WRKY Transcription Factor Response to High-Temperature Stress
Abstract
:1. Introduction
2. Characteristics of WRKY TFs
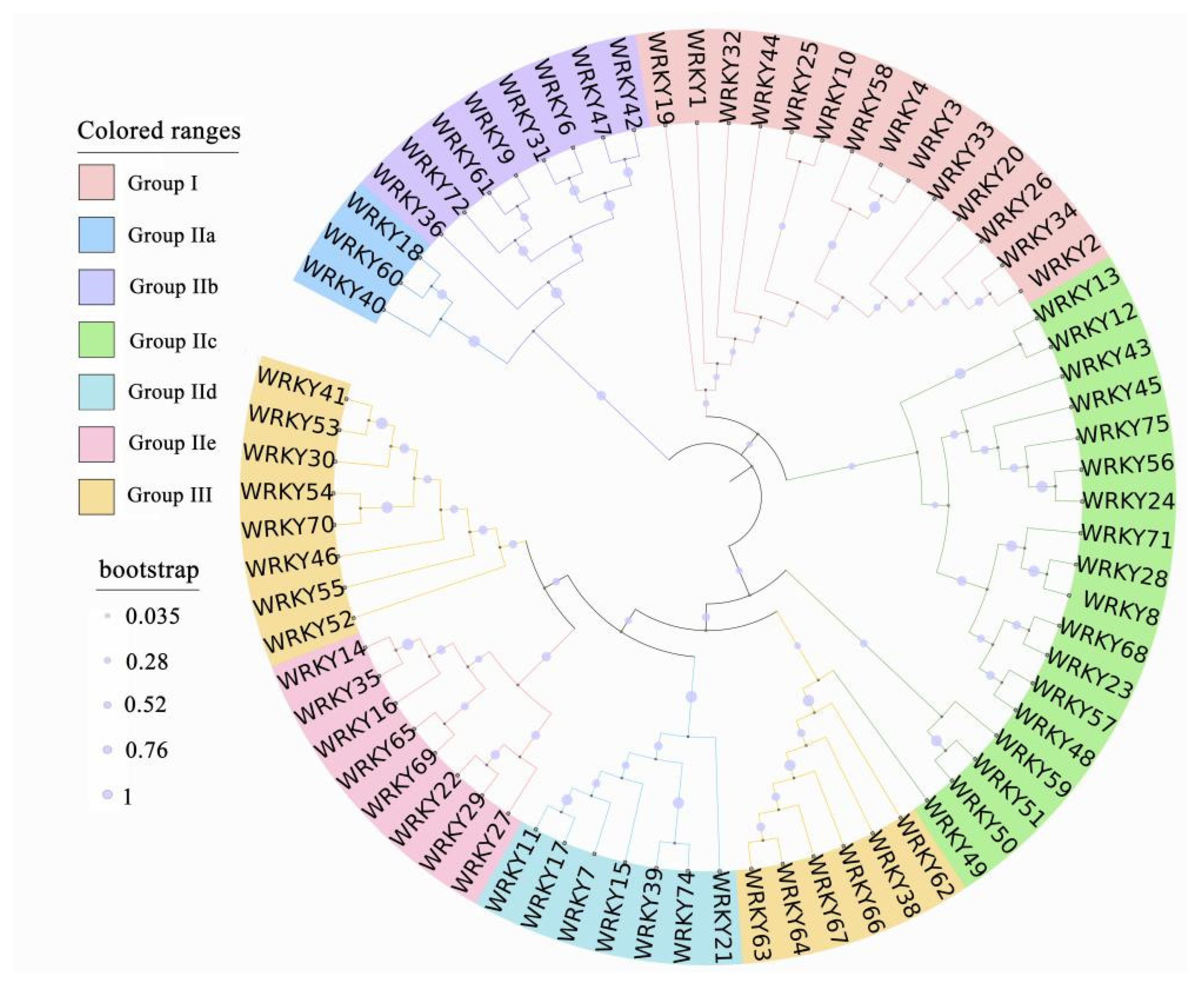
2.1. Structure and Classification
2.2. Origin and Evolution
3. Regulatory Mechanism of WRKY TFs
3.1. Transcription-Level Regulation
3.1.1. WRKY Expression Patterns
3.1.2. WRKY TFs Interact with Downstream Target Genes
3.2. Protein-Level Regulation
3.2.1. WRKY TFs Interact with Other WRKY Proteins
3.2.2. WRKY-VQ Motif Protein Interaction
3.2.3. WRKY TFs Interact with Other Proteins
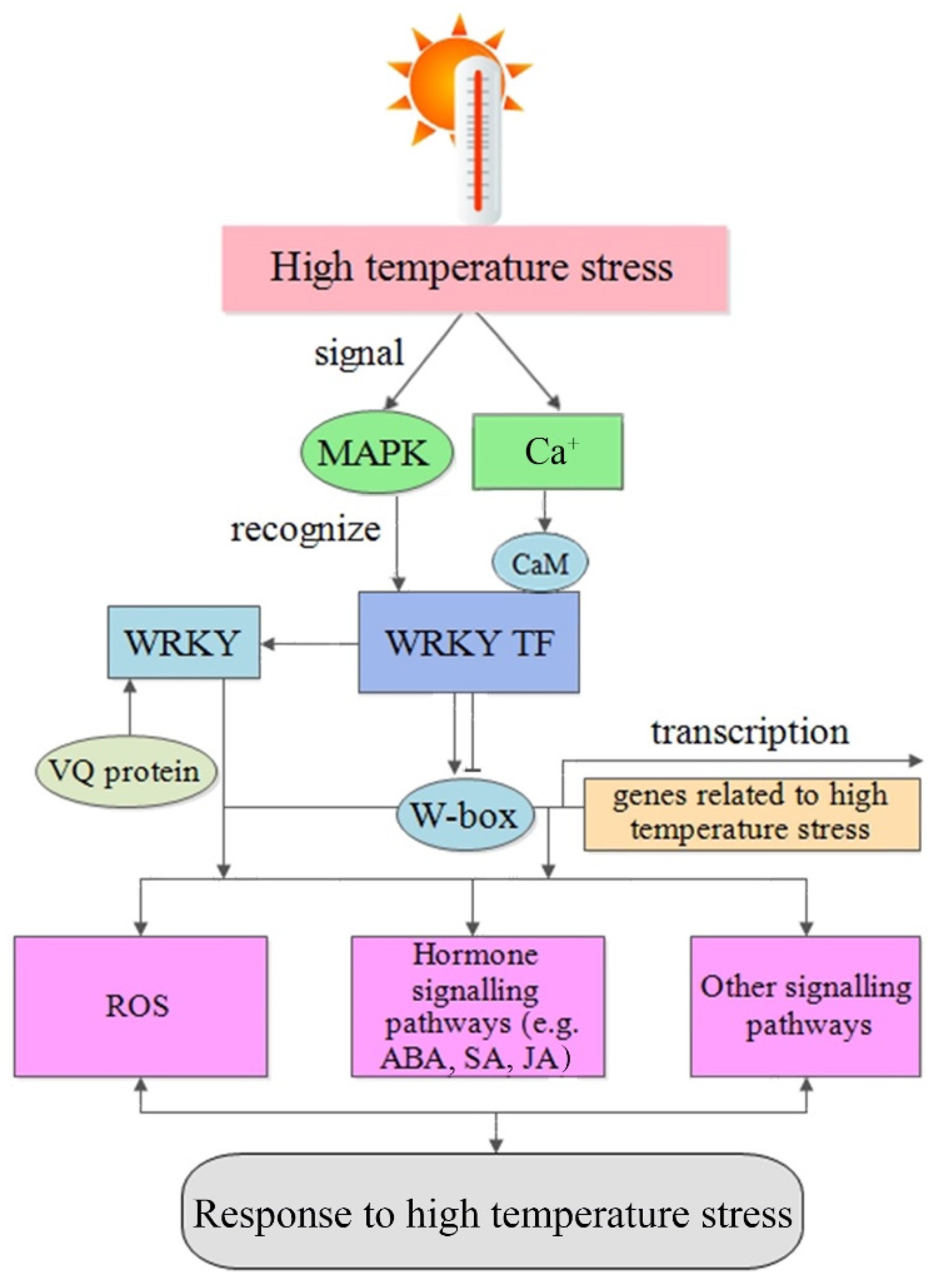
4. WRKY TFs in Response to High Temperature
4.1. Role of WRKY TFs under High-Temperature Stress Conditions
4.2. Role of WRKY TFs in Signaling in Response to High Temperature
4.2.1. ROS-Mediated Signaling Pathway
4.2.2. Plant Hormone-Mediated Signaling Pathway
4.2.3. MAPK-Mediated Signaling Pathway
5. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goraya, G.K.; Kaur, B.; Asthir, B.; Bala, S.; Kaus, G.; Faroog, M. Rapid Injuries of High Temperature in Plants. J. Plant Biol. 2017, 60, 298–305. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S. Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grover, A.; Mittal, D.; Negi, M.; Lavania, D. Generating high temperature tolerant transgenic plants: Achievements and challenges. Plant Sci. 2013, 205, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Gao, K.; Ren, H.; Tang, W. Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 2018, 60, 757–779. [Google Scholar] [CrossRef]
- Rivero, R.M.; Mittler, R.; Shulaev, V.; Blumwald, E.; Suzuki, N. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar]
- Maa, B. Thermotolerance in plants: Potential physio-biochemical and molecular markers for crop improvement—Science Direct. Environ. Exp. Bot. 2021, 186, 104454. [Google Scholar]
- Nijabat, A.; Bolton, A.; Mahmood-Ur-Rehman, M.; Shah, A.I.; Simon, P. Cell Membrane Stability and Relative Cell Injury in Response to Heat Stress during Early and Late Seedling Stages of Diverse Carrot (Daucus carota L.) Germplasm. Hort Sci. Publ. Am. Soc. Hortic. Sci. 2020, 55, 1–7. [Google Scholar] [CrossRef]
- Soengas, P.; Rodríguez, V.M.; Velasco, P.; Cartea, M.E. Effect of temperature stress on antioxidant defenses in Brassica oleracea. ACS Omega 2018, 3, 5237–5243. [Google Scholar] [CrossRef]
- Abdelrahman, M.; El-Sayed, M.; Jogaiah, S.; Burritt, D.J.; Tran, L. The “STAY-GREEN” trait and phytohormone signaling networks in plants under heat stress. Plant Cell Rep. 2017, 36, 1–17. [Google Scholar] [CrossRef]
- Jiang, Y.; Zeng, B.; Zhao, H.; Zhang, M.; Xie, S.; Lai, J. Genome-wide Transcription Factor Gene Prediction and their Expressional Tissue-Specificities in Maize, F. J. Integr. Plant Biol. 2012, 54, 616–630. [Google Scholar] [CrossRef]
