Comparison of Silicon-Evoked Responses on Arsenic Stress between Different Dular Rice Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Rice Cultivation
2.2. As Concentration Measurement
2.3. Dry Weight and Si Concentration Measurement
2.4. Soluble Protein Contents Measurement
2.5. Transcriptomic Analyses
2.6. Quantitative RT-PCR
2.7. Statistical Analyses
3. Results
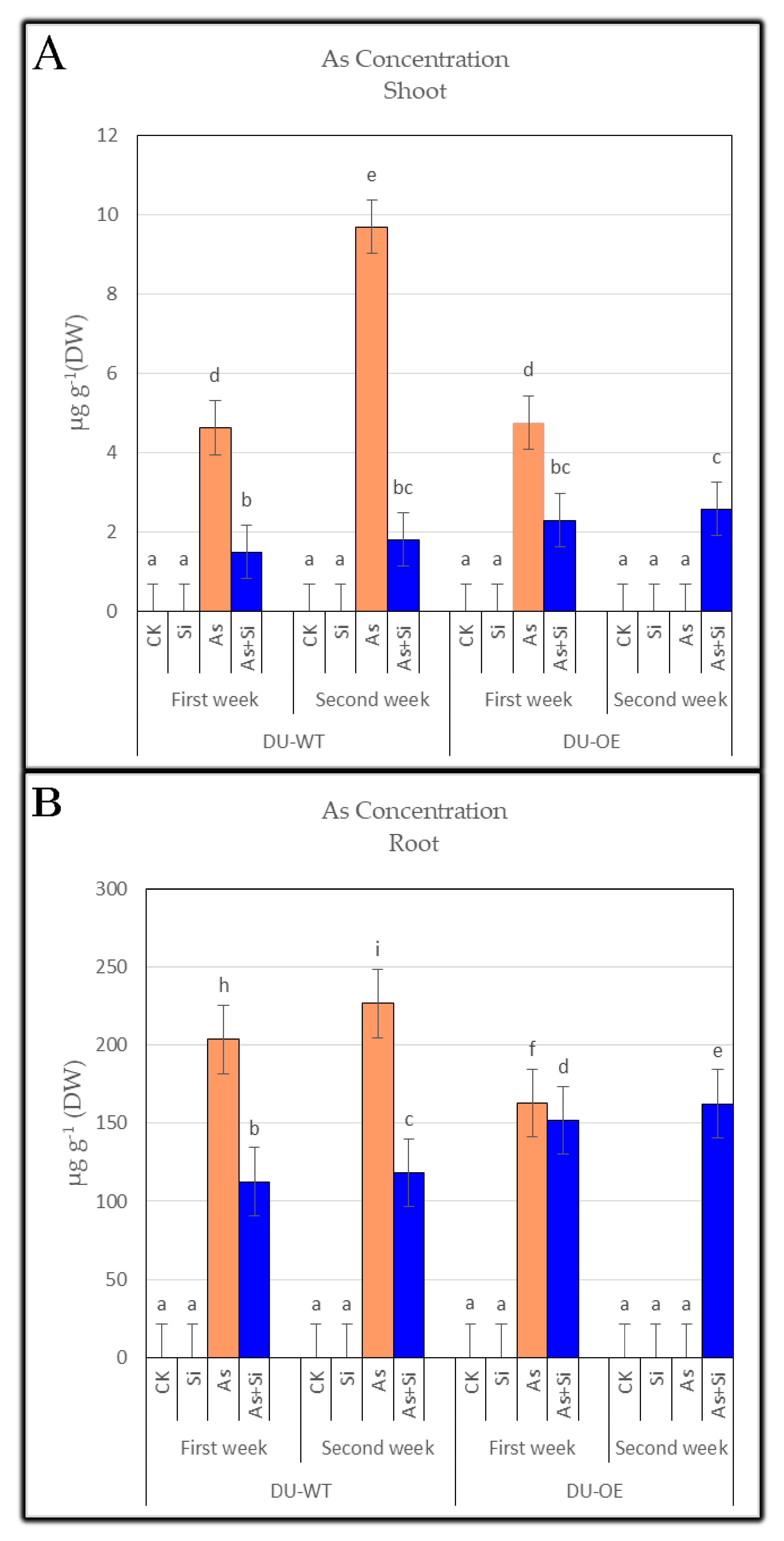
3.1. Arsenic Concentration of Shoots and Roots
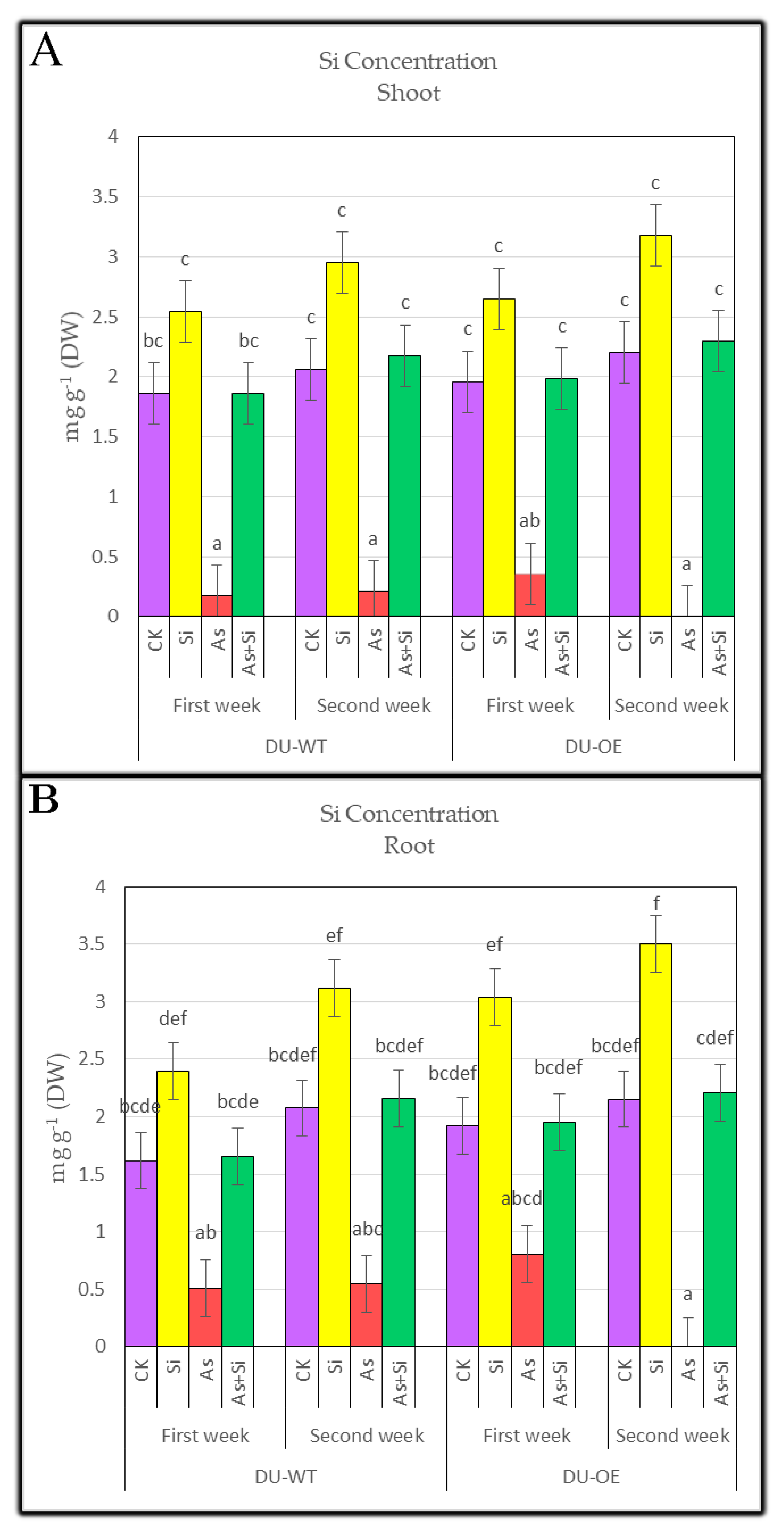
3.2. Silicon Concentration of Shoots and Roots
3.3. Dry Weight of Shoots and Roots
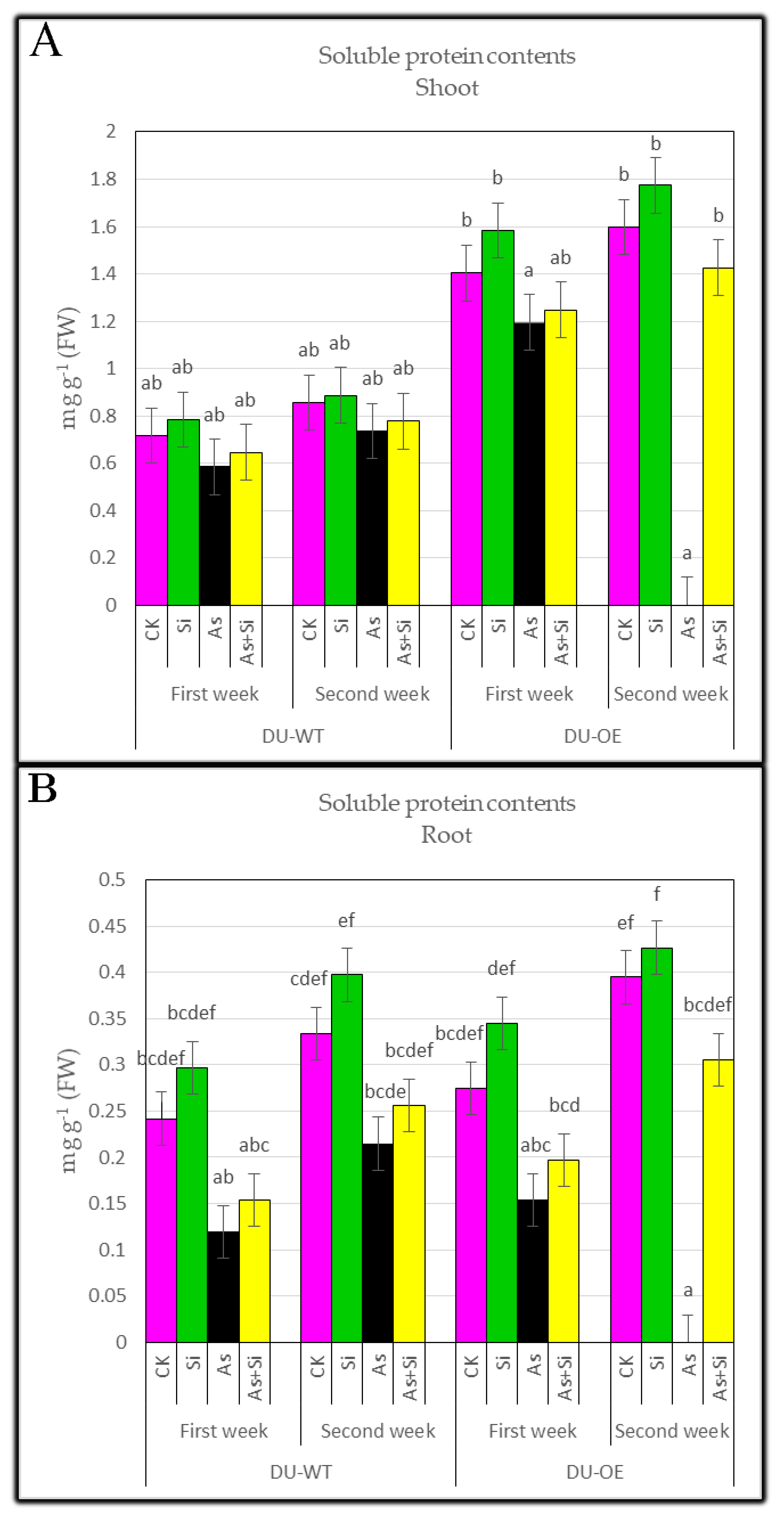
3.4. Soluble Protein Contents of Shoots and Roots
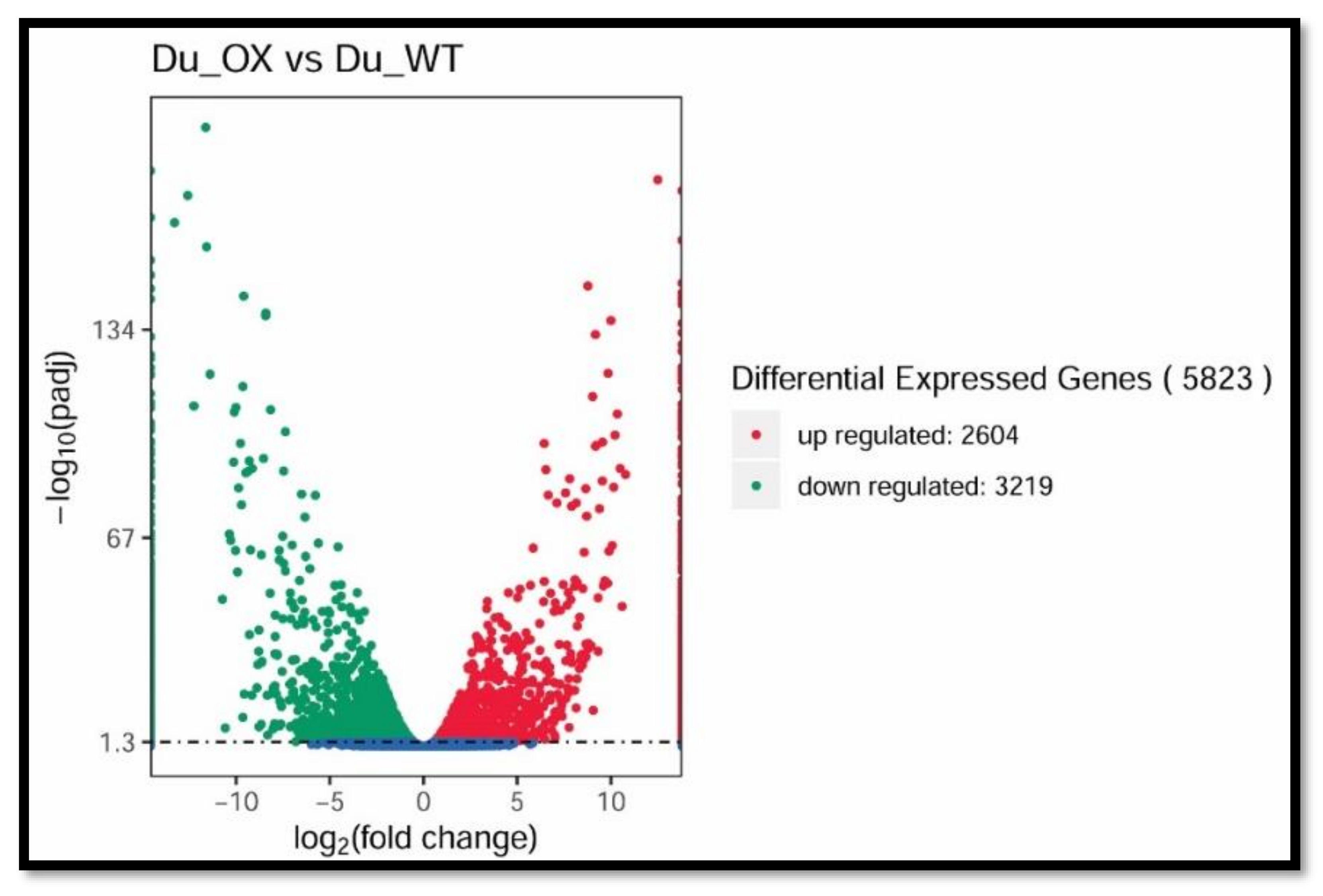
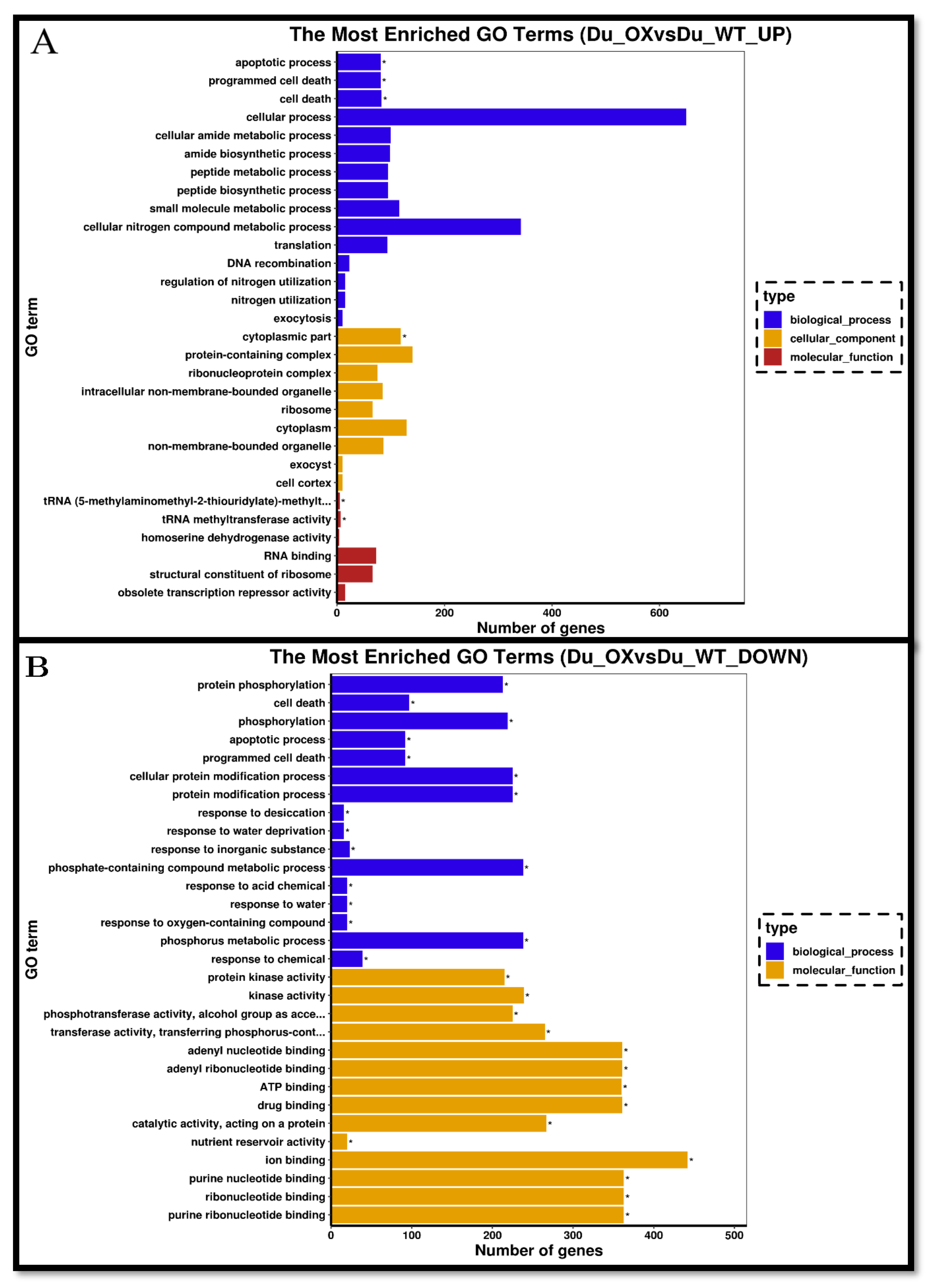
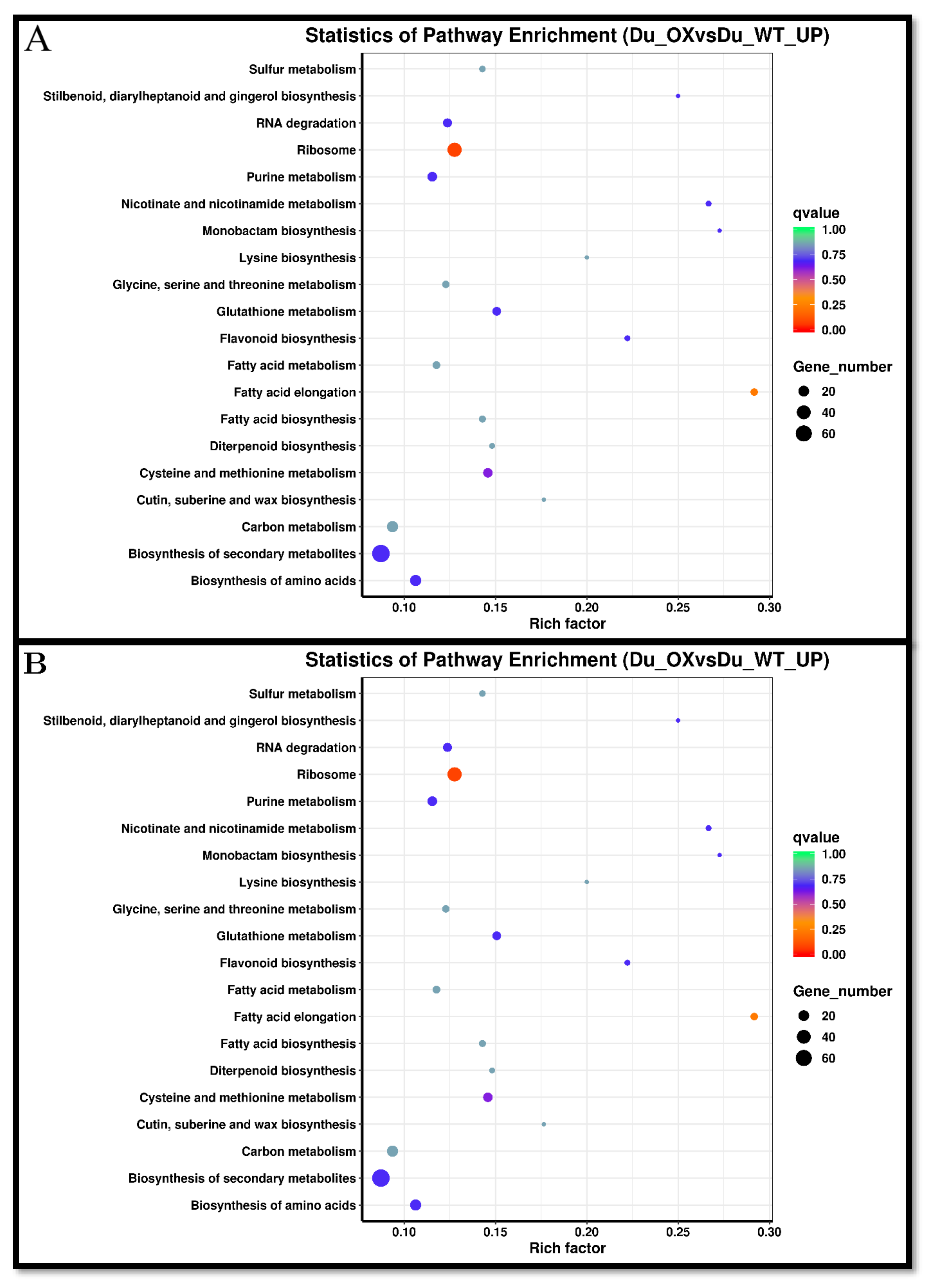
3.5. Transcriptome Analysis of Rice Roots
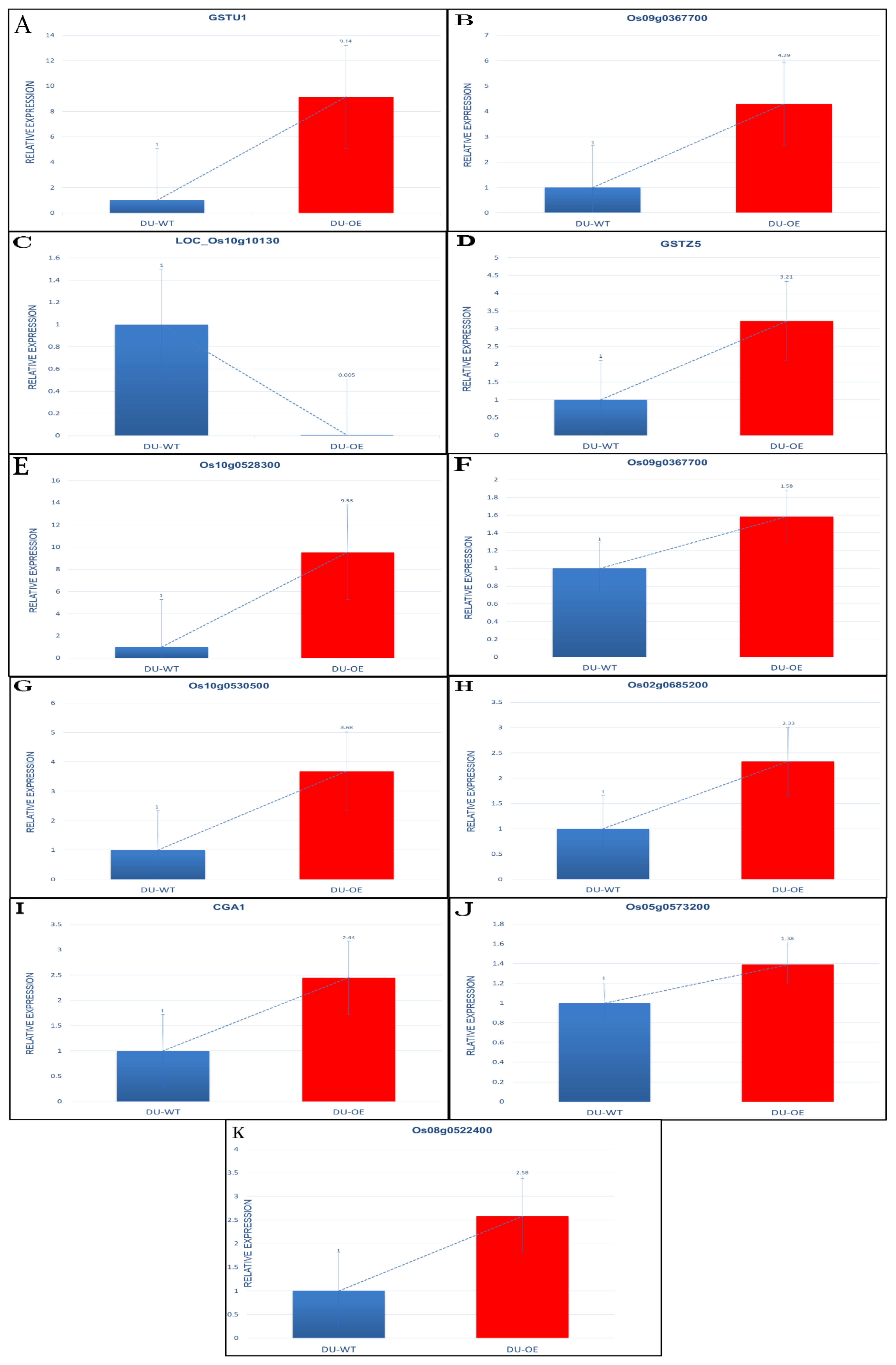
3.6. Expression of Some Genes in DU-OE Exposed to Si and As
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zazouli, M.A.; Bandpei, A.M.; Maleki, A.; Saberian, M.; Izanloo, H. Determination of cadmium and lead contents in black tea and tea liquor from Iran. Asian J. Chem. 2010, 22, 1387. [Google Scholar]
- Shokrzadeh, M.; Fathalinezhad, F.; Khoshvishkaie, E. Concentrations of Heavy Metals (Cr, Cd, Pb) in Three Types of Rice in Astaneh-Ashrafieh, Iran. J. Maz. Univ. Med. Sci. 2017, 26, 196–200. [Google Scholar]
- Bhattacharya, P.; Samal, A.C.; Majumdar, J.; Santra, S. Accumulation of arsenic and its distribution in rice plant (Oryza sativa L.) in Gangetic West Bengal, India. Paddy Water Environ. 2009, 8, 63–70. [Google Scholar] [CrossRef]
