Transcriptome Analysis of Potato Infected with the Necrotrophic Pathogen Alternaria solani
Abstract
:1. Introduction
2. Results
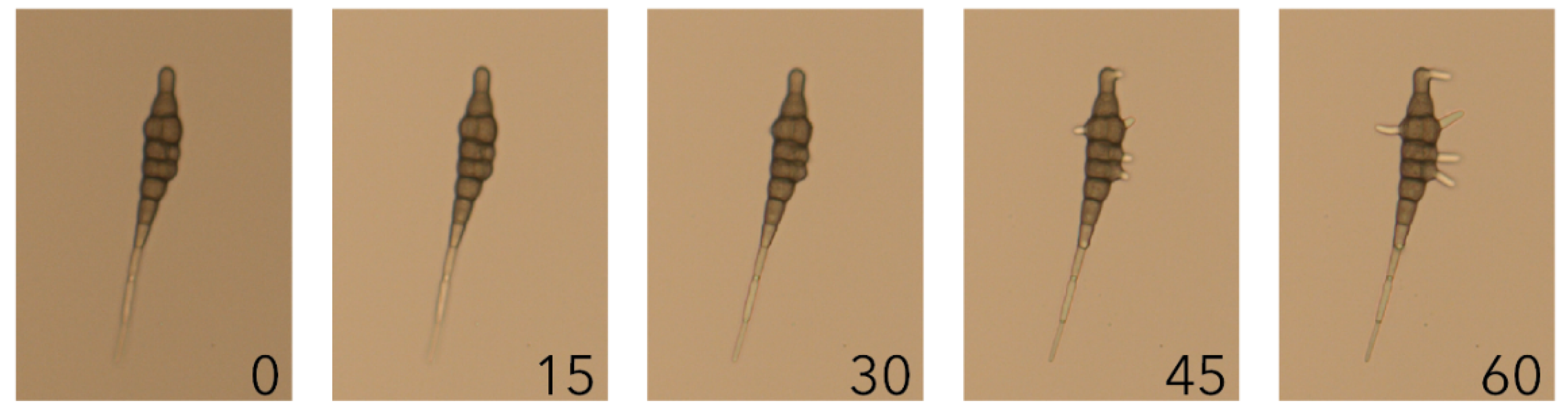
2.1. Alternaria solani Infection Progress over a Time Course of 48 Hours
2.2. Mapping of Reads to Potato and A. solani Reference Genomes
2.3. Differential Expression of Plant Transcripts during Infection
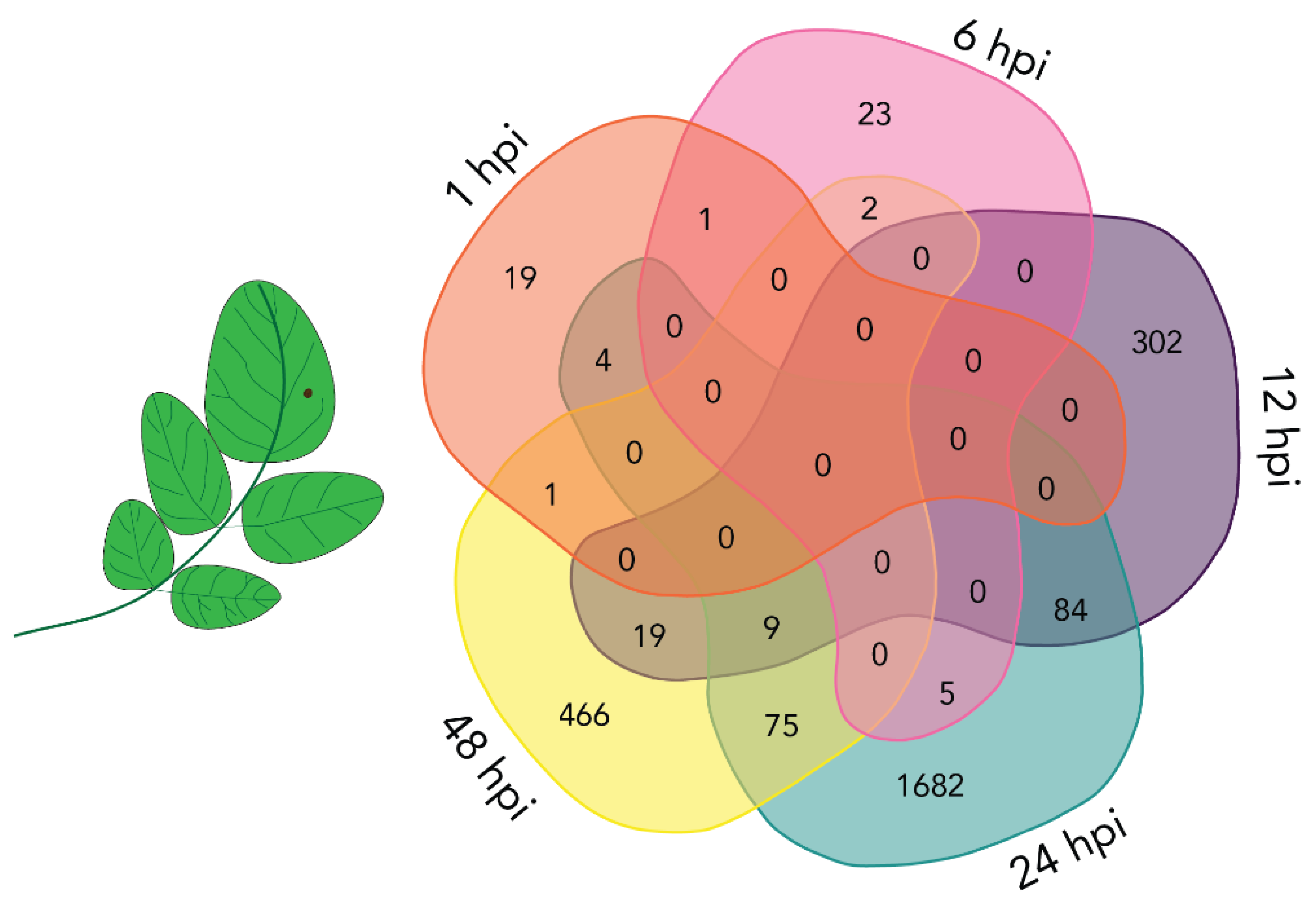
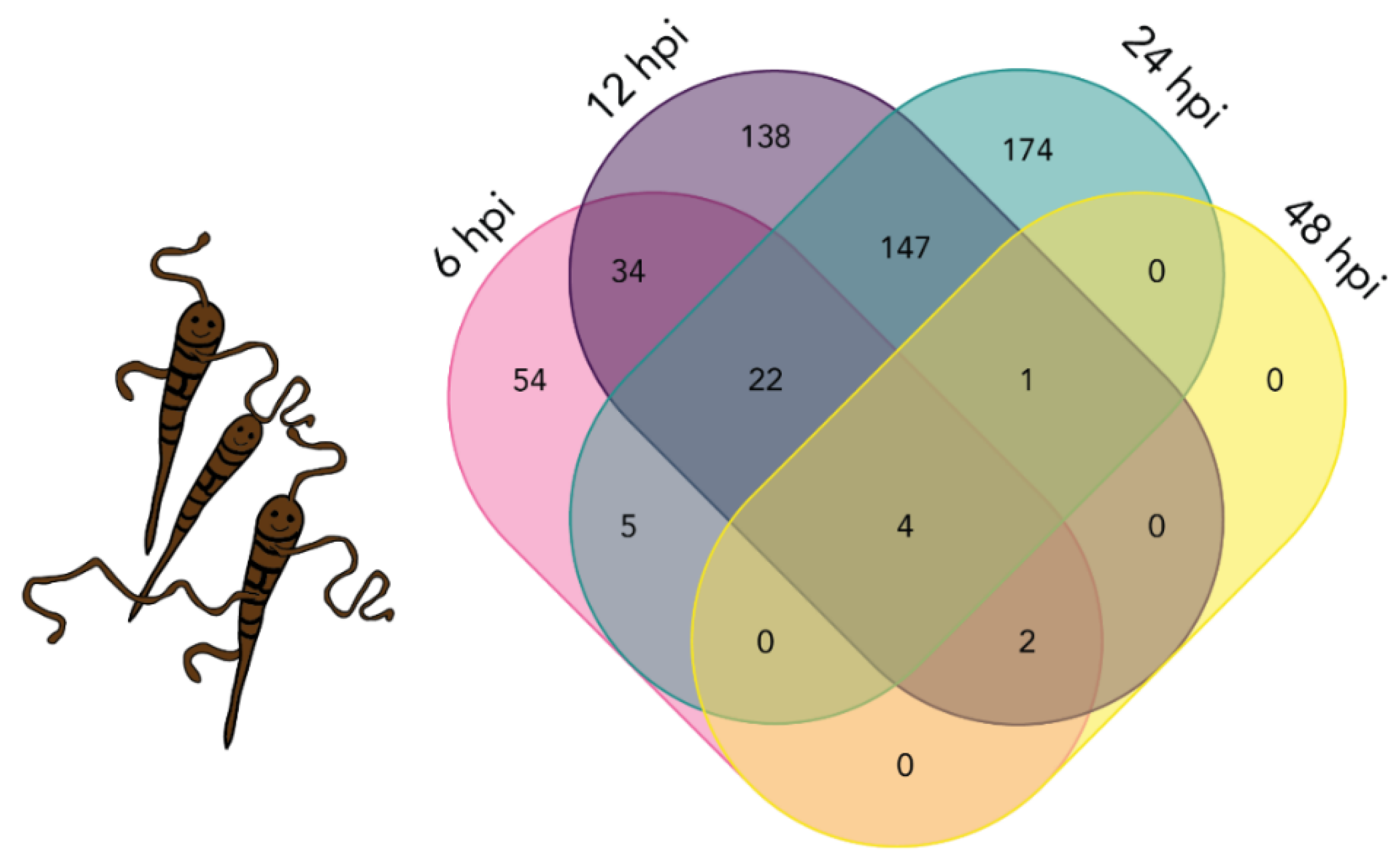
2.4. Mostly Unique Plant DETs at Different Time Points
2.5. Gene Ontology Enrichment Analysis
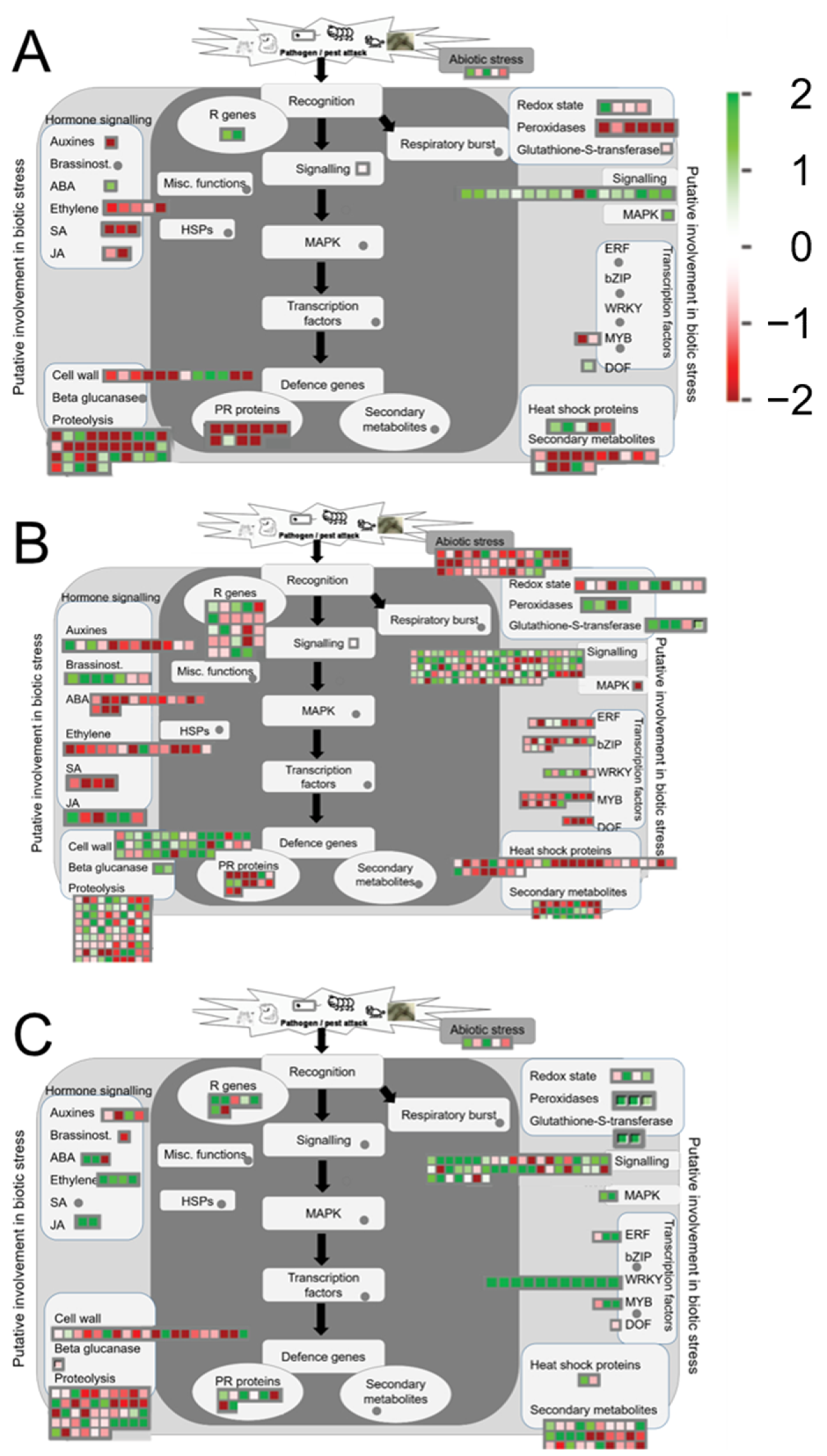
2.6. Differential Expression of Biotic Stress Related Plant Transcripts
2.7. Differential Expression of A. solani Transcripts during Infection
2.8. Quantitative Real Time PCR Gene Expression Analysis Correlates Well with RNA Sequencing Data
3. Discussion
