OsGGC2, Gγ Subunit of Heterotrimeric G Protein, Regulates Plant Height by Functionally Overlapping with DEP1 in Rice
Abstract
:1. Introduction
2. Results
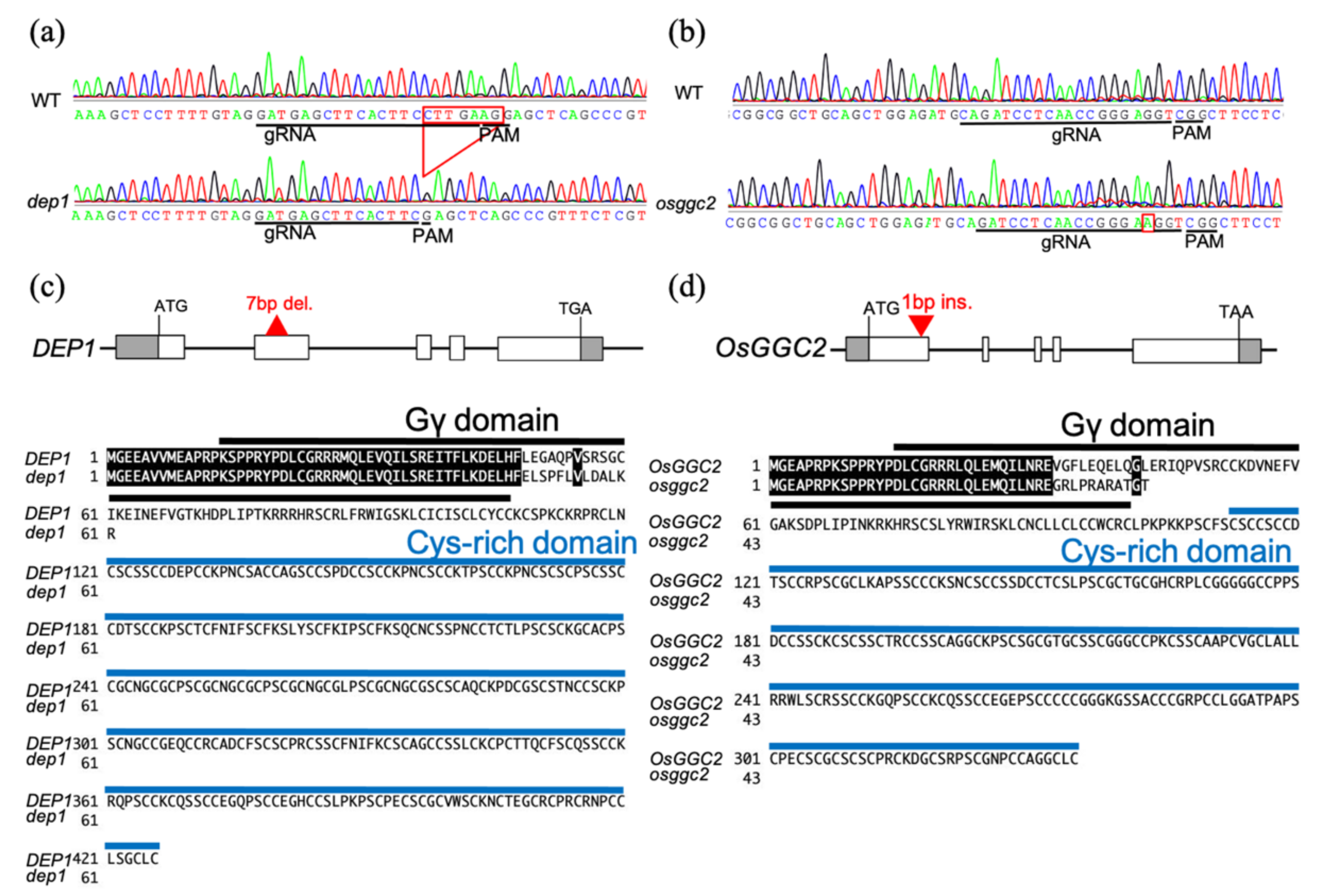
2.1. Loss-of-Function Mutants of DEP1 and OsGGC2
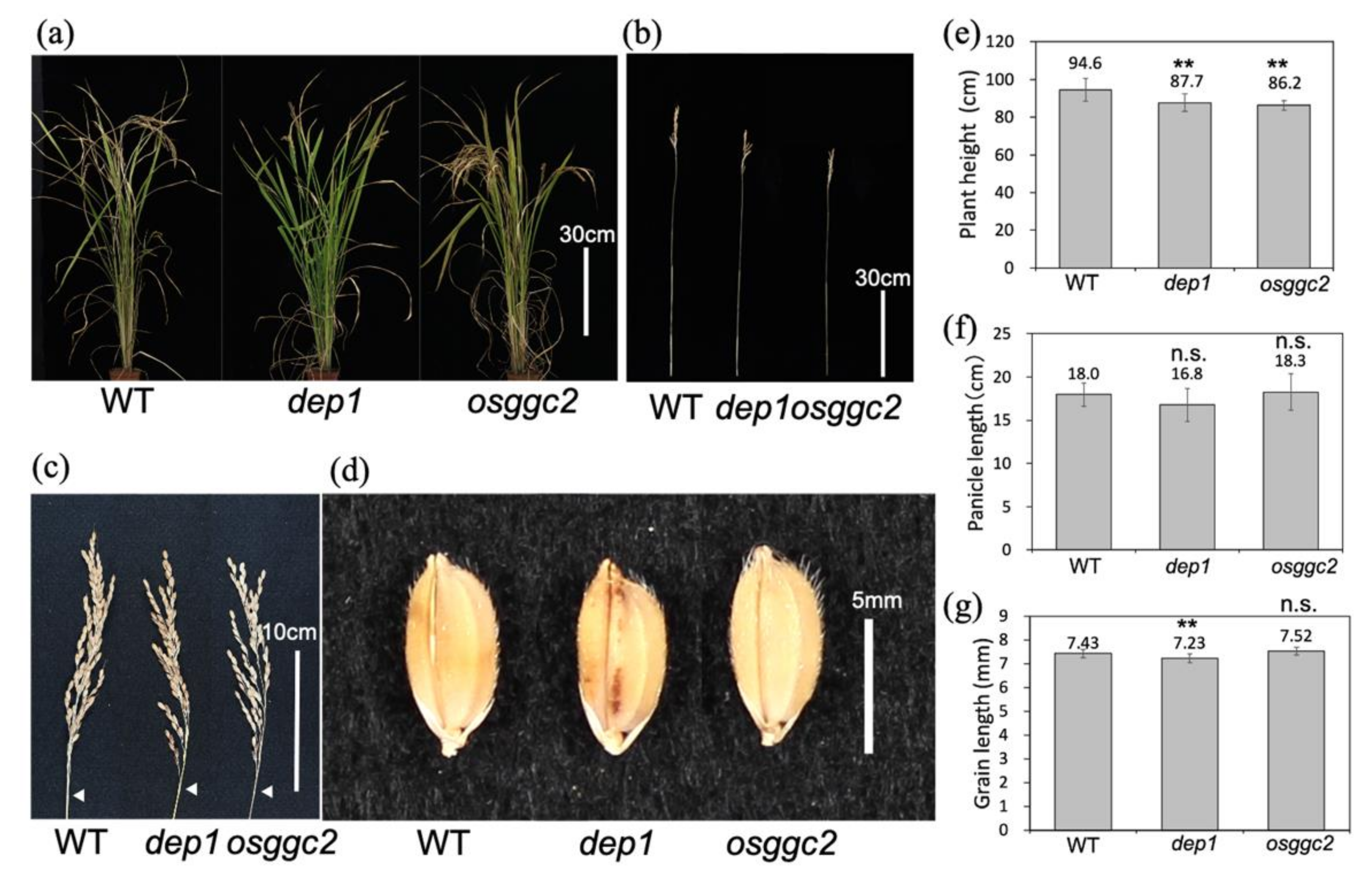
2.2. dep1 and osggc2 Show a Similar Semi-Dwarf Phenotype
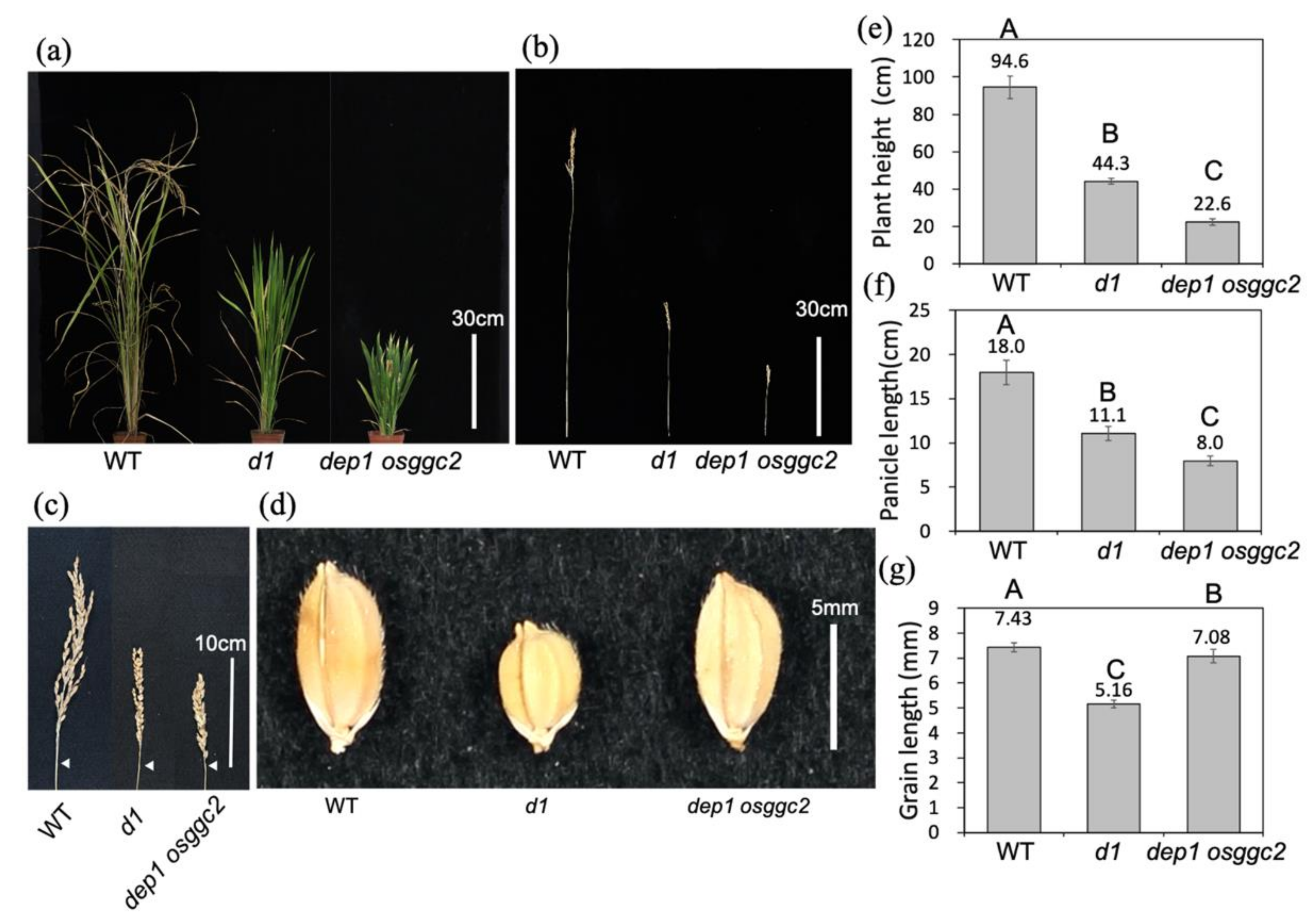
2.3. d1 Is Almost Epistatic to the Dwarf Phenotypes of dep1 and osggc2
2.4. OsGGC2 and DEP1 Redundantly Regulate Plant Height and Panicle Length in Rice
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Production of dep1 and osggc2 Mutants
4.3. Production of Double Mutants
4.4. Phenotype Evaluation
4.5. DNA and Amino Acid Sequences
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Offermanns, S. Mammalian G-protein function in vivo: New insights through altered gene expression. Rev. Physiol. Biochem. Pharmacol. 2000, 140, 63–133. [Google Scholar] [PubMed]
- Gomperts, B.D.; Kramer, I.J.M.; Tatham, P.E.R. Signal Transduction; Elsevier Inc.: Amsterdam, The Netherlands, 2002. [Google Scholar]
- Wettschureck, N.; Offermanns, S. Mammalian G proteins and their cell type specific functions. Physiol. Rev. 2005, 85, 1159–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milligan, G.; Kostenis, E. Heterotrimeric G-proteins: A short history. Br. J. Pharmacol. 2006, 147, S46–S55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, S. Plant receptor-like kinase signaling through heterotrimeric G-proteins. J. Exp. Bot. 2020, 71, 1742–1751. [Google Scholar] [CrossRef]