- Xu, Y.-P.; Xu, H.; Wang, B.; Su, X.-D. Crystal structures of N-terminal WRKY transcription factors and DNA complexes. Protein Cell 2020, 11, 208–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Saiyinduleng; Chang, Q.; Cheng, C.; Zheng, Z.; Yu, S. Identification of yellowhorn (Xanthoceras sorbifolium) WRKY transcription factor family and analysis of abiotic stress response model. J. For. Res. 2020, 32, 987–1004. [Google Scholar] [CrossRef]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Ülker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Liang, G.; Yu, D. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 2012, 5, 1375–1388. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Jia, H.; Chen, X.; Hao, L.; An, H.; Guo, X. The Cotton WRKY Transcription Factor GhWRKY17 Functions in Drought and Salt Stress in Transgenic Nicotiana benthamiana Through ABA Signaling and the Modulation of Reactive Oxygen Species Production. Plant Cell Physiol. 2014, 55, 2060–2076. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.Y.; Tian, A.G.; Zou, H.F.; Xie, Z.M.; Lei, G.; Huang, J.; Wang, C.M.; Wang, H.W.; Zhang, J.S.; Chen, S.Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Kilian, J.; Whitehead, D.; Horak, J.; Wanke, D.; Weinl, S.; Batistic, O.; D’Angelo, C.; Bornberg-Bauer, E.; Kudla, J.; Harter, K. The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2010, 50, 347–363. [Google Scholar] [CrossRef]
- Eulgem, T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Ence. 2005, 10, 71–78. [Google Scholar] [CrossRef]
- Aha, B.; Anc, B.; Mika, B.; Mz, D.; Ma, E.; Ma, F.; Mfag, B.; Zl, B.; Ar, H.; Sm, A. Molecular regulation of pepper innate immunity and stress tolerance: An overview of WRKY TFs. Microb. Pathog. 2019, 135, 103610. [Google Scholar]
- Jiang, J.; Ma, S.; Ye, N.; Ming, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2016, 59, 86. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, Z.-L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef] [Green Version]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef] [Green Version]
- Rushton, P.J.; Macdonald, H.; Huttly, A.K.; Lazarus, C.M.; Hooley, R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol. Biol. 1995, 29, 691–702. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Tomo, Y. Solution Structure of an Arabidopsis WRKY DNA Binding Domain. Plant Cell 2005, 17, 944–956. [Google Scholar] [CrossRef] [Green Version]
- Duan, M.-R.; Nan, J.; Liang, Y.-H.; Mao, P.; Lu, L.; Li, L.; Wei, C.; Lai, L.; Li, Y.; Su, X.-D. DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res. 2007, 35, 1145–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciolkowski, I.; Wanke, D.; Birkenbihl, R.P.; Somssich, I.E. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 2008, 68, 81–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, H.; Wang, S. WRKY-type transcription factors: A significant factor in rice-pathogen interactions. Sci. Sin. Vitae 2014, 44, 784–793. [Google Scholar] [CrossRef] [Green Version]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Watanabe, S.; Tateno, M.; Seki, M.; Shinozaki, K.; Yokoyama, S. Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains. Plant Physiol. Biochem. PPB 2008, 46, 394–401. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.; Samaha, R. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Wu, K.-L.; Guo, Z.-J.; Wang, H.