic and act network under alkaline salt stress. BMC Genom. 2021, 22, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Teakle, N.L.; Tyerman, S.D. Mechanisms of Cl-transport contributing to salt tolerance. Plant Cell Environ. 2010, 33, 566–589. [Google Scholar] [CrossRef] [PubMed]
- Eduardo, B. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 2000, 12, 431–434. [Google Scholar] [CrossRef]
- Park, H.J.; Kim, W.-Y.; Yun, D.-J. A new insight of salt stress signaling in plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef]
- Shumilina, J.; Kusnetsova, A.; Tsarev, A.; Janse van Rensburg, H.C.; Medvedev, S.; Demidchik, V.; Van den Ende, W.; Frolov, A. Glycation of plant proteins: Regulatory roles and interplay with sugar signalling? Int. J. Mol. Sci. 2019, 20, 2366. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2015, 243, 281–296. [Google Scholar] [CrossRef]
- Bird, K.A.; VanBuren, R.; Puzey, J.R.; Edger, P.P. The causes and consequences of subgenome dominance in hybrids and recent polyploids. New Phytol. 2018, 220, 87–93. [Google Scholar] [CrossRef]
- Parisod, C.; Holderegger, R.; Brochmann, C. Evolutionary consequences of autopolyploidy. New Phytol. 2010, 186, 5–17. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 2016, 30, 159–165. [Google Scholar] [CrossRef]
- Chen, P.L.; Chen, J.; Sun, M.; Yan, H.D.; Feng, G.Y.; Wu, B.C.; Zhang, X.Q.; Wang, X.S.; Huang, L.K. Comparative transcriptome study of switchgrass (Panicum virgatum L.) homologous autopolyploid and its parental amphidiploid responding to consistent drought stress. Biotechnol. Biofuels 2020, 13, 170. [Google Scholar] [CrossRef]
- Oustric, J.; Morillon, R.; Luro, F.; Herbette, S.; Lourkisti, R.; Giannettini, J.; Berti, L.; Santini, J. Tetraploid Carrizo citrange rootstock (Citrus sinensis Osb. × Poncirus trifoliata L. Raf.) enhances natural chilling stress tolerance of common clementine (Citrus clementina Hort. ex Tan). J. Plant Physiol. 2017, 214, 108–115. [Google Scholar] [CrossRef]
- Liu, B.B.; Sun, G.L. microRNAs contribute to enhanced salt adaptation of the autopolyploidHordeum bulbosumcompared with its diploid ancestor. Plant J. 2017, 91, 57–69. [Google Scholar] [CrossRef]
- Sun, M.; Huang, D.J.; Zhang, A.L.; Khan, I.; Yan, H.D.; Wang, X.S.; Zhang, X.Q.; Zhang, J.; Huang, L.K. Transcriptome analysis of heat stress and drought stress in pearl millet based on Pacbio full-length transcriptome sequencing. BMC Plant Biol. 2020, 20, 323–337. [Google Scholar] [CrossRef]
- Yan, H.D.; Sun, M.; Zhang, Z.R.; Jin, Y.R.; Zhang, A.; Lin, C.; Wu, B.C.; He, M.; Xu, B.; Wang, J.; et al. Pangenomic analysis identifies structural variation associated with heat tolerance in pearl millet. Nat. Genet. 2023, 55, 507–518. [Google Scholar] [CrossRef]
- Sun, M.; Yan, H.D.; Zhang, A.; Jin, Y.R.; Lin, C.; Luo, L.; Wu, B.C.; Fan, Y.H.; Tian, S.L.; Cao, X.F.; et al. Milletdb: A multi-omics database to accelerate the research of functional genomics and molecular breeding of millets. Plant Biotechnol. J. 2023, 21, 2348–2357. [Google Scholar] [CrossRef]