3.1. Differential Expression of Solanum tuberosum Transcripts
3.2. Gene Ontology Enrichment of Drug Metabolic Processes at 12 hpi
3.3. Gene Ontology Enrichment of Photosynthesis Related Plant Transcripts at 48 hpi
3.4. Differential Expression of Hormone Biosynthesis Related Transcripts during A. solani Infection
3.5. A. solani DETs Overlapping in all Four Time Points
3.6. A. solani DETs Reveal Potential Pathogenicity Factors
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Alternaria solani Maintenance and Inoculum Preparation
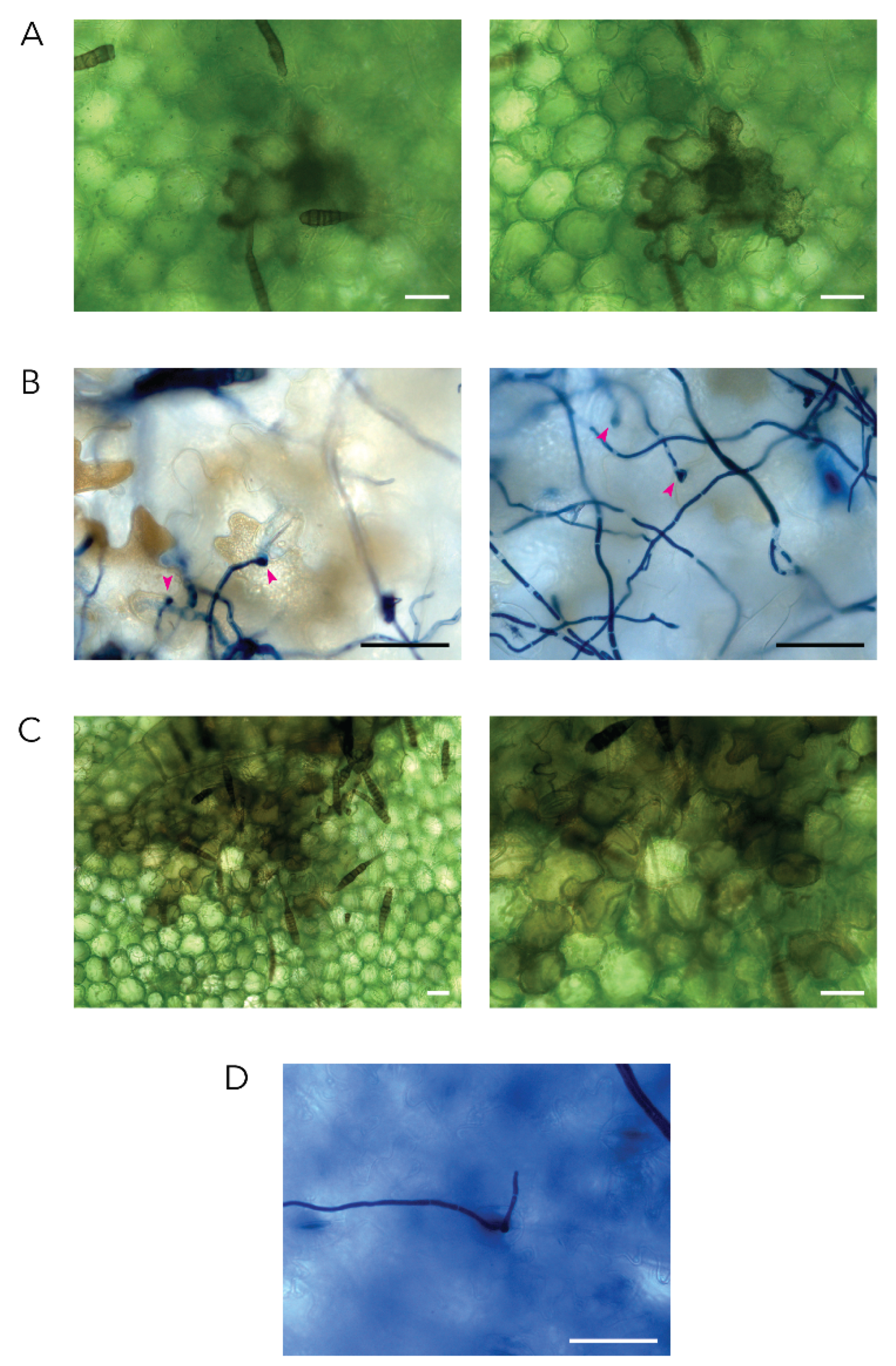
4.3. Microscopy
4.4. Plant Inoculation with Alternaria solani
4.5. RNA Preparation and Sequencing
4.6. Expression Analysis from RNA Sequencing
4.7. Validation of RNA Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Edin, E.; Andersson, B. The Early Blight Situation in Sweden—Species Abundance and Strobilurin Sensitivity. PPO Spec. Rep. 2014, 16, 83–84. [Google Scholar]