-H.; Li, J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef] [Green Version]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. MGG 1994, 244, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Deslandes, L.; Olivier, J.; Theulieres, F.; Hirsch, J.; Feng, D.X.; Bittner-Eddy, P.; Beynon, J.; Marco, Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA 2002, 99, 2404–2409. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Palmqvist, S.; Olsson, H.; Borén, M.; Ahlandsberg, S.; Jansson, C. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell Online 2003, 15, 2076–2092. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.Y.; Lee, S.H.; Park, H.C.; Bae, C.G.; Cheong, Y.H.; Choi, Y.J.; Han, C.D.; Lee, S.Y.; Lim, C.O.; Cho, M.J. Identification of rice blast fungal elicitor-responsive genes by differential display analysis. Mol. Plant-Microbe Interact. 2000, 13, 470–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyer, K.; Binder, A.; Boller, T.; Collinge, M. Identification of potato genes induced during colonization by Phytophthora infestans. Mol. Plant Pathol. 2010, 2, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Dellagi, A.; Heilbronn, J.; Avrova, A.O.; Montesano, M.; Palva, E.T.; Stewart, H.E.; Toth, I.K.; Cooke, D.; Lyon, G.D.; Birch, P. A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp. atroseptica and Phytophthora infestans and is coregulated with class I endochitinase expression. Mol. Plant Microbe Interact. 2000, 13, 1092–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandrova, K.S.; Conger, B.V. Isolation of two somatic embryogenesis-related genes from orchardgrass (Dactylis glomerata). Plant Sci. 2002, 162, 301–307. [Google Scholar] [CrossRef]
- Qi, S.; Yuhang, S.; Du Miying, D.J. Progress on Plant WRKY Transcription Factor. Chin. Agric. Sci. Bull. 2007, 23, 94. [Google Scholar]
- Zentgraf, H.U. Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 2001, 213, 469–473. [Google Scholar]
- Chen, W.; Provart, N.J.; Glazebrook, J.; Katagiri, F.; Chang, H.-S.; Eulgem, T.; Mauch, F.; Luan, S.; Zou, G.; Whitham, S.A. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 2002, 14, 559–574. [Google Scholar] [CrossRef]
- Jiang, Y.; Duan, Y.; Yin, J.; Ye, S.; Zhu, J.; Zhang, F.; Lu, W.; Fan, D.; Luo, K. Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 2014, 22, 6629–6644. [Google Scholar] [CrossRef]
- Villano, C.; Esposito, S.; D’Amelia, V.; Garramone, R.; Carputo, D. WRKY genes family study reveals tissue-specific and stress-responsive TFs in wild potato species. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, D.; Yang, C.; Kong, N.; Shi, Z.; Peng, Z.; Nan, Y.; Nie, T.; Wang, R.; Ma, H. Genome-wide identification of the potato WRKY transcription factor family. PLoS ONE 2017, 12, e0181573. [Google Scholar] [CrossRef] [Green Version]
- Gong, L.; Zhang, H.; Gan, X.; Zhang, L.; Chen, Y.; Nie, F.; Shi, L.; Li, M.; Guo, Z.; Zhang, G. Transcriptome Profiling of the Potato (Solanum tuberosum L.) Plant under Drought Stress and Water-Stimulus Conditions. PLoS ONE 2015, 10, e0128041. [Google Scholar] [CrossRef]
- Li, S.; Fu, Q.; Huang, W.; Yu, D. Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 2009, 28, 683–693. [Google Scholar] [CrossRef]
