Genome-Wide Identification of the Pectate Lyase Gene Family in Potato and Expression Analysis under Salt Stress
Abstract
:1. Introduction
2. Results
2.1. Chromosomal Localization of StPL Genes
2.2. Physicochemical Properties and Subcellular Localization of the StPL Proteins
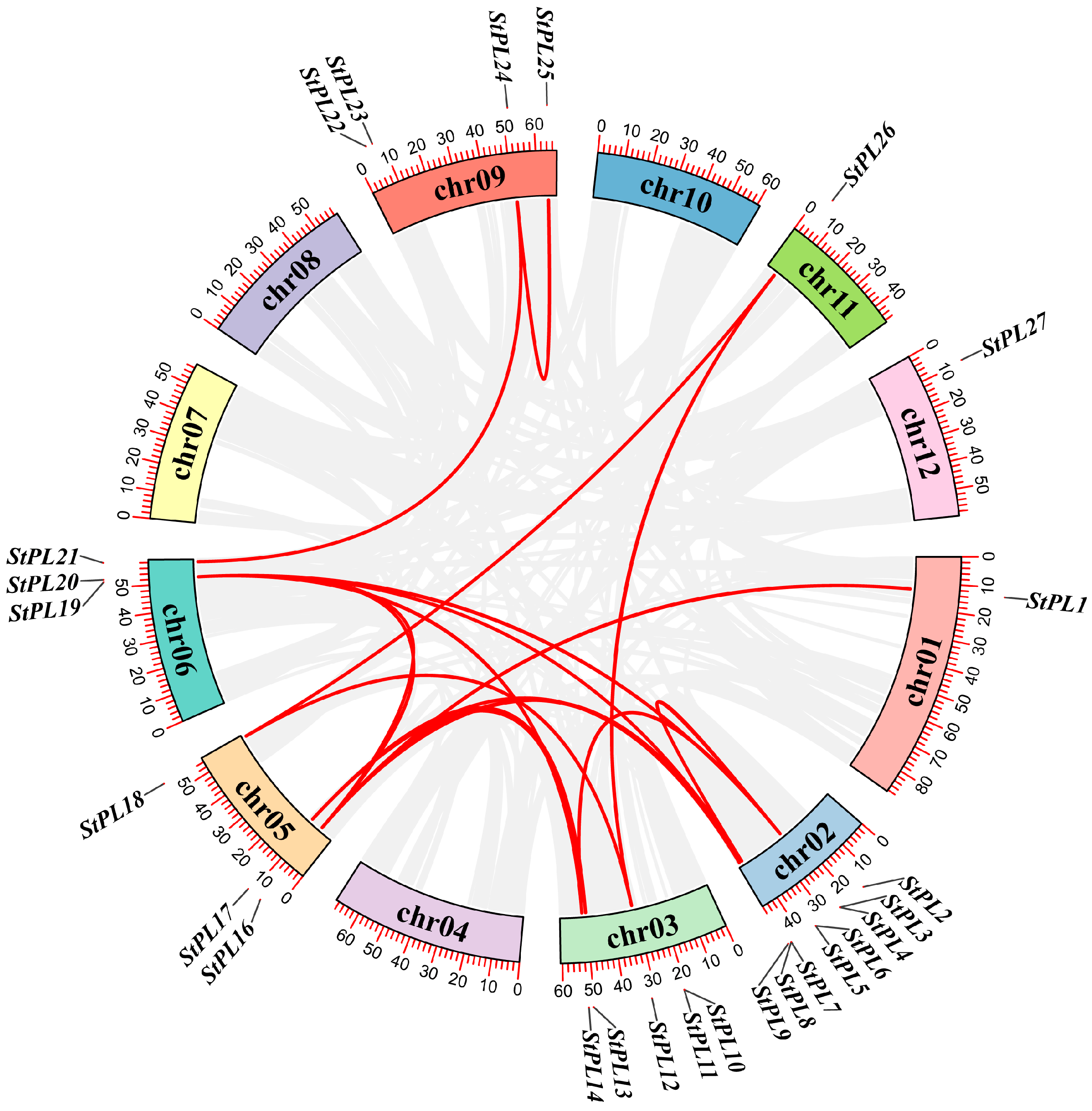
2.3. Gene Duplication and Collinearity Analysis of StPL Gene Family Members
2.4. Gene Duplication and Collinearity Analysis of PL Gene Family Members in Potato and Arabidopsis