- Landschoot, S.; Vandecasteele, M.; De Baets, B.; Höfte, M.; Audenaert, K.; Haesaert, G. Identification of A. Arborescens, A. Grandis, and A. Protenta as New Members of the European Alternaria Population on Potato. Fungal Biol. 2017, 121, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Leiminger, J.H.; Hausladen, H. Early Blight Control in Potato Using Disease-Orientated Threshold Values. Plant Dis. 2012, 96, 124–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odilbekov, F.; Edin, E.; Mostafanezhad, H.; Coolman, H.; Grenville-Briggs, L.J.; Liljeroth, E. Within-Season Changes in Alternaria Solani Populations in Potato in Response to Fungicide Application Strategies. Eur. J. Plant Pathol. 2019, 155, 953–965. [Google Scholar] [CrossRef] [Green Version]
- Odilbekov, F.; Selga, C.; Ortiz, R.; Chawade, A.; Liljeroth, E. QTL Mapping for Resistance to Early Blight in a Tetraploid Potato Population. Agronomy 2020, 10, 728. [Google Scholar] [CrossRef]
- Dita, M.A.; Brommonschenkel, S.H.; Matsuoka, K.; Mizubuti, E.S.G. Histopathological Study of the Alternaria solani Infection Process in Potato Cultivars with Different Levels of Early Blight Resistance. J. Phytopathol 2007, 155, 462–469. [Google Scholar] [CrossRef]
- Govrin, E.M.; Levine, A. The Hypersensitive Response Facilitates Plant Infection by the Necrotrophic Pathogen Botrytis Cinerea. Curr. Biol. 2000, 10, 751–757. [Google Scholar] [CrossRef] [Green Version]
- Balint-Kurti P: The Plant Hypersensitive Response: Concepts, Control and Consequences. Mol. Plant Pathol. 2019, 20, 1163–1178.