- Bhattacharya, P.; Samal, A.C.; Majumdar, J.; Banerjee, S.; Santra, S. In vitro assessment on the impact of soil arsenic in the eight rice varieties of West Bengal, India. J. Hazard. Mater. 2013, 262, 1091–1097. [Google Scholar] [CrossRef]
- Srivastava, S.; Tripathi, R.D.; Dhankhera, O.P.; Upadhyay, M.K. Arsenic Transport, Metabolism and Toxicity in Plants. Int. J. Plant Environ. 2016, 2, 17–28. [Google Scholar] [CrossRef]
- Awasthi, S.; Chauhan, R.; Srivastava, S.; Tripathi, R.D. The Journey of Arsenic from Soil to Grain in Rice. Front. Plant Sci. 2017, 8, 1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nookabkaew, S.; Rangkadilok, N.; Mahidol, C.; Promsuk, G.; Satayavivad, J. Determination of Arsenic Species in Rice from Thailand and Other Asian Countries Using Simple Extraction and HPLC-ICP-MS Analysis. J. Agric. Food Chem. 2013, 61, 6991–6998. [Google Scholar] [CrossRef] [PubMed]
- Duan, G.-L.; Hu, Y.; Liu, W.-J.; Kneer, R.; Zhao, F.-J.; Zhu, Y.-G. Evidence for a role of phytochelatins in regulating arsenic accumulation in rice grain. Environ. Exp. Bot. 2011, 71, 416–421. [Google Scholar] [CrossRef]
- Vandevenne, F.; Struyf, E.; Clymans, W.; Meire, P. Agricultural silica harvest: Have humans created a new loop in the global silica cycle? Front. Ecol. Environ. 2012, 10, 243–248. [Google Scholar] [CrossRef]
- Song, Z.; Wang, H.; Strong, P.J.; Shan, S. Increase of available soil silicon by Si-rich manure for sustainable rice production. Agron. Sustain. Dev. 2014, 34, 813–819. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.-Y.; Wen, S.-L.; Chen, P.; Zhang, L.; Cen, K.; Sun, G.-X. Mitigation of cadmium and arsenic in rice grain by applying different silicon fertilizers in contaminated fields. Environ. Sci. Pollut. Res. 2016, 23, 3781–3788. [Google Scholar] [CrossRef] [PubMed]