- Liu, Y.Y. Identification and Analysis of Wheat WRKY Transcription Factors and Preliminary Study on Heat-Resistant Genes; Northwest A & F University: Xianyang, China, 2019. (In Chinese) [Google Scholar]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [Green Version]
- He, G.H.; Xu, J.Y.; Wang, Y.X.; Liu, J.M.; Li, P.S.; Chen, M.; Ma, Y.Z.; Xu, Z.S. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016, 16, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef] [Green Version]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef]
- Park, C.Y.; Lee, J.H.; Yoo, J.H.; Moon, B.C.; Choi, M.S.; Kang, Y.H.; Lee, S.M.; Kim, H.S.; Kang, K.Y.; Chung, W.S. WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 2005, 579, 1545–1550. [Google Scholar] [CrossRef]
- Han, X.; Wang, H.P.; Jin-Jing, P.; Yan-Ru, H.U.; Chen, X.L.; Di-Qiu, Y.U. Arabidopsis WRKY8 Transcription Factor-Associated Genes VQ10 and VQ11 are Responsive to Multiple Abiotic Stresses. Plant Divers. Resour. 2015, 37, 760–766. [Google Scholar]
- Qian-Tang, F.U. Expression profiles of AtWRKY25, AtWRKY26 and AtWRKY33 under abiotic stresses. Hereditas 2010, 32, 848–856. [Google Scholar]
- El-Esawi, M.A.; Al-Ghamdi, A.A.; Ali, H.M.; Ahmad, M. Overexpression of AtWRKY30 Transcription Factor Enhances Heat and Drought Stress Tolerance in Wheat (Triticum aestivum L.). Genes 2019, 10, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Xiang, Z.; Chen, L.; Huang, W.; Yu, D. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells 2010, 29, 475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Z.J.; Yan, J.Y.; Li, G.X.; Wu, Z.C.; Zhang, S.Q.; Zheng, S.J. WRKY 41 controls Arabidopsis seed dormancy via direct regulation of ABI 3 transcript levels not downstream of ABA. Plant J. 2014, 79, 810–823. [Google Scholar] [CrossRef]
- Suzuki, N.; Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Mittler, R. Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol. 2005, 139, 1313–1322. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Pang, S.; Lu, Z.; Jin, B. Function and Mechanism of WRKY Transcription Factors in Abiotic Stress Responses of Plants. Plants 2020, 9, 1515. [Google Scholar] [CrossRef]
- Qi, L.I.; Ye, L.I.; Niu, F.F.; Guo, X.H.; Zhao, X.J.; Xiang-Min, W.U.; Yang, B.; Jiang, Y.Q. Characterization and Stress-resistance Functional Identification of Transcription Factor Gene WRKY72 in Arabidopsis thaliana. J. Agric. Biotechnol. 2019, 2, 191–203. [Google Scholar]
- Dang, F.; Lin, J.; Xue, B.; Chen, Y.; He, S. CaWRKY27 Negatively Regulates H2O2-Mediated Thermotolerance in Pepper (Capsicum annuum). Front. Plant Sci. 2018, 9, 1633. [Google Scholar] [CrossRef]
- Dang, F.F.; Wang, Y.N.; Yu, L.; Eulgem, T.; Lai, Y.; Liu, Z.Q.; Wang, X.; Qiu, A.L.; Zhang, T.X.; Lin, J. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef]
- Wu, Z.J.; Li, X.H.; Liu, Z.W.; Li, H.; Wang, Y.X.; Zhuang, J. Transcriptome-wide identification of Camellia sinensis WRKY transcription factors in response to temperature stress. Mol. Genet. Genom. 2016, 291, 255–269. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, H.; Yang, X.; Li, Q.; Ling, J.; Wang, H.; Gu, X.; Huang, S.; Jiang, W. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Biochem. 2016, 108, 478–487. [Google Scholar] [CrossRef]