- Brouwer, S.M.; Odilbekov, F.; Burra, D.D.; Lenman, M.; Hedley, P.E.; Grenville-Briggs, L.; Alexandersson, E.; Liljeroth, E.; Andreasson, E. Intact Salicylic Acid Signalling is Required for Potato Defence Against the Necrotrophic Fungus Alternaria Solani. Plant Mol. Biol. 2020, 104, 1–19. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A Modular and Extensible Implementation of the RAST Algorithm for Building Custom Annotation Pipelines and Annotating Batches of Genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Ramšak, Ž.; Baebler, Š.; Rotter, A.; Korbar, M.; Mozetič, I.; Usadel, B.; Gruden, K. Gomapman: Integration, Consolidation and Visualization of Plant Gene Annotations within the Mapman Ontology. Nucleic Acids Res. 2014, 42, D1167–D1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The Interpro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2020, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Massa, A.N.; Childs, K.L.; Lin, H.; Bryan, G.J.; Giuliano, G.; Buell, C.R. The Transcriptome of the Reference Potato Genome Solanum Tuberosum Group Phureja Clone DM1-3 516R44. PLoS ONE 2011, 6, e26801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large-Scale Biological Data Sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Ogawa, T.; Ishikawa, K.; Harada, K.; Fukusaki, E.; Yoshimura, K.; Shigeoka, S. Overexpression of an ADP-Ribose Pyrophosphatase, Atnudx2, Confers Enhanced Tolerance to Oxidative Stress in Arabidopsis Plants. Plant J. 2009, 57, 289–301. [Google Scholar] [CrossRef]
- Ge, X.; Xia, Y. The Role of Atnudt7, a Nudix Hydrolase, in the Plant Defense Response. Plant Signal. Behav. 2008, 3, 119–120. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, K.; Yoshimura, K.; Ogawa, T.; Shigeoka, S. Distinct Regulation of Arabidopsis ADP-Ribose/NADH Pyrophosphohydrolases, Atnudx6 And 7, In Biotic and Abiotic Stress Responses. Plant Signal. Behav. 2010, 5, 839–841. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, J.P.; Dong, X. Functional Characterization of a Nudix Hydrolase AtNUDX8 upon Pathogen Attack Indicates a Positive Role in Plant Immune Responses. PLoS ONE 2014, 9, e114119. [Google Scholar]
- Dong, S.; Wang, Y. Nudix Effectors: A Common Weapon in the Arsenal of Plant Pathogens. PLoS Pathog. 2016, 12, e1005704. [Google Scholar] [CrossRef] [Green Version]
- Sajjaanantakul, T.; Pitifer, L.A. Chapter 8—Pectinesterase. In The Chemistry and Technology of Pectin; Walter, R.H., Ed.; Academic Press: San Diego, CA, USA, 1991; pp. 135–164. [Google Scholar]
- Houston, K.; Tucker, M.R.; Chowdhury, J.; Shirley, N.; Little, A. The Plant Cell Wall: A Complex and Dynamic Structure As Revealed by the Responses of Genes under Stress Conditions. Front. Plant Sci. 2016, 7, 984. [Google Scholar] [CrossRef] [Green Version]
- Raiola, A.; Lionetti, V.; Elmaghraby, I.; Immerzeel, P.; Mellerowicz, E.J.; Salvi, G.; Cervone, F.; Bellincampi, D. Pectin Methylesterase is Induced in Arabidopsis Upon Infection and is Necessary for a Successful Colonization by Necrotrophic Pathogens. Mol. Plant-Microbe Interact. MPMI 2011, 24, 432–440. [Google Scholar] [CrossRef] [Green Version]
- Bilgin, D.D.; Zavala, J.A.; Zhu, J.; Clough, S.J.; Ort, D.R.; DeLucia, E.H. Biotic Stress Globally Downregulates Photosynthesis Genes. Plant Cell Environ. 2010, 33, 1597–1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Han, X.; Feng, D.; Yuan, D.; Huang, L.-J. Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering? Int. J. Mol. Sci. 2019, 20, 671. [Google Scholar] [CrossRef] [Green Version]
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and Diversity of the 2–Oxoglutarate-Dependent Dioxygenase Superfamily in Plants. Plant J. 2014, 78, 328–343. [Google Scholar] [CrossRef]