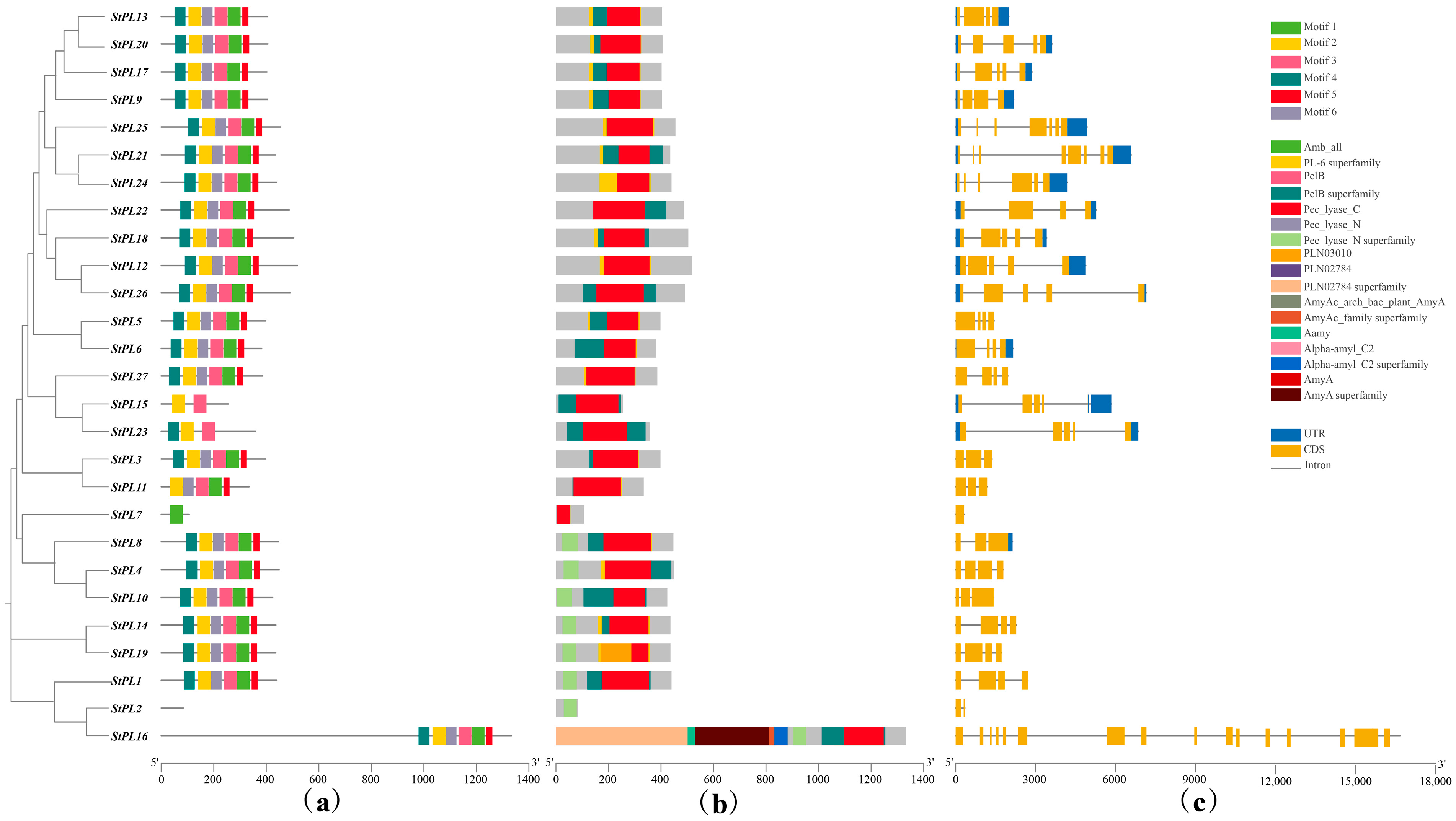
2.5. Phylogenetic Tree Analysis of StPL Gene Family Members
2.6. StPL Gene Family Members’ Gene Structure and Conserved Motif Analysis
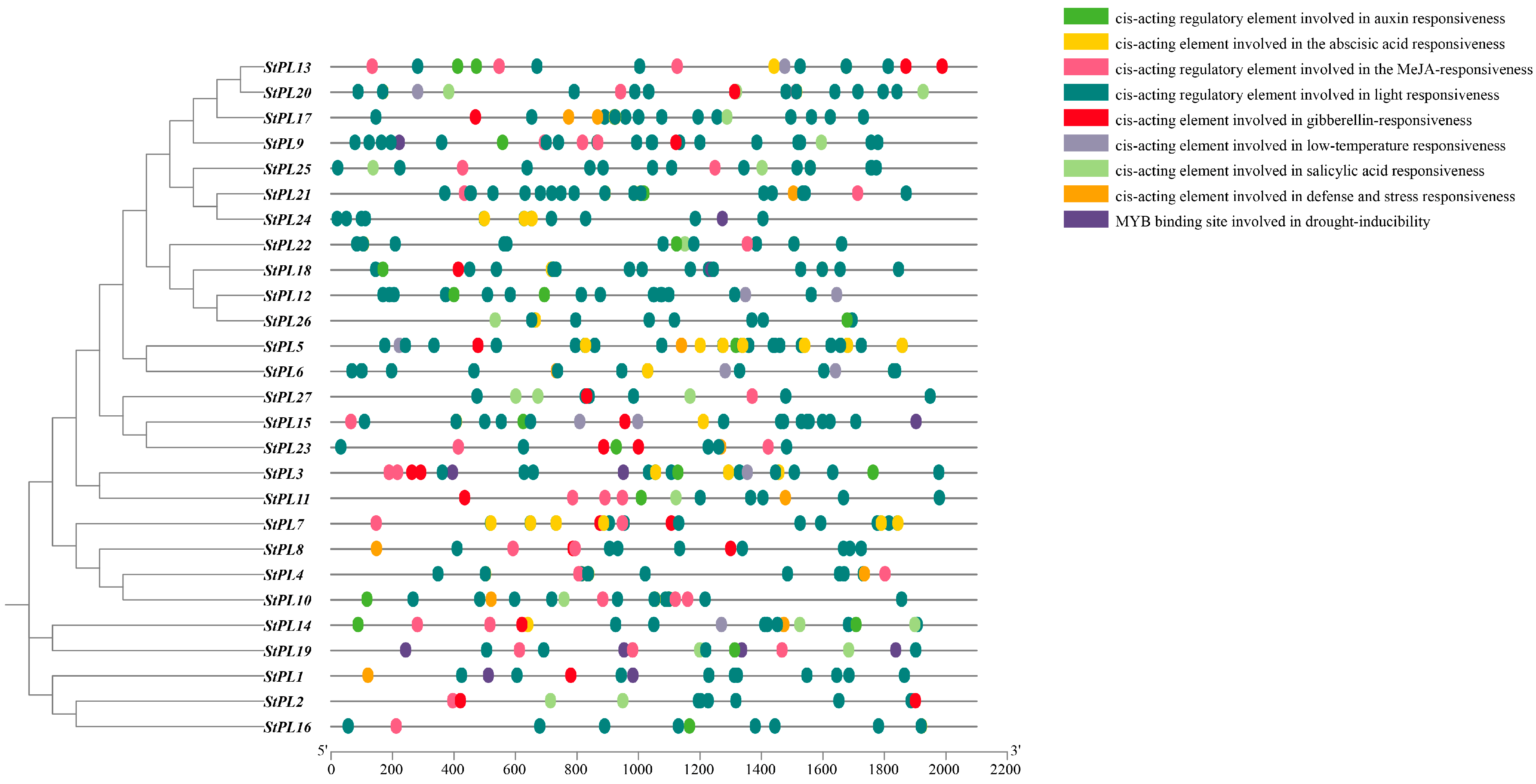
2.7. Analysis of the Cis-Acting Elements in the Promoter of StPL Gene Family Members
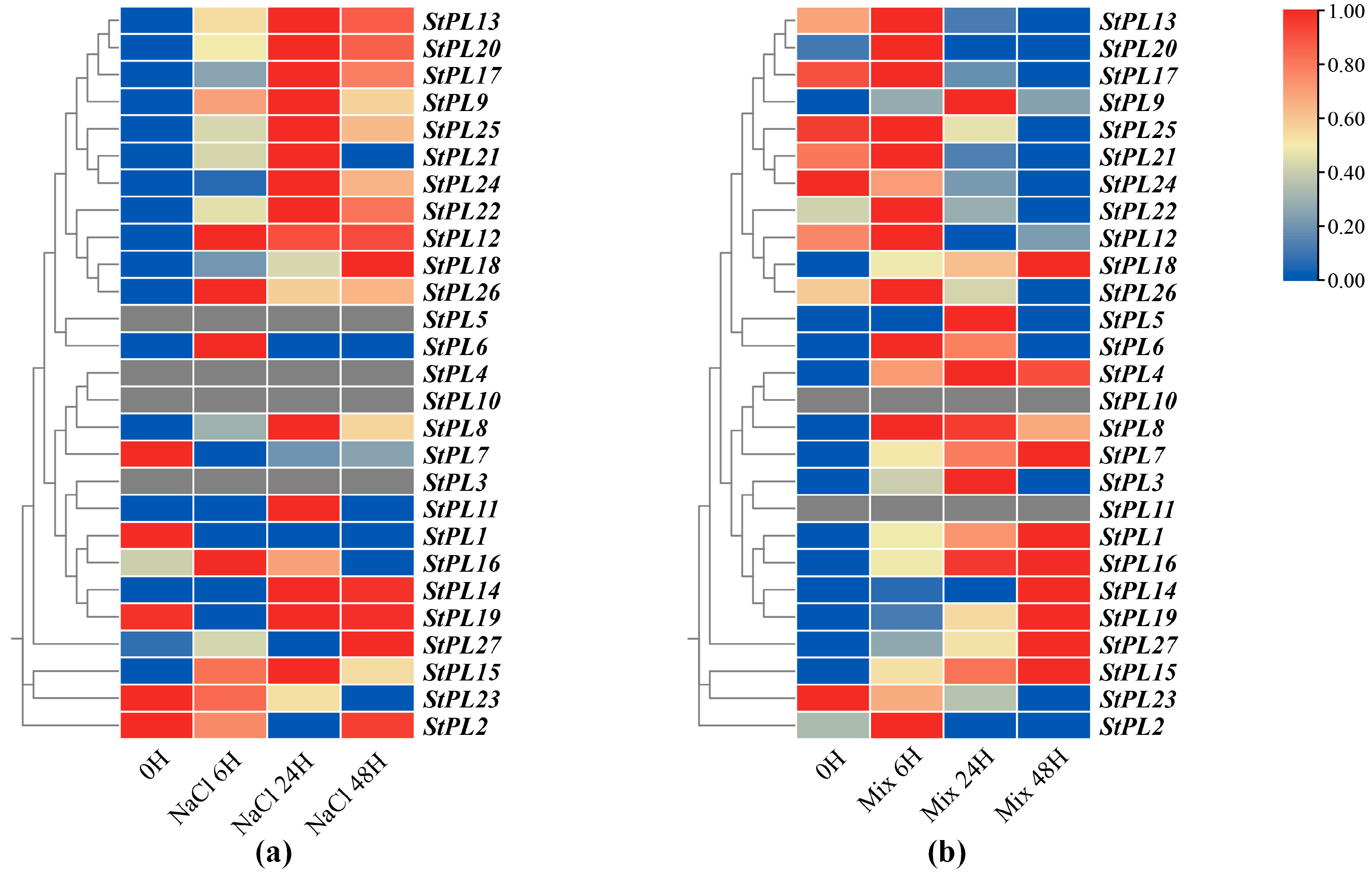
2.8. Expression Pattern Analysis of StPL Gene Family Members under Salt Stress
2.9. Construction of Overexpression Vector of Potato StPL18 Gene and Subcellular Localization in Tobacco
3. Discussion
3.1. Analysis of Pectate Lyase Gene Family Members
3.2. Expression Pattern of Pectate Lyase Gene Family Members under Salt Stress
4. Materials and Methods
4.1. Identification of Members of the Potato PL Gene Family
4.2. Chromosomal Localization of Potato StPL Genes
4.3. Analysis of Physicochemical Properties of Potato StPL Protein and Prediction of Subcellular Localization
4.4. Phylogenetic Tree and Gene Structure Analysis of the Potato StPL Gene
4.5. Gene Collinearity Analysis
4.6. Analysis of Potato StPL Promoter Cis-Acting Elements
4.7. Analysis of the Expression Pattern of StPL Family Genes in Potato Seedlings under Single and Mixed Salts
4.8. Subcellular Localization of StPL Genes
- (1)
- Pick Agrobacterium containing the corresponding vectors and single colonies of Agrobacterium containing the P19 plasmid. Inoculate them into 10 mL of LB liquid medium with the corresponding resistance and incubate them for 24 h at 28 °C and 220 rpm until the OD600 value greater than 0.6.