- Wu, B.C.; Zhu, J.; Ma, X.X.; Jia, J.Y.; Luo, D.; Ding, Q.; Wang, X.S.; Huang, L.K. Comparative analysis of switchgrass chloroplast genomes provides insights into identification, phylogenetic relationships and evolution of different ecotypes. Ind. Crops Prod. 2023, 205, 117570. [Google Scholar] [CrossRef]
- Song, Q.X.; Chen, Z.J. Epigenetic and developmental regulation in plant polyploids. Curr. Opin. Plant Biol. 2015, 24, 101–109. [Google Scholar] [CrossRef]
- Tu, Y.; Jiang, A.M.; Gan, L.; Hossain, M.; Zhang, J.M.; Peng, B.; Xiong, Y.G.; Song, Z.J.; Cai, D.T.; Xu, W.F.; et al. Genome duplication improves rice root resistance to salt stress. Rice 2014, 7, 15. [Google Scholar] [CrossRef]
- Dong, Y.P.; Fan, G.Q.; Zhao, Z.L.; Xu, E.K.; Deng, M.J.; Wang, L.M.; Niu, S.Y. Transcriptome-wide profiling and expression analysis of two accessions of Paulownia australis under salt stress. Tree Genet. Genomes 2017, 13, 97. [Google Scholar] [CrossRef]
- Fan, G.Q.; Wang, L.M.; Deng, M.J.; Zhao, Z.L.; Dong, Y.P.; Zhang, X.S.; Li, Y.S. Changes in transcript related to osmosis and intracellular ion homeostasis in Paulownia tomentosa under salt stress. Front. Plant Sci. 2016, 7, 384. [Google Scholar] [CrossRef]
- Chao, D.Y.; Dilkes, B.; Luo, H.; Douglas, A.; Yakubova, E.; Lahner, B.; Salt, D.E. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 2013, 341, 658–667. [Google Scholar] [CrossRef]
- Wu, B.; Munkhtuya, Y.; Li, J.J.; Hu, Y.N.; Zhang, Q.; Zhang, Z.W. Comparative transcriptional profiling and physiological responses of two contrasting oat genotypes under salt stress. Sci. Rep. 2018, 8, 16248. [Google Scholar] [CrossRef]
- Arrigo, N.; Barker, M.S. Rarely successful polyploids and their legacy in plant genomes. Curr. Opin. Plant Biol. 2012, 15, 140–146. [Google Scholar] [CrossRef]
- Ma, Y.Q.; An, Y.; Shui, J.F.; Sun, Z.J. Adaptability evaluation of switchgrass (Panicum virgatum L.) cultivars on the Loess Plateau of China. Plant Sci. 2011, 181, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.; Cui, X.; Liu, H.Y.; Li, X.; Li, M.Q.; Zhang, Y.W. Proline biosynthesis enzyme genes confer salt tolerance to switchgrass (Panicum virgatum L.) in cooperation with polyamines metabolism. Front. Plant Sci. 2020, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.M.; Zhang, X.Z.; Miao, J.M.; Huang, L.K.; Frazier, T.; Zhao, B.Y. Evaluation of salinity tolerance and genetic diversity of thirty-three switchgrass (Panicum virgatum) populations. BioEnergy Res. 2014, 7, 1329–1342. [Google Scholar] [CrossRef]
- Kim, J.; Liu, Y.M.; Zhang, X.Z.; Zhao, B.Y.; Childs, K.L. Analysis of salt-induced physiological and proline changes in 46 switchgrass (Panicum virgatum) lines indicates multiple response modes. Plant Physiol. Biochem. 2016, 105, 203–212. [Google Scholar] [CrossRef]
- Wang, Q.Z.; Wu, C.H.; Xie, B.; Liu, Y.; Cui, J.; Chen, G.; Zhang, Y.W. Model analysing the antioxidant responses of leaves and roots of switchgrass to NaCl-salinity stress. Plant Physiol. Biochem. 2012, 58, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.S.; Zhu, Y.; Fan, X.F.; Hou, X.C.; Zhao, C.Q.; Zhang, S.; Wu, J.Y. Generation of octoploid switchgrass in three cultivars by colchicine treatment. Ind. Crops Prod. 2017, 107, 20–21. [Google Scholar] [CrossRef]