- Wang, J.Y. Moleculor Mechanism of Cotton GhWRKY21 Transcription Factor Regulating Plant Drought and High Temperature Resistance; Shandong Agricultural University: Tai’an, China, 2020. (In Chinese) [Google Scholar]
- Giacomelli, J.I.; Weigel, D.; Chan, R.L.; Manavella, P.A. Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol. 2012, 195, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.Y.; Zhang, W.H.; Sun, Y.D.; Zhang, T.T.; Zhai, M.Z. Two novel WRKY genes from Juglans regia JrWRKY6 and JrWRKY53 are involved in ABA-dependent stress responses. Biol. Plant. 2017, 61, 611–621. [Google Scholar] [CrossRef]
- Hana, M.; Marie, H.; Jana, D.; Veronika, T.; Ondřej, N.; Zuzana, L.; Václav, M.; Daniel, H.; Tomáš, H.; Tom, P.I. Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. J. Exp. Bot. 2013, 64, 2805–2815. [Google Scholar]
- Lan, A.; Huang, J.; Zhao, W.; Peng, Y.; Chen, Z.; Kang, D. A salicylic acid-induced rice (Oryza sativa L.) transcription factor OsWRKY77 is involved in disease resistance of Arabidopsis thaliana. Plant Biol. 2013, 15, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Ma, K.; Lu, Z.; Chen, G.; Cui, J.; Tong, P.; Wang, L.; Teng, N.; Jin, B. Transcriptomic and Metabolomic Analysis of the Heat-Stress Response of Populus tomentosa Carr. Forests 2019, 10, 383. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Tao, F.; An, F.; Zou, Y.; Tian, W.; Chen, X.; Xu, X.; Hu, X. Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling plant resistance to Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 2017, 18, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.T.; Ru, J.N.; Liu, Y.W.; Li, M.; Zhao, D.; Yang, J.F.; Fu, J.D.; Xu, Z.S. Maize WRKY Transcription Factor ZmWRKY106 Confers Drought and Heat Tolerance in Transgenic Plants. Int. J. Mol. Ences 2018, 19, 3046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, Y.; Yang, Y.; Zhou, Y.; Zhou, J.; Fan, B.; Yu, J.-Q.; Chen, Z. Protein–protein interactions in the regulation of WRKY transcription factors. Mol. Plant 2013, 6, 287–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besseau, S.; Jing, L.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, W.; Liu, Z.; Yi-Maer, A.Y.; Xu, Z. Both JrWRKY2 and JrWRKY7 of Juglans regia mediate responses to abiotic stresses and abscisic acid through formation of homodimers and interaction. Plant Biol. 2017, 19, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Kwon, S.I.; Choi, C.; Lee, H.; Ahn, I.; Park, S.R.; Bae, S.C.; Lee, S.C.; Hwang, D.J. Expression analysis of rice VQ genes in response to biotic and abiotic stresses. Gene 2013, 529, 208–214. [Google Scholar] [CrossRef]
- Yamasaki, K. Structural Basis for Sequence-specific DNA Recognition by an Arabidopsis WRKY Transcription Factor. J. Biol. Chem. 2012, 287, 7683–7691. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Yang, Y.; Zhou, X.; Chi, Y.; Fan, B.; Chen, Z. Structural and Functional Characterization of the VQ Protein Family and VQ Protein Variants from Soybean. Sci. Rep. 2016, 6, 34663. [Google Scholar] [CrossRef] [Green Version]