- (2)
- Centrifuge at 5000 rpm for 2 min and discard the liquid medium.
- (3)
- Resuspend the organisms by adding 700 μL of tobacco infestation buffer, mix well, and take 200 μL of the bacterial solution to measure the OD600 value.
- (4)
- Mix Agrobacterium with different vectors in a 2 mL centrifuge tube so that the OD600 value of Agrobacterium containing the P19 plasmid is adjusted to 0.3, and other Agrobacterium is adjusted to 0.5. Leave the mixture at room temperature for about 3 h. Afterward, carefully pipette the well-grown Agrobacterium from the center of the centrifuge tube and discard the rest.
- (5)
- Take the tobacco leaves (tobacco has about four true leaves and is 10 cm tall) with good growth conditions and inject Agrobacterium from the back of the leaves using a 1 mL syringe until the leaves are completely wet.
- (6)
- Incubate in the dark for 24 h and then transfer to dim light culture for 36 h.
- (7)
- For co-localization, the marker plasmid was transformed into Agrobacterium, operating in suspension with the constructed vector plasmid Agrobacterium, and mixed at an equivalent ratio before injection into tobacco leaves.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Camire, M.; Kubow, S.; Donnelly, D. Potatoes and human health. Crit. Rev. Food Sci. 2009, 49, 823–840. [Google Scholar] [CrossRef] [PubMed]
- McGill, C.R.; Kurilich, A.C.; Davignon, J. The role of potatoes and potato components in cardiometabolic health: A review. Ann. Med. 2013, 45, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Lutaladio, N.; Castaldi, L. Potato: The hidden treasure. J. Food Compos. Anal. 2009, 22, 491–493. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhao, H.; Liu, Z.; He, J.; Liu, W. Research progress and development of mechanized potato planters: A review. Agriculture 2021, 11, 521. [Google Scholar] [CrossRef]
- Zhang, B.; Gao, Y.; Zhang, L.; Zhou, Y. The plant cell wall: Biosynthesis, construction, and functions. J. Integr. Plant Biol. 2021, 63, 251–272. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.T.; Kieber, J.J. Dynamic construction, perception, and remodeling of plant cell walls. Annu. Rev. Plant Biol. 2020, 71, 39–69. [Google Scholar] [CrossRef] [PubMed]
- Harholt, J.; Suttangkakul, A.; Vibe Scheller, H. Biosynthesis of Pectin. Plant Physiol. 2010, 153, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Yadav, P.K.; Yadav, D.; Yadav, K.D.S. Pectin lyase: A review. Process Biochem. 2009, 44, 1–10. [Google Scholar] [CrossRef]
- Celia, M.; Âguez, M.Â.-R.; Orchard, J.; Seymour, G. Pectate lyases, cell wall degradation and fruit softening. J. Exp. Bot. 2002, 53, 2115–2119. [Google Scholar] [CrossRef] [PubMed]
- Starr, M.; Moran, F. Eliminative split of pectic substances by phytopathogenic soft-rot Bacteria. Science 1962, 135, 920–921. [Google Scholar] [CrossRef]
- Payasi, A.; Sanwal, G.G. Pectate lyase activity during ripening of banana fruit. Phytochemistry 2003, 63, 243–248. [Google Scholar] [CrossRef]
- Lionetti, V.; Cervone, F.; Bellincampi, D. Methyl esterification of pectin plays a role during plant-pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 2012, 169, 1623–1630. [Google Scholar] [CrossRef]