- Koo, Y.J.; Kim, M.A.; Kim, E.H.; Song, J.T.; Jung, C.; Moon, J.-K.; Kim, J.-H.; Seo, H.S.; Song, S.I.; Kim, J.-K.; et al. Overexpression of Salicylic Acid Carboxyl Methyltransferase Reduces Salicylic Acid-Mediated Pathogen Resistance in Arabidopsis Thaliana. Plant Mol. 2007, 64, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Jang, M.; Srivastava, A.; Jang, J.-H.; Soung, N.-K.; Ko, S.-K.; Kang, D.-O.; Ahn, J.S.; Kim, B.Y. A Pectate Lyase-Coding Gene Abundantly Expressed during Early Stages of Infection Is Required for Full Virulence in Alternaria brassicicola. PLoS ONE 2015, 10, e0127140. [Google Scholar] [CrossRef] [Green Version]
- Vélëz, H.; Glassbrook, N.J.; Daub, M.E. Mannitol Biosynthesis is Required for Plant Pathogenicity by lternaria Alternata. Fems. Microbiol. Lett. 2008, 285, 122–129. [Google Scholar] [PubMed] [Green Version]
- Calmes, B.; Guillemette, T.; Teyssier, L.; Siegler, B.; Pigné, S.; Landreau, A.; Iacomi, B.; Richomme, P.; Lemoine, R.; Simoneau, P. Role of Mannitol Metabolism in the Pathogenicity of the Necrotrophic Fungus Alternaria Brassicicola. Front. Plant Sci. 2013, 4, 131. [Google Scholar] [CrossRef] [Green Version]
- Meena, M.; Upadhyay, R.S.; Prasad, V.; Zehra, A.; Gupta, V.K. Mannitol Metabolism during Pathogenic Fungal–Host Interactions under Stressed Conditions. Front. Microbiol. 2015, 6, 1019. [Google Scholar] [CrossRef] [Green Version]
- Kustrzeba-Wójcicka, I.; Siwak, E.; Terlecki, G.; Wolańczyk-Mędrala, A.; Mędrala, W. Alternaria Alternata and its Allergens: A Comprehensive Review. Clin. Rev. Allerg Immunol. 2014, 47, 354–365. [Google Scholar] [CrossRef]
- Abdul, W.; Aliyu, S.R.; Lin, L.; Sekete, M.; Chen, X.; Otieno, F.J.; Yang, T.; Lin, Y.; Norvienyeku, J.; Wang, Z. Family-Four Aldehyde Dehydrogenases Play an Indispensable Role in the Pathogenesis of Magnaporthe Oryzae. Front. Plant Sci. 2018, 9, 980. [Google Scholar] [CrossRef] [Green Version]
- Pigné, S.; Zykwinska, A.; Janod, E.; Cuenot, S.; Kerkoud, M.; Raulo, R.; Bataillé-Simoneau, N.; Marchi, M.; Kwasiborski, A.; N’Guyen, G.; et al. A Flavoprotein Supports Cell Wall Properties in the Necrotrophic Fungus Alternaria Brassicicola. Fungal Biol. Biotechnol. 2017, 4, 1. [Google Scholar] [CrossRef]
- Tournu, H.; Fiori, A.; Dijck, P.V. Relevance of Trehalose in Pathogenicity: Some General Rules, Yet Many Exceptions. PLoS Pathog. 2013, 9, e1003447. [Google Scholar] [CrossRef] [Green Version]
- Sützl, L.; Foley, G.; Gillam, E.M.J.; Bodén, M.; Haltrich, D. The GMC Superfamily of Oxidoreductases Revisited: Analysis and Evolution of Fungal GMC Oxidoreductases. Biotechnol. Biofuels 2019, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, L.; Hong, Y.; Zhang, Y.; Liu, S.; Li, D.; Zhang, H.; Song, F. Co-Silencing of Tomato S-Adenosylhomocysteine Hydrolase Genes Confers Increased Immunity against Pseudomonas Syringae Pv. Tomato DC3000 and Enhanced Tolerance to Drought Stress. Front. Plant Sci. 2015, 6, 717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, S.; Li, R.; Shi, L.; Wang, J.; Shang, J.; Zhu, P.; Chen, B. Functional Analysis of an S-Adenosylhomocysteine Hydrolase Homolog of Chestnut Blight Fungus. Fems. Microbiol. Lett. 2012, 336, 64–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Edwards, M.C. Genome-Wide Analysis of Small Secreted Cysteine-Rich Proteins Identifies Candidate Effector Proteins Potentially Involved in Fusarium graminearum—Wheat Interactions. Phytopathol 2016, 106, 166–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iftikhar, S.; Shahid, A.A.; Halim, S.A.; Wolters, P.J.; Vleeshouwers, V.G.A.A.; Khan, A.; Al-Harrasi, A.; Ahmad, S. Discovering Novel Alternaria Solani Succinate Dehydrogenase Inhibitors by in Silico Modeling and Virtual Screening Strategies to Combat Early Blight. Front. Chem. 2017, 5, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Littlejohn, G.R.; Mansfield, J.C.; Christmas, J.T.; Witterick, E.; Fricker, M.D.; Grant, M.R.; Smirnoff, N.; Everson, R.M.; Moger, J.; Love, J. An Update: Improvements in Imaging Perfluorocarbon-Mounted Plant Leaves with Implications for Studies of Plant Pathology, Physiology, Development and Cell Biology. Front. Plant Sci. 2014, 5, 140. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Bautista, N.; Domínguez-Núñez, J.A.; Moreno, M.M.C.; Berrocal-Lobo, M. Plant Tissue Trypan Blue Staining During Phytopathogen Infection. Bio-Protocol 2016, 6, e2078. [Google Scholar] [CrossRef] [Green Version]