- Ding, H.; Yuan, G.; Mo, S.; Qian, Y.; Ge, C. Genome-wide analysis of the plant-specific VQ motif-containing proteins in tomato (Solanum lycopersicum) and characterization of SlVQ6 in thermotolerance. Plant Physiol. Biochem. 2019, 143, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhou, Y.; Yang, Y.; Chi, Y.-J.; Zhou, J.; Chen, J.-Y.; Wang, F.; Fan, B.; Shi, K.; Zhou, Y.-H. Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 2012, 159, 810–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, Z.; Wang, F.; Zheng, Z.; Fan, B.; Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. Cell Mol. Biol. 2011, 66, 953–968. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.N.; Kuang, J.F.; Wei, S.; Chen, J.; Hui, X.; Lu, W.J.; Chen, J.W.; Chen, J.Y. Expression profiles of a banana fruit linker histone H1 gene MaHIS1 and its interaction with a WRKY transcription factor. Plant Cell Rep. 2012, 31, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Perruc, E.; Charpenteau, M.; Ramirez, B.C.; Jauneau, A.; Ranty, B. A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J. 2010, 38, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Zentgraf, U.; Laun, T.; Ying, M. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 2010, 89, 133–137. [Google Scholar] [CrossRef]
- Liu, Z.; Shi, L.; Yang, S.; Qiu, S.; Ma, X.; Cai, J.; Guan, D.; Wang, Z.; He, S. A conserved double-W box in the promoter of CaWRKY40 mediates autoregulation during response to pathogen attack and heat stress in pepper. Mol. Plant Pathol. 2021, 22, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Torres-Zabala, M.D.; Truman, W.; Bennett, M.H.; Lafforgue, G.; Mansfield, J.W.; Rodriguez Egea, P.; Bögre, L.; Grant, M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signaling pathway to cause disease. EMBO J. 2007, 26, 1434–1443. [Google Scholar] [CrossRef]
- Shen, Q.-H.; Saijo, Y.; Mauch, S.; Biskup, C.; Bieri, S.; Keller, B.; Seki, H.; Ülker, B.; Somssich, I.E.; Schulze-Lefert, P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 2007, 315, 1098–1103. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.R.; Dubey, K.; Arora, K.; Dalal, M.; Rai, G.K.; Mishra, D.; Praveen, S. Characterizing the putative mitogen-activated protein kinase ( MAPK ) and their protective role in oxidative stress tolerance and carbon assimilation in wheat under terminal heat stress. Biotechnol. Rep. 2021, 29, e00597. [Google Scholar] [CrossRef]
- Sierla, M.; Waszczak, C.; Vahisalu, T.; Kangasjärvi, J. Reactive oxygen species in the regulation of stomatal movements. Plant Physiol. 2016, 171, 1569–1580. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Venegas, R.; Abdallat, A.A.; Guo, M.; Alfano, J.R.; Avramova, Z. Epigenetic Control of a Transcription Factor at the Cross Section of Two Antagonistic Pathways. Epigenetics 2007, 2, 106–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvucci, M.E.; Osteryoung, K.W.; Crafts-Brandner, S.J.; Vierling, E. Exceptional sensitivity of Rubisco activase to thermal denaturation in vitro and in vivo. Plant Physiol. 