- Temple, B.R.S.; Jones, A.M. The Plant Heterotrimeric G-Protein Complex. Annu. Rev. Plant Biol. 2007, 58, 249–266. [Google Scholar] [CrossRef] [Green Version]
- Urano, D.; Chen, J.G.; Botella, J.R.; Jones, A.M. Heterotrimeric G protein signaling in the plant kingdom. Open Biol. 2013, 3, 120–186. [Google Scholar] [CrossRef] [Green Version]
- Urano, D.; Miura, K.; Wu, Q.; Iwasaki, Y.; Jackson, D.; Jones, A.M. Plant morphology of heterotrimeric G protein mutants. Plant Cell Physiol. 2016, 57, 437–445. [Google Scholar] [CrossRef]
- Maruta, N.; Trusov, Y.; Jones, A.M.; Botella, J.R. Heterotrimeric G Proteins in Plants: Canonical and Atypical Galpha Subunits. Int. J. Mol. Sci. 2021, 22, 11841. [Google Scholar] [CrossRef]
- Ashikari, M.; Wu, J.; Yano, M.; Sasaki, T.; Yoshimura, A. Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the alpha-subunit of GTP-binding protein. Proc. Natl. Acad. Sci. USA 1999, 96, 10284–10289. [Google Scholar] [CrossRef] [Green Version]
- Fujisawa, Y.; Kato, T.; Ohki, S.; Ishikawa, A.; Kitano, H.; Sasaki, T.; Asahi, T.; Iwasaki, Y. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc. Natl. Acad. Sci. USA 1999, 96, 7575–7580. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Jiang, N.; Xu, Z.; Xu, Q. Heterotrimeric G protein are involved in the regulation of multiple agronomic traits and stress tolerance in rice. BMC Plant Biol. 2020, 20, 90. [Google Scholar] [CrossRef] [Green Version]