- Liang, D.; Andersen, C.B.; Vetukuri, R.R.; Dou, D.; Grenville-Briggs, L.J. Horizontal Gene Transfer and Tandem Duplication Shape the Unique CAZyme Complement of the Mycoparasitic Oomycetes Pythium oligandrum and Pythium periplocum. Front. Microbiol. 2020, 11, 2609. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and Accurate Filtering of Ribosomal Rnas in Metatranscriptomic Data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- FastQC. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 15 October 2020).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11.14.11–11.14.19. [Google Scholar] [CrossRef] [Green Version]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential Expression Analysis for Sequence Count Data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-Based Differential Expression Analysis of RNA Sequencing Data Using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with Deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, H.X.; Pryor, B.; Peever, T.; Lawrence, C.B. The Alternaria Genomes Database: A Comprehensive Resource for a Fungal Genus Comprised of Saprophytes, Plant Pathogens, and Allergenic Species. BMC Genom. 2015, 16, 239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize Implements and Enhances Circular Visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI Search and Sequence Analysis Tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 Improves Signal Peptide Predictions Using Deep Neural Networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.M. The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005. [Google Scholar]
- Kumar, S.; Singh, R.; Kashyap, P.L.; Srivastava, A.K. Rapid Detection and Quantification of Alternaria Solani in Tomato. Sci. Hortic. 2013, 151, 184–189. [Google Scholar] [CrossRef]
- Avrova, A.O.; Venter, E.; Birch, P.R.J.; Whisson, S.C. Profiling and Quantifying Differential Gene Transcription in Phytophthora Infestans Prior to and During the Early Stages of Potato Infection. Fungal Genet. Biol. 2003, 40, 4–14. [Google Scholar] [CrossRef]

| Time Point (Hours Post Inoculation) | |||||
|---|---|---|---|---|---|
| 1 | 6 | 12 | 24 | 48 | |
| Total reads (Million) | 27.33 | 28.23 | 31.06 | 29.55 | 35.17 |
| Mapping to S. tuberosum reference genome (%) | 87.2 | 88.5 | 86.6 | 86.6 | 88.9 |
| Unmapped to S. tuberosum mapping to A. solani reference genome (%) | 1.64 | 4.17 | 1.63 | 0.27 | 0.05 |
| Transcript ID | log2 Fold Change | Gene Description |
|---|---|---|
| PGSC0003DMT400020751 | 7.74 | Hydrolase |
| PGSC0003DMT400029007 | 7.65 | ATPase |
| PGSC0003DMT400021742 | 7.65 | Mutt domain protein |
| PGSC0003DMT400078504 | −7.56 | ALG2-interacting protein X |
| PGSC0003DMT400026165 | −7.57 | Stress regulated protein |
| PGSC0003DMT400076667 | −7.66 | WRKY transcription factor |
| PGSC0003DMT400006755 | −7.81 | Protein disulfide isomerase |
| PGSC0003DMT400061097 | −7.89 | Phosphomethylpyrimidine kinase |
| PGSC0003DMT400079490 | −8.45 | Chromatin remodeling complex subunit |
| PGSC0003DMT400073533 | −8.57 | Sugar transporter |
| Transcript ID | Log2 Fold Change | Gene Description RASTtk-v1.073 | Gene Description Alternaria alternata | |||
|---|---|---|---|---|---|---|
| 6 hpi | 12 hpi | 24 hpi | 48 hpi | |||
| mRNA_3291 | −2.68 | −8.17 | −4.52 | −4.52 | Pectate lyase (EC 4.2.2.2) | pectate lyase precursor |
| mRNA_8720 | −2.54 | −4.34 | −8.14 | −6.31 | Inositol-1-phosphate synthase (EC 5.5.1.4) | Myo-inositol-1-phosphate synthase |
| mRNA_10760 | 4.86 | 6.02 | 5.78 | 6.74 | hypothetical protein | NADP-dependent mannitol dehydrogenase |
| mRNA_10750 | 5.62 | 7.78 | 7.53 | 6.69 | Lead, cadmium, zinc and mercury transporting ATPase (EC 3.6.3.3) (EC 3.6.3.5) | potassium/sodium eff |
| Time Point | Transcript ID | Log2 Fold Change | Gene Description RASTtk-v1.073 | Gene Description Alternaria alternata | Predicted Signal Peptide/Non-Cytoplamic Domain |