- Wing, R.; Yamaguchi, J.; Larabell, S.K.; Ursin, V.; McCormick, S. Molecular and genetic characterization of two pollen-expressed genes that have sequence similarity to pectate lyases of the plant pathogen Erwinia. Plant Mol. Biol. 1990, 14, 17–28. [Google Scholar] [CrossRef]
- Dubey, A.K.; Yadav, S.; Kumar, M.; Singh, V.K.; Sarangi, B.K.; Yadav, D. In Silico characterization of pectate lyase protein sequences from different source organisms. Enzym. Res. 2010, 2010, 950230. [Google Scholar] [CrossRef]
- Zheng, Y.; Yan, J.; Wang, S.; Xu, M.; Huang, K.; Chen, G.; Ding, Y. Genome-wide identification of the pectate lyase-like (PLL) gene family and functional analysis of two PLL genes in rice. Mol. Genet. Genom. 2018, 293, 1317–1331. [Google Scholar] [CrossRef]
- Sun, L.; Nocker, S.v. Analysis of promoter activity of members of the PECTATE LYASE-LIKE (PLL) gene family in cell separation in Arabidopsis. BMC Plant Biol. 2010, 10, 152. [Google Scholar] [CrossRef]
- Jiang, J.; Yao, L.; Miao, Y.; Cao, J. Genome-wide characterization of the pectate lyase-like (PLL) genes in Brassica rapa. Mol. Genet. Genom. 2013, 288, 601–614. [Google Scholar] [CrossRef]
- Yang, L.; Huang, W.; Xiong, F.; Xian, Z.; Su, D.; Ren, M.; Li, Z. Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 2017, 15, 1544–1555. [Google Scholar] [CrossRef]
- Palusa, S.G.; Golovkin, M.; Shin, S.B.; Richardson, D.N.; Reddy, A.S.N. Organ-specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in Arabidopsis. New Phytol. 2007, 174, 537–550. [Google Scholar] [CrossRef]
- Vogel, J.P.; Raab, T.K.; Somerville, C.R.; Somerville, S.C. Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 2004, 40, 968–978. [Google Scholar] [CrossRef]
- Vogel, J.; Raab, T.; Schiff, C.; Somerville, S. PMR6, a pectate lyase–like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 2002, 14, 2095–2106. [Google Scholar] [CrossRef] [PubMed]
- Uluisik, S.; Chapman, N.H.; Smith, R.; Poole, M.; Adams, G.; Gillis, R.B.; Besong, T.M.D.; Sheldon, J.; Stiegelmeyer, S.; Perez, L.; et al. Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. 2016, 34, 950–952. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Yang, R.; Zhang, L.; Wei, Q.; Zhang, Y.; Wang, Y.; Shi, Y. A review of potato salt tolerance. Int. J. Mol. Sci. 2023, 24, 10726. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, M.; Biller, S.; Stanley, K.; Kajstura, T.J.; Prusty, R. Expression profiling of auxin-treated Arabidopsis roots: Toward a molecular analysis of lateral root emergence. Plant Cell Physiol. 2006, 47, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Feng, S.; Tang, D.; Zhang, L.; Li, Y.; Kear, P.; Huang, S.; Zhang, C. The mutation of a PECTATE LYASE-LIKE gene is responsible for the Yellow Margin phenotype in potato. Theor. Appl. Genet. 2020, 133, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2022, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Bazzini, A. Translation and mRNA stability control. Annu. Rev. Biochem. 2023, 92, 227–245. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Li, M.; Chen, J.; Liu, P.; Li, Z. Effects of MeJA on Arabidopsis metabolome under endogenous JA deficiency. Sci. Rep. 2016, 6, 37674. [Google Scholar] [CrossRef]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and mechanism of Jasmonic Acid in plant responses to abiotic and biotic stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef]