- Utsunomiya, Y.; Samejima, C.; Takayanagi, Y.; Izawa, Y.; Yoshida, T.; Sawada, Y.; Fujisawa, Y.; Kato, H.; Iwasaki, Y. Suppression of the rice heterotrimeric G protein beta-subunit gene, RGB1, causes dwarfism and browning of internodes and lamina joint regions. Plant J. 2011, 67, 907–916. [Google Scholar] [CrossRef]
- Trusov, Y.; Chakravorty, D.; Botella, J.R. Diversity of heterotrimeric G-protein gamma subunits in plants. BMC Res. Notes 2012, 5, 608. [Google Scholar] [CrossRef] [Green Version]
- Botella, J.R. Can heterotrimeric G proteins help to feed the world? Trends Plant Sci. 2012, 17, 563–568. [Google Scholar] [CrossRef]
- Miao, J.; Yang, Z.; Zhang, D.; Wang, Y.; Xu, M.; Zhou, L.; Wang, J.; Wu, S.; Yao, Y.; Du, X.; et al. Mutation of RGG2, which encodes a type B heterotrimeric G protein gamma subunit, increases grain size and yield production in rice. Plant Biotechnol. J. 2019, 17, 650–664. [Google Scholar] [CrossRef] [Green Version]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef]
- Takano-Kai, N.; Jiang, H.; Powell, A.; McCouch, S.; Takamure, I.; Furuya, N.; Doi, K.; Yoshimura, A. Multiple and independent origins of short seeded alleles of GS3 in rice. Breed. Sci. 2013, 63, 77–85. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Wang, L.; Mao, H.; Shao, L.; Li, X.; Xiao, J.; Ouyang, Y.; Zhang, Q. A G-protein pathway determines grain size in rice. Nat. Commun. 2018, 9, 851. [Google Scholar] [CrossRef] [Green Version]