- Hedayatifar, R.; Falahi, E.; Birjandi, M. Determination of Cadmium and Lead levels in high consumed rice (Oryza Sativa L.) cultivated in Lorestan province and its comparison with national standards. Yafteh 2011, 12, 15–22. [Google Scholar]
- Hoogenkamp, H.; Kumagai, H.; Wanasundara, J. Rice Protein and Rice Protein Products. In Sustainable Protein Sources; Elsevier: Amsterdam, The Netherlands, 2017; pp. 47–65. [Google Scholar]
- Pan, W.; Wu, C.; Xue, S.; Hartley, W. Arsenic dynamics in the rhizosphere and its sequestration on rice roots as affected by root oxidation. J. Environ. Sci. 2014, 26, 892–899. [Google Scholar] [CrossRef]
- Wu, C.; Zou, Q.; Xue, S.; Mo, J.; Pan, W.; Lou, L.; Wong, M.H. Effects of silicon (Si) on arsenic (As) accumulation and speciation in rice (Oryza sativa L.) genotypes with different radial oxygen loss (ROL). Chemosphere 2015, 138, 447–453. [Google Scholar] [CrossRef]
- Seyfferth, A.L.; Fendorf, S. Silicate Mineral Impacts on the Uptake and Storage of Arsenic and Plant Nutrients in Rice (Oryza sativa L.). Environ. Sci. Technol. 2012, 46, 13176–13183. [Google Scholar] [CrossRef]
- Kumar, N.; Dubey, A.K.; Upadhyay, A.K.; Gautam, A.; Ranjan, R.; Srikishna, S.; Sahu, N.; Behera, S.K.; Mallick, S. GABA accretion reduces Lsi-1 and Lsi-2 gene expressions and modulates physiological responses in Oryza sativa to provide tolerance towards arsenic. Sci. Rep. 2017, 7, 8786. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.F.; Yamaji, N.; Mitani, N.; Xu, X.