- Shomer, I.; Novacky, A.; Pike, S.; Yermiyahu, U.; Kinraide, T. Electrical potentials of plant cell walls in response to the ionic environment. Plant Physiol. 2003, 133, 411–422. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, W.; Long, S.; Zhao, C. Maintenance of cell wall integrity under high salinity. Int. J. Mol. Sci. 2021, 22, 3260. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Lee, B.-h.; Dellinger, M.; Cui, X.; Zhang, C.; Wu, S.; Nothnagel, E.; Zhu, J.K. A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 2010, 63, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Dabravolski, S.A.; Isayenkov, S.V. The regulation of plant cell wall organisation under salt stress. Front. Plant Sci. 2023, 14, 1118313. [Google Scholar] [CrossRef] [PubMed]
- Gigli-Bisceglia, N.; Zelm, E.v.; Huo, W.; Lamers, J.; Testerink, C. Arabidopsis root responses to salinity depend on pectin modification and cell wall sensing. Development 2020, 149, dev200363. [Google Scholar] [CrossRef] [PubMed]
- Leng, Y.; Yang, Y.; Ren, D.; Huang, L.; Dai, L.; Wang, Y.; Chen, L.; Tu, Z.; Gao, Y.; Li, X.; et al. A Rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol. 2017, 174, 1151–1166. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Gao, D.S. Endoplasmic reticulum proteins quality control and the unfolded protein response: The regulative mechanism of organisms against stress injuries. BioFactors 2014, 40, 569–585. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools—An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2019, 48, D265–D268. [Google Scholar] [CrossRef]
- Wilkins, M.; Gasteiger, E.; Bairoch, A.; Sanchez, J.E.; Williams, K.L.; Appel, R.; Hochstrasser, D. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Frazee, A.C.; Pertea, G.; Jaffe, A.; Langmead, B.; Salzberg, S.; Leek, J. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015, 33, 243–246. [Google Scholar] [CrossRef]
- Mravec, J.; Skůpa, P.; Bailly, A.; Hoyerová, K.; Krecek, P.; Bielach, A.; Petrášek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.; et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef]




| Gene ID | Transcript ID | Molecular Mass (Da) | Amino Acid Length (aa) | Theoretical pI | Instability Index | Stability | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| StPL1 | Soltu.DM.01G009330.1 | 49,343.26 | 439 | 8.24 | 40.13 | unstable | 81.55 | −0.349 | Chloroplast |
| StPL2 | Soltu.DM.02G003590.1 | 9534.89 | 83 | 5.27 | 48.63 | unstable | 92.89 | −0.125 | Chloroplast |
| StPL3 | Soltu.DM.02G009950.1 | 44,342.64 | 397 | 8.67 | 36.01 | stable | 81.08 | −0.344 | Peroxisome |
| StPL4 | Soltu.DM.02G009960.1 | 50,669.45 | 448 | 7.65 | 37.59 | stable | 70.09 | −0.45 | Extracellular |
| StPL5 | Soltu.DM.02G020450.1 | 44,279.08 | 397 | 6.86 | 33.21 | stable | 77.36 | −0.321 | Extracellular |