- Kim, S.; Rayburn, A.L.; Voigt, T.; Parrish, A.; Lee, D.K. Salinity effects on germination and plant growth of prairie cordgrass and switchgrass. BioEnergy Res. 2011, 5, 225–235. [Google Scholar] [CrossRef]
- Wang, L.F.; Cao, S.; Wang, P.T.; Lu, K.N.; Song, Q.X.; Zhao, F.J.; Chen, Z.J. DNA hypomethylation in tetraploid rice potentiates stress-responsive gene expression for salt tolerance. Proc. Natl. Acad. Sci. USA 2021, 118, e2023981118. [Google Scholar] [CrossRef]
- Fahad, S.; Hussain, S.; Matloob, A.; Khan, F.A.; Khaliq, A.; Saud, S.; Hassan, S.; Shan, D.; Khan, F.; Ullah, N.; et al. Phytohormones and plant responses to salinity stress: A review. Plant Growth Regul. 2014, 75, 391–404. [Google Scholar] [CrossRef]
- Li, C.X.; Zhang, W.L.; Yuan, M.; Jiang, L.N.; Sun, B.; Zhang, D.J.; Shao, Y.; Liu, A.Q.; Liu, X.Q.; Ma, J.H. Transcriptome analysis of osmotic-responsive genes in ABA-dependent and -independent pathways in wheat (Triticum aestivum L.) roots. PeerJ 2019, 7, e6519. [Google Scholar] [CrossRef]
- Mattingly, K.Z.; Hovick, S.M. Autopolyploids of Arabidopsis thaliana are more phenotypically plastic than their diploid progenitors. Ann. Bot. 2023, 131, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Chen, S.M.; Chen, Y.; Guan, Z.Y.; Yin, D.M.; Chen, F.D. In vitro induced tetraploid of Dendranthema nankingense (Nakai) Tzvel. shows an improved level of abiotic stress tolerance. Sci. Hortic. 2011, 127, 411–419. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- de Azevedo Neto, A.D.; Prisco, J.T.; Enéas-Filho, J.; Abreu, C.E.B.d.; Gomes-Filho, E. Effect of salt stress on antioxidative enzymes and lipid peroxidation in leaves and roots of salt-tolerant and salt-sensitive maize genotypes. Environ. Exp. Bot. 2006, 56, 87–94. [Google Scholar] [CrossRef]
- Ana, B.C.; Janice, d.A.E.; Suresh, I.; Tom, C.; Marc, V.M.; Allan, B.C. Effects of osmoprotectant upon NaCl stress in rice. Plant Physiol. 1997, 115, 159–169. [Google Scholar] [CrossRef]
- Pan, Y.; Wu, L.J.; Yu, Z.L. Effect of salt and drought stress on antioxidant enzymes activities and SOD isoenzymes of liquorice (Glycyrrhiza uralensis Fisch). Plant Growth Regul. 2006, 49, 157–165. [Google Scholar] [CrossRef]
- Wei, T.L.; Wang, Y.; Liu, J.H. Comparative transcriptome analysis reveals synergistic and disparate defense pathways in the leaves and roots of trifoliate orange (Poncirus trifoliata) autotetraploids with enhanced salt tolerance. Hortic. Res. 2020, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.J.; Ma, X.L.; Wan, P.; Liu, L.Y. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef]