-Y.; Su, Y.-H.; McGrath, S.; Zhao, F.-J. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. USA 2008, 105, 9931–9935. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Fang, C.; Li, Y.; Lin, W.; He, J.; Lin, R.; Lin, W. Cadmium-stress mitigation through gene expression of rice and silicon addition. Plant Growth Regul. 2017, 81, 91–101. [Google Scholar] [CrossRef]
- Dubey, A.K.; Kumar, N.; Ranjan, R.; Gautam, A.; Pande, V.; Sanyal, I.; Mallick, S. Application of glycine reduces arsenic accumulation and toxicity in Oryza sativa L. by reducing the expression of silicon transporter genes. Ecotoxicol. Environ. Saf. 2018, 148, 410–417. [Google Scholar] [CrossRef]
- Cock, J.; Yoshida, S.; Forno, D.A. Laboratory Manual for Physiological Studies of Rice; International Rice Research Institute: Los Baños, Philippines, 1976. [Google Scholar]
- Meharg, A.A.; Jardine, L. Arsenite transport into paddy rice (Oryza sativa) roots. New Phytol. 2003, 157, 39–44. [Google Scholar] [CrossRef] [Green Version]
- Dai, W.-M.; Zhang, K.-Q.; Duan, B.-W.; Sun, C.-X.; Zheng, K.-L.; Cai, R.; Zhuang, J.-Y. Rapid determination of silicon content in rice. Rice Sci. 2005, 12, 145–147. [Google Scholar]
- Hallmark, C.; Wilding, L.; Smeck, N.; Silicon, J. Methods of Soil Analysis: Part 2 Chemical Microbiological Properties; American Society of Agronomy: Madison, WI, USA, 1983; Volume 9, pp. 263–273. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Chomczynski, P.; Mackey, K. Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques 1995, 19, 942–945. [Google Scholar]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ashfaque, F.; Inam, A.; Iqbal, S.; Sahay, S. Response of silicon on metal accumulation, photosynthetic inhibition and oxidative stress in chromium-induced mustard (Brassica juncea L.). S. Afr. J. Bot. 2017, 111, 153–160. [Google Scholar] [CrossRef]
- Boorboori, M.; Lin, W.; Fang, C. The effects of arsenic and silicon on the oxidative and non-oxidative enzymes in the seedlings of three different rice (Oryza sativa L.) varieties in different growth periods. Appl. Ecol. Environ. Res. 2020, 18, 5263–5278. [Google Scholar] [CrossRef]
- Liu, W.-J.; McGrath, S.; Zhao, F.-J. Silicon has opposite effects on the accumulation of inorganic and methylated arsenic species in rice. Plant Soil 2014, 376, 423–431. [Google Scholar] [CrossRef]
- Limmer, M.; Mann, J.; Amaral, D.C.; Vargas, R.; Seyfferth, A.L. Silicon-rich amendments in rice paddies: Effects on arsenic uptake and biogeochemistry. Sci. Total Environ. 2018, 624, 1360–1368. [Google Scholar] [CrossRef]
- Zhang, S.; Geng, L.; Fan, L.; Zhang, M.; Zhao, Q.; Xue, P.; Liu, W. Spraying silicon to decrease inorganic arsenic accumulation in rice grain from arsenic-contaminated paddy soil. Sci. Total Environ. 2020, 704, 135239. [Google Scholar] [CrossRef]
- Li, G.; Zheng, M.; Tang, J.; Shim, H.; Cai, C. Effect of silicon on arsenic concentration and speciation in different rice tissues. Pedosphere 2018, 28, 511–520. [Google Scholar] [CrossRef]
- Bhattacharya, P. Assessment of Arsenic Accumulation by Different Varieties of Rice (Oryza sativa L.) Irrigated with Arsenic-contaminated Groundwater in West Bengal (India). Environ. Pollut. Prot. 2017, 2, 92–99. [Google Scholar] [CrossRef]