- Mikami, M.; Toki, S.; Endo, M. Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol. Biol. 2015, 88, 561–572. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Xu, R.; Duan, P.; Li, Y. Control of grain size in rice. Plant Reprod. 2018, 31, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Qian, Q.; Xu, T.; Zhang, Y.; Dong, G.; Gao, T.; Xie, Q.; Xue, Y. The U-box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G α subunit to regulate Brassinosteroid-mediated growth in rice. PLoS Genet. 2013, 9, e1003391. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaya, G.; Segami, S.; Fujita, M.; Morinaka, Y.; Iwasaki, Y.; Miura, K. OsGGC2, Gγ Subunit of Heterotrimeric G Protein, Regulates Plant Height by Functionally Overlapping with DEP1 in Rice. Plants 2022, 11, 422. https://doi.org/10.3390/plants11030422
Chaya G, Segami S, Fujita M, Morinaka Y, Iwasaki Y, Miura K. OsGGC2, Gγ Subunit of Heterotrimeric G Protein, Regulates Plant Height by Functionally Overlapping with DEP1 in Rice. Plants. 2022; 11(3):422. https://doi.org/10.3390/plants11030422
Chicago/Turabian StyleChaya, Genki, Shuhei Segami, Moeka Fujita, Yoichi Morinaka, Yukimoto Iwasaki, and Kotaro Miura. 2022. "OsGGC2, Gγ Subunit of Heterotrimeric G Protein, Regulates Plant Height by Functionally Overlapping with DEP1 in Rice" Plants 11, no. 3: 422. https://doi.org/10.3390/plants11030422