- Dai, W.S.; Wang, M.; Gong, X.Q.; Liu, J.H. The transcription factor FcWRKY40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 2018, 219, 972–989. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Zhang, J.S.; Wu, Y.H.; Wang, T.; Wang, K.; Liang, Q.Y.; Bai, Z.Y.; Liu, Q.L.; Pan, Y.Z.; Jiang, B.B.; Zhang, L. Comparative analysis of the Chrysanthemum leaf transcript profiling in response to salt stress. PLoS ONE 2016, 11, e0159721. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Gamir, J.; Ancillo, G.; Legaz, F.; Primo-Millo, E.; Forner-Giner, M.A. Influence of salinity on pip gene expression in citrus roots and its relationship with root hydraulic conductance, transpiration and chloride exclusion from leaves. Environ. Exp. Bot. 2012, 78, 163–166. [Google Scholar] [CrossRef]
- Kaleem, F.; Shabir, G.; Aslam, K.; Rasul, S.; Manzoor, H.; Shah, S.M.; Khan, A.R. An overview of the genetics of plant response to salt stress: Present status and the way forward. Appl. Biochem. Biotechnol. 2018, 186, 306–334. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.J.; Zhou, Y.H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.Q. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Liu, J.; Zhai, L.M.; Gan, Z.Y.; Zhang, G.F.; Yang, S.H.; Wang, Y.; Wu, T.; Zhang, X.Z.; Xu, X.F.; et al. Natural variation in cytokinin maintenance improves salt tolerance in apple rootstocks. Plant Cell Environ. 2019, 42, 424–436. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 2006, 225, 353–364. [Google Scholar] [CrossRef]
- R Khan, M.I.; A Khan, N. Salicylic acid and jasmonates: Approaches in abiotic stress tolerance. J. Plant Biochem. Physiol. 2013, 1, e113. [Google Scholar] [CrossRef]
- Krishna, P. Brassinosteroid-mediated stress responses. J. Plant Growth Regul. 2003, 22, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.L.; Xiao, Z.X.; Li, M.W.; Wong, F.L.; Yung, W.S.; Ku, Y.S.; Wang, Q.W.; Wang, X.; Xie, M.; Yim, A.K.-Y.; et al. Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ. 2019, 42, 98–114. [Google Scholar] [CrossRef]
- Yoon, J.Y.; Hamayun, M.; Lee, S.K.; Lee, I.J. Methyl jasmonate alleviated salinity stress in soybean. J. Crop Sci. Biotechnol. 2009, 12, 63–68. [Google Scholar] [CrossRef]
- Seo, H.; Kriechbaumer, V.; Park, W.J. Modern quantitative analytical tools and biosensors for functional studies of auxin. J. Plant Biol. 2016, 59, 93–104. [Google Scholar] [CrossRef]
- Sorce, C.; Montanaro, G.; Bottega, S.; Spanò, C. Indole-3-acetic acid metabolism and growth in young kiwifruit berry. Plant Growth Regul. 2017, 82, 505–515. [Google Scholar] [CrossRef]
- Liu, J.H.; Zhao, Z.L.; Li, Y.S.; Liu, H.F.; Zhai, X.Q.; Deng, M.J.; Dong, Y.P.; Fan, G.Q. Genome-wide expression analysis of salt-stressed diploid and autotetraploid Paulownia tomentosa. PLoS ONE 2017, 12, e0185455. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Hadiarto, T.; Tran, L.-S.P. Progress studies of drought-responsive genes in rice. Plant Cell Rep. 2010, 30, 297–310. [Google Scholar] [CrossRef]
- Iqbal, M.; Ashraf, M.; Jamil, A. Seed enhancement with cytokinins: Changes in growth and grain yield in salt stressed wheat plants. Plant Growth Regul. 2006, 50, 29–39. [Google Scholar] [CrossRef]
- Blazquez, M.A.; Nishiyama, R.; Le, D.T.; Watanabe, Y.; Matsui, A.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS ONE 2012, 7, e32124. [Google Scholar] [CrossRef] [PubMed]