|---|---|---|---|---|---|
| 6 hpi | mRNA_9018 | 7.98 | Aldehyde dehydrogenase (EC 1.2.1.3) | aldehyde dehydrogenase | |
| mRNA_10227 | 6.96 | Acetyl-CoA hydrolase/transferase family protein | hypothetical protein | ||
| mRNA_11966 | 6.60 | hypothetical protein | hypothetical protein | x/x | |
| mRNA_9568 | 6.57 | hypothetical protein | hypothetical protein | ||
| mRNA_5212 | 6.51 | Malate dehydrogenase (EC 1.1.1.37) | malate dehydrogenase-like protein | ||
| mRNA_9008 | −4.90 | hypothetical protein | hypothetical protein | x/x | |
| mRNA_914 | −5.75 | hypothetical protein | S-adenosyl-L-methionine-dependent methyltransferase | ||
| mRNA_4527 | −5.99 | hypothetical protein | hypothetical protein | ||
| mRNA_9891 | −6.42 | monooxygenase FAD-binding protein | monooxygenase | ||
| mRNA_8569 | −6.92 | hypothetical protein | x/x | ||
| 12 hpi | mRNA_9018 | 10.21 | Aldehyde dehydrogenase (EC 1.2.1.3) | aldehyde dehydrogenase | |
| mRNA_29 | 8.93 | Trehalase (EC 3.2.1.28) | trehalase | x/x | |
| mRNA_10328 | 8.82 | hypothetical protein | hypothetical protein | ||
| mRNA_11100 | 8.70 | hypothetical protein | opsin-1 | ||
| mRNA_5135 | 8.70 | hypothetical protein | GroES-like protein | ||
| mRNA_4571 | −5.43 | hypothetical protein | hypothetical protein | x/x | |
| mRNA_10884 | −5.46 | hypothetical protein | hypothetical protein | ||
| mRNA_8569 | −5.79 | hypothetical protein | x/x | ||
| mRNA_714 | −5.97 | Glycerol-3-phosphate dehydrogenase [NAD(P)%2B] (EC 1.1.1.94) | NAD-dependent glycerol-3-phosphate dehydrogenase | ||
| mRNA_3291 | −8.17 | Pectate lyase (EC 4.2.2.2) | pectate lyase precursor | x/x | |
| 24 hpi | mRNA_6735 | 10.92 | hypothetical protein | heme oxygenase-like protein | |
| mRNA_6435 | 9.47 | hypothetical protein | hypothetical protein | ||
| mRNA_7342 | 9.07 | hypothetical protein | DUF1761-domain-containing protein | ||
| mRNA_5135 | 8.90 | hypothetical protein | GroES-like protein | ||
| mRNA_2512 | 8.14 | Oxidoreductase, GMC family | glucose-methanol-choline oxidoreductase-like protein | x/x | |
| mRNA_9110 | −5.20 | Adenosylhomocysteinase (EC 3.3.1.1) | S-adenosyl-L-homocysteine hydrolase | ||
| mRNA_8537 | −6.08 | hypothetical protein | general substrate transporter | ||
| mRNA_3291 | −4.52 | Pectate lyase (EC 4.2.2.2) | pectate lyase precursor | x/x | |
| mRNA_4806 | −6.74 | cell wall surface anchor family protein | |||
| mRNA_8720 | −8.14 | Inositol-1-phosphate synthase (EC 5.5.1.4) | Myo-inositol-1-phosphate synthase | ||
| 48 hpi | mRNA_10760 | 6.74 | hypothetical protein | NADP-dependent mannitol dehydrogenase | |
| mRNA_10750 | 6.69 | Lead, cadmium, zinc and mercury transporting ATPase (EC 3.6.3.3) (EC 3.6.3.5) | potassium/sodium eff | ||
| mRNA_10227 | 6.53 | Acetyl-CoA hydrolase/transferase family protein | hypothetical protein | ||
| mRNA_5802 | 5.76 | hypothetical protein | peptidyl-prolyl cis-trans isomeras-like protein | ||
| mRNA_3291 | −4.52 | Pectate lyase (EC 4.2.2.2) | pectate lyase precursor | x/x | |
| mRNA_4806 | −4.94 | cell wall surface anchor family protein | |||
| mRNA_8720 | −6.31 | Inositol-1-phosphate synthase (EC 5.5.1.4) | Myo-inositol-1-phosphate synthase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brouwer, S.M.; Brus-Szkalej, M.; Saripella, G.V.; Liang, D.; Liljeroth, E.; Grenville-Briggs, L.J. Transcriptome Analysis of Potato Infected with the Necrotrophic Pathogen Alternaria solani. Plants 2021, 10, 2212. https://doi.org/10.3390/plants10102212
Brouwer SM, Brus-Szkalej M, Saripella GV, Liang D, Liljeroth E, Grenville-Briggs LJ. Transcriptome Analysis of Potato Infected with the Necrotrophic Pathogen Alternaria solani. Plants. 2021; 10(10):2212. https://doi.org/10.3390/plants10102212
Chicago/Turabian StyleBrouwer, Sophie M., Maja Brus-Szkalej, Ganapathi V. Saripella, Dong Liang, Erland Liljeroth, and Laura J. Grenville-Briggs. 2021. "Transcriptome Analysis of Potato Infected with the Necrotrophic Pathogen Alternaria solani" Plants 10, no. 10: 2212. https://doi.org/10.3390/plants10102212