2001, 127, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhang, L.; Yu, D. Wounding-induced WRKY8 is involved in basal defense in Arabidopsis. Mol. Plant-Microbe Interact. 2010, 23, 558–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.; Kim, C.Y.; Lee, J.; Lee, S.K.; Jeon, J.S. OsWRKY42 Represses OsMT1d and Induces Reactive Oxygen Species and Leaf Senescence in Rice. Mol. Cells 2014, 37, 532. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.F.; Wei, W.; Zhou, Q.Y.; Tian, A.G.; Chen, S.Y. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef]
- Babitha, K.C.; Ramu, S.V.; Pruthvi, V.; Mahesh, P. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 2013, 22, 327–341. [Google Scholar] [CrossRef]
- Jiang, W.; Yu, D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009, 9, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, Y.; Lu, Y.; Liu, Z.Q.; Cao, Z.; Zhang, D.P. The Mg-Chelatase H Subunit of Arabidopsis Antagonizes a Group of WRKY Transcription Repressors to Relieve ABA-Responsive Genes of Inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.-Y.; Wang, X.-F.; Wu, F.-Q.; Du, S.-Y.; Cao, Z.; Shang, Y.; Wang, X.-L.; Peng, C.-C.; Yu, X.-C.; Zhu, S.-Y. The Mg-chelatase H subunit is an abscisic acid receptor. Nature 2006, 443, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Finatto, T.; Viana, V.E.; Woyann, L.G.; Busanello, C.; Maia, L.C.D.; Oliveira, A.C.D. Can WRKY transcription factors help plants to overcome environmental challenges? Genet. Mol. Biol. 2018, 41, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Blanco, F.A.; Zanetti, M.E.; Casalongué, C.A.; Daleo, G.R. Molecular characterization of a potato MAP kinase transcriptionally regulated by multiple environmental stresses. Plant Physiol. Biochem. 2006, 44, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Dat, J.F. Changes in Salicylic Acid and Antioxidants during Induced Thermotolerance in Mustard Seedlings. Plant Physiol. 1998, 118, 1455–1461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.Y.; Zhang, S. Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J. 2010, 38, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.M.; Zou, C.; Shu, Y.N.; Liu, S.S. WRKY Transcription Factors in Nicotiana tabacum Modulate Plant Immunity against Whitefly via Interacting with MAPK Cascade Pathways. Insects 2020, 12, 16. [Google Scholar] [CrossRef]
- Sangwan, V.; Orvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa, R.S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 2010, 31, 629–638. [Google Scholar] [CrossRef] [Green Version]
- Rao, K.P.; Vani, G.; Kumar, K.; Sinha, A.K. Rhythmic expression of mitogen activated protein kinase activity in rice. Mol. Cells 2009, 28, 417. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.K.; Rakwal, R.; Iwahashi, H. Isolation of novel rice (Oryza sativa L.) multiple stress responsive MAP kinase gene, OsMSRMK2, whose mRNA accumulates rapidly in response to environmental cues. Biochem. Biophys. Res. Commun. 2002, 294, 1009–1016. [Google Scholar] [CrossRef]
- Tsuneaki, A.; Guillaume, T.; Joulia, P.; Willmann, M.R.; Wan-Ling, C.; Lourdes, G.G.; Thomas, B.; Ausubel, F.M.; Jen, S. MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 2009, 415, 977–983. [Google Scholar]