- Geng, A.; Wang, X.; Wu, L.; Wang, F.; Wu, Z.; Yang, H.; Chen, Y.; Wen, D.; Liu, X. Silicon improves growth and alleviates oxidative stress in rice seedlings (Oryza sativa L.) by strengthening antioxidant defense and enhancing protein metabolism under arsanilic acid exposure. Ecotoxicol. Environ. Saf. 2018, 158, 266–273. [Google Scholar] [CrossRef]
- Pontigo, S.; Godoy, K.; Jiménez, H.; Gutiérrez-Moraga, A.; Mora, M.L.; Cartes, P. Silicon-mediated alleviation of aluminum toxicity by modulation of Al/Si uptake and antioxidant performance in ryegrass plants. Front. Plant Sci. 2017, 8, 642. [Google Scholar] [CrossRef] [Green Version]
- Joy, E.; Broadley, M.; Young, S.D.; Black, C.R.; Chilimba, A.D.; Ander, E.L.; Barlow, T.S.; Watts, M.J. Soil type influences crop mineral composition in Malawi. Sci. Total Environ. 2015, 505, 587–595. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarty, D.; Trivedi, P.K.; Misra, P.; Tiwari, M.; Shri, M.; Shukla, D.; Kumar, S.; Rai, A.; Pandey, A.; Nigam, D.; et al. Comparative transcriptome analysis of arsenate and arsenite stresses in rice seedlings. Chemosphere 2009, 74, 688–702. [Google Scholar] [CrossRef]
- Tripathi, R.D.; Tripathi, P.; Dwivedi, S.; Dubey, S.; Chatterjee, S.; Chakrabarty, D.; Trivedi, P.K. Arsenomics: Omics of arsenic metabolism in plants. Front. Physiol. 2012, 3, 275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Wu, S.; Yu, R.; Cang, T.; Chen, L.; Zhao, X.; Cai, L.; Wu, C. Identification and expression pattern of ag lutathione S-transferase in E chinochloa crus-galli. Weed Res. 2013, 53, 314–321. [Google Scholar] [CrossRef]