| StPL6 | Soltu.DM.02G020460.1 | 42,099.02 | 381 | 9.11 | 29.28 | stable | 79.37 | −0.267 | Extracellular |
| StPL7 | Soltu.DM.02G031990.1 | 11,791.5 | 105 | 8.58 | 35.78 | stable | 75.14 | −0.098 | Extracellular |
| StPL8 | Soltu.DM.02G032040.1 | 49,905.67 | 446 | 8.31 | 28.17 | stable | 76.3 | −0.345 | Vacuole |
| StPL9 | Soltu.DM.02G033640.1 | 44,508.98 | 403 | 7.65 | 38.52 | stable | 77.67 | −0.382 | Extracellular |
| StPL10 | Soltu.DM.03G008540.1 | 48,094.64 | 423 | 8.59 | 34.61 | stable | 70.76 | −0.522 | Cytoplasmic |
| StPL11 | Soltu.DM.03G008580.1 | 36,799.61 | 333 | 6.22 | 31.4 | stable | 86.4 | −0.266 | Cytoplasmic |
| StPL12 | Soltu.DM.03G011300.1 | 56,927.17 | 517 | 6.38 | 31.8 | stable | 83.35 | −0.269 | Plasma membrane |
| StPL13 | Soltu.DM.03G025090.1 | 44,178.86 | 403 | 6.96 | 35.52 | stable | 78.88 | −0.242 | Extracellular |
| StPL14 | Soltu.DM.03G027030.1 | 48,610.72 | 435 | 9.51 | 34.68 | stable | 79.1 | −0.32 | Vacuole |
| StPL15 | Soltu.DM.04G007160.1 | 28,596.42 | 254 | 8.67 | 27.68 | stable | 80.24 | −0.416 | Cytoplasmic |
| StPL16 | Soltu.DM.05G006330.1 | 150,181.53 | 1332 | 6.05 | 43.29 | unstable | 78.68 | −0.475 | Mitochondrial |
| StPL17 | Soltu.DM.05G010370.1 | 44,248.03 | 401 | 6.93 | 33.57 | stable | 77.83 | −0.3 | Extracellular |
| StPL18 | Soltu.DM.05G026200.1 | 55,513.46 | 503 | 5.9 | 39.15 | stable | 83.7 | −0.162 | Membrane |
| StPL19 | Soltu.DM.06G026130.1 | 48,636.13 | 435 | 8.78 | 35.83 | stable | 76.67 | −0.391 | Vacuole |
| StPL20 | Soltu.DM.06G026970.1 | 44,496.3 | 405 | 7.64 | 31.07 | stable | 84.25 | −0.218 | Vacuole |
| StPL21 | Soltu.DM.06G033950.1 | 48,183.37 | 434 | 6.8 | 45.92 | unstable | 80.94 | −0.231 | Chloroplast |
| StPL22 | Soltu.DM.09G005200.1 | 53,964.23 | 486 | 5.48 | 37.44 | stable | 75.99 | −0.324 | Vacuole |
| StPL23 | Soltu.DM.09G007270.1 | 40,151.06 | 357 | 7.92 | 36.37 | stable | 68.52 | −0.602 | Peroxisome |
| StPL24 | Soltu.DM.09G018370.1 | 48,797.13 | 439 | 7.68 | 36.78 | stable | 79.54 | −0.274 | Chloroplast |
| StPL25 | Soltu.DM.09G028450.1 | 50,015.35 | 454 | 7.28 | 34.07 | stable | 74.56 | −0.299 | Extracellular |
| StPL26 | Soltu.DM.11G004550.1 | 53,705.64 | 490 | 5.95 | 29.56 | stable | 84.37 | −0.125 | Plasma membrane |
| StPL27 | Soltu.DM.12G009730.1 | 42,657.73 | 385 | 9.52 | 28.18 | stable | 73.61 | −0.382 | Vacuole |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Liu, T.; Wu, W.; Shi, W.; Shi, J.; Mo, F.; Du, C.; Wang, C.; Yang, Z. Genome-Wide Identification of the Pectate Lyase Gene Family in Potato and Expression Analysis under Salt Stress. Plants 2024, 13, 1322. https://doi.org/10.3390/plants13101322
Wang Z, Liu T, Wu W, Shi W, Shi J, Mo F, Du C, Wang C, Yang Z. Genome-Wide Identification of the Pectate Lyase Gene Family in Potato and Expression Analysis under Salt Stress. Plants. 2024; 13(10):1322. https://doi.org/10.3390/plants13101322
Chicago/Turabian StyleWang, Zhiqi, Tao Liu, Wenbo Wu, Wenting Shi, Jian Shi, Fengyan Mo, Chong Du, Chaonan Wang, and Zhongmin Yang. 2024. "Genome-Wide Identification of the Pectate Lyase Gene Family in Potato and Expression Analysis under Salt Stress" Plants 13, no. 10: 1322. https://doi.org/10.3390/plants13101322