- Shi, W. The Expressional and Functional Characterization of CaMAPK1 and Its Correlationship to CaWRKY17 and CaWRKY40 in Pepper’s Response to Ralatonia solanacearum Inoculation; Fujian Agriculture and Forestry University: Fujian, China, 2016. (In Chinese) [Google Scholar]
| 1 | Yeast two-hybrid |

| No. | Gene | Plant Species | Research Method | References |
|---|---|---|---|---|
| 1 | AtWRKY7 | Arabidopsis thaliana | Site-directed mutagenesis | [56] |
| 2 | AtWRKY8 | Arabidopsis thaliana | Yeast two-hybrid | [57] |
| 3 | AtWRKY25 AtWRKY26 AtWRKY33 | Arabidopsis thaliana | Constitutive expression, T-DNA insertion mutants | [58] |
| 4 | AtWRKY30 | Triticum aestivum | Overexpression | [59] |
| 5 | AtWRKY39 | Arabidopsis thaliana | Overexpression, knockout mutation | [60] |
| 6 | AtWRKY41 | Arabidopsis thaliana | Gene silencing | [61] |
| 7 | AtWRKY46 | Arabidopsis thaliana | Overexpression | [62] |
| 8 | AtWRKY54 | Arabidopsis thaliana | Expression analysis, transcriptome and metabolome analysis | [63] |
| 9 | AtWRKY72 | Arabidopsis thaliana | Yeast two-hybrid bimolecular fluorescence complementation, T-DNA insertion mutant | [64] |
| 10 | CaWRKY27 | Capsicum annuum | Overexpression, gene silencing | [65] |
| 11 | CaWRKY40 | Capsicum annuum | Overexpression, gene silencing | [66] |
| 12 | CsWRKYs | Camellia sinensis | Transcriptomics analysis | [67] |
| 13 | CsWRKY46 | Cucumis sativus | Overexpression, VIGS | [68] |
| 14 | GhWRKY21 | Gossypium hirsutum | VIGS, overexpression, yeast one-hybrid | [69] |
| 15 | HaWRKY6 | Helianthus annuus | Expression analysis, northern blotting | [70] |
| 16 | JrWRKY6 JrWRKY53 | Juglans regia | Expression analysis | [71] |
| 17 | NtWRKY6 | Nicotiana tabacum | Ectopic expression | [72] |
| 18 | OsWRKY11 | Oryza sativa | Fusion with HSP101 promoter | [53] |
| 19 | OsWRKY77 | Oryza sativa | Overexpression | [73] |
| 20 | PtWRKY13 PtWRKY50 | Populus tomentosa | Expression analysis, transcriptome and metabolome | [74] |
| 21 | TaWRKY1 TaWRKY33 | Triticum aestivum | Transcriptomics analysis | [52] |
| 22 | TaWRKY30 | Triticum aestivum | Overexpression | [59] |
| 23 | TaWRKY70 | Triticum aestivum | Gene silencing | [75] |
| 24 | ZmWRKY106 | Zea mays | Expression analysis, transcriptome analysis | [76] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Z.; Luan, Y.; Meng, J.; Sun, J.; Tao, J.; Zhao, D. WRKY Transcription Factor Response to High-Temperature Stress. Plants 2021, 10, 2211. https://doi.org/10.3390/plants10102211
Cheng Z, Luan Y, Meng J, Sun J, Tao J, Zhao D. WRKY Transcription Factor Response to High-Temperature Stress. Plants. 2021; 10(10):2211. https://doi.org/10.3390/plants10102211
Chicago/Turabian StyleCheng, Zhuoya, Yuting Luan, Jiasong Meng, Jing Sun, Jun Tao, and Daqiu Zhao. 2021. "WRKY Transcription Factor Response to High-Temperature Stress" Plants 10, no. 10: 2211. https://doi.org/10.3390/plants10102211
APA StyleCheng, Z., Luan, Y., Meng, J., Sun, J., Tao, J., & Zhao, D. (2021). WRKY Transcription Factor Response to High-Temperature Stress. Plants, 10(10), 2211. https://doi.org/10.3390/plants10102211