- Hardin, C.F.; Fu, C.; Hisano, H.; Xiao, X.; Shen, H.; Stewart, C.N.; Parrott, W.; Dixon, R.A.; Wang, Z.-Y. Standardization of switchgrass sample collection for cell wall and biomass trait analysis. BioEnergy Res. 2013, 6, 755–762. [Google Scholar] [CrossRef]
- Anjorin, F.B. Leaf growth traits and photosynthetic pigments of maize as influenced by water deficit stress. Not. Sci. Biol. 2020, 12, 366–375. [Google Scholar] [CrossRef]
- Kramer, P.J. Water deficits and plant growth. Water Relat. Plants 1983, 342–389. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Li, B.; Colin, N.D. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323–393. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 2007, 1, 54. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]




| Ploidy | Treatments | Height (cm) | Stem Thick (mm) | Leaf Length (cm) | Leaf Width (cm) | Biomass (kg) |
|---|---|---|---|---|---|---|
| 4× | Control | 126.00 ± 2.74 a | 5.62 ± 0.23 b | 55.00 ± 2.12 b | 1.55 ± 0.11 b | 5.65 ± 0.19 a |
| Salt | 79.25 ± 1.92 d | 4.55 ± 0.26 c | 41.25 ± 1.92 c | 1.30 ± 0.07 c | 2.59 ± 0.17 c | |
| 8× | Control | 115.25 ± 6.76 b | 6.64 ± 0.30 a | 69.25 ± 2.68 a | 2.00 ± 0.07 a | 5.45 ± 0.34 a |
| Salt | 86.75 ±1.48 c | 5.29 ± 0.24 b | 53.00 ± 3.39 b | 1.60 ± 0.07 b | 3.09 ±0.19 b |
| Gene Name | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) |
|---|---|---|
| CYP-5 (reference gene) | CACTACAAGGGAAGCACATTCCA | TTCACCACCCCTTCCATCAC |
| Pavir.4NG135600.1.v5.1 | CAATGAGCCTTGGAGTTTCA | CCAATGCTACATCCCGAATT |
| Pavir.9NG711500.1.v5.1 | CGGCTCCAGAACTACCTCTC | TCCACCATGTAGGACTCCAC |
| Pavir.9KG317457.1.v5.1 | ACAACATCGAGTCAGGGCAA | TCAGGTAGGGGATGATCTGC |
| Pavir.7KG383000.1.v5.1 | CACATTGATGGGCAAAACTC | ATTCGACAAAGCACGAGCAG |
| Pavir.9KG337100.1.v5.1 | TGGTTGACTATCCGTCTGCT | GAAAGCGTTCCATCGTAGTC |
| Pavir.1NG500500.1.v5.1 | CGCTCCATAACTCCTACGAT | CAGTCCCATTCTCCATCCTC |
| Pavir.2NG572200.1.v5.1 | ATTTCCTCTTCGTCAAGTCG | GGTTCTTGAAGTGGTTGCTC |
| Pavir.2KG593700.1.v5.1 | GACTTCAACTTCAACACCCC | GCGTCGTACATCTCCATCAG |
| Pavir.9KG401100.1.v5.1 | GGGAGGTTTATTGAGGATGA | GTTTCAGATCCCTATGGCAT |
| Pavir.1KG111060.1.v5.1 | GCTATGGAGAAGGTGGTTGA | TAAGTTTTGCCACCTCAGCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, J.; Fan, Y.; Zhang, H.; Teng, W.; Teng, K.; Wu, J.; Fan, X.; Wang, S.; Yue, Y. Octoploids Show Enhanced Salt Tolerance through Chromosome Doubling in Switchgrass (Panicum virgatum L.). Plants 2024, 13, 1383. https://doi.org/10.3390/plants13101383
Ye J, Fan Y, Zhang H, Teng W, Teng K, Wu J, Fan X, Wang S, Yue Y. Octoploids Show Enhanced Salt Tolerance through Chromosome Doubling in Switchgrass (Panicum virgatum L.). Plants. 2024; 13(10):1383. https://doi.org/10.3390/plants13101383
Chicago/Turabian StyleYe, Jiali, Yupu Fan, Hui Zhang, Wenjun Teng, Ke Teng, Juying Wu, Xifeng Fan, Shiwen Wang, and Yuesen Yue. 2024. "Octoploids Show Enhanced Salt Tolerance through Chromosome Doubling in Switchgrass (Panicum virgatum L.)" Plants 13, no. 10: 1383. https://doi.org/10.3390/plants13101383