- Radyukina, N.L.; Shashukova, A.V.; Makarova, S.S.; Kuznetsov, V.V. Exogenous proline modifies differential expression of superoxide dismutase genes in UV-B-irradiated Salvia officinalis plants. Russ. J. Plant Physiol. 2011, 58, 51–59. [Google Scholar] [CrossRef]
- Ma, J.F.; Tamai, K.; Yamaji, N.; Mitani, N.; Konishi, S.; Katsuhara, M.; Ishiguro, M.; Murata, Y.; Yano, M. A silicon transporter in rice. Nature 2006, 440, 688–691. [Google Scholar] [CrossRef]
- Ranjan, R.; Kumar, N.; Dubey, A.K.; Gautam, A.; Pandey, S.N.; Mallick, S. Diminution of arsenic accumulation in rice seedlings co-cultured with Anabaena sp.: Modulation in the expression of lower silicon transporters, two nitrogen dependent genes and lowering of antioxidants activity. Ecotoxicol. Environ. Saf. 2018, 151, 109–117. [Google Scholar] [CrossRef]

| Gene Name | Protein Name | Primer Sequences | Amplicon (bp) |
|---|---|---|---|
| GSTU1-F | Probable glutathione S-transferase GSTU1 | GTGAGTTTGTTGTTACCGTTGA | 84 |
| GSTU1-R | GGACTCGTCAGTGTTGTACATA | ||
| Os01g0369700-F | Os09g0367700 protein | GTTCGGTGAGATTCCAGTACTG | 86 |
| Os01g0369700-R | TGTACTTGCGAAAGATGTACCT | ||
| LOC_Os03g17460-F | IN2-1 protein, putative, expressed | CACCTTGATCTGCATTTGTCAA | 89 |
| LOC_Os03g17460-R | ATGGGCAAATGTTGACATGTAC | ||
| GSTZ5-F | Protein IN2-1 homolog B | AAGATTGTCGCGATTGATCTTG | 95 |
| GSTZ5-R | TGATTGTTGTGCTCAAGTGAAG | ||
| Os10g0528300-F | Glutathione S-transferase | CAAGATCTTCGACGAGGAGAAG | 123 |
| Os10g0528300-R | CTCATCTTAGCGAACTCGACC | ||
| Os09g0367700-F | Os09g0367700 protein | TGGTTCCACGCCTACGAGA | 126 |
| Os09g0367700-R | CTTCTCAGGATCAGTAAGCGTC | ||
| Os10g0530500-F | Os10g0530500 protein | ACAACATGTTCCCTGGAATGG | 166 |
| Os10g0530500-R | TCGACGTACCCGATGGAGTC | ||
| Os02g0685200-F | Os02g0685200 protein | CGGTGGGTTCTCGAATAACTC | 193 |
| Os02g0685200-R | CGTGGTTGCAATTGACATCTTA | ||
| CGA1-F | CYTOKININ-RESPONSIVE GATA TRANSCRIPTION FACTOR 1 | CACAGAACCCGATATCCAAGG | 197 |
| CGA1-R | GCCCTCATCAAATTAACGGTAC | ||
| Os05g0573200-F | Isocitrate dehydrogenase [NADP] | CTAAATGGCACTGTGTTCAGAG | 252 |
| Os05g0573200-R | GGACTCGTCAGTGTTGTACATA | ||
| Os08g0522400-F | Os08g0522400 protein | CAAAGACAAGCTTTCACCGTAA | 282 |
| Os08g0522400-R | CAGAAAAGAACGCTGCCTTTAA | ||
| actin1(Os03g0718100)-F | CTTCATAGGAATGGAAGCTGCGGGTA | 26 | |
| actin1(Os03g0718100)-R | CGACCACCTTGATCTTCATGCTGCTA |
| Sampling Time | Parameter | Genotypes | As | Si | Genotypes × As | Genotypes × Si | As × Si | Genotypes × As × Si |
|---|---|---|---|---|---|---|---|---|
| 1st week | Shoot As concentration | 42.01 ** | 16595.59 ** | 2453.05 ** | 118.54 ** | 50.85 ** | 904.34 ** | 15.45 ** |
| Root As concentration | 22.18 ** | 10391.57 ** | 711.33 ** | 5.58 ** | 46.15 ** | 207.94 ** | 92.80 ** | |
| Shoot dry weight | 17.21 ** | 55.04 ** | 22.71 ** | 0.70 | 1.42 | 2.11 | 0.26 | |
| Root dry weight | 3.21 | 43.13 ** | 23.79 ** | 0.02 | 0.38 | 1.24 | 0.13 | |
| 2nd week | Shoot As concentration | 863.84 ** | 12289.71 ** | 2397.37 ** | 611.74 ** | 799.68 ** | 1161.99 ** | 371.13 ** |
| Root As concentration | 32.77 ** | 12514.50 ** | 866.25 ** | 12.56 ** | 148.88 ** | 217.80 ** | 123.61 ** | |
| Shoot dry weight | 2.87 ** | 45.87 ** | 28.03 ** | 0.06 | 0.25 | 1.20 | 0.30 | |
| Root dry weight | 0.86 | 44.26 ** | 26.90 ** | 0.02 | 0.06 | 1.66 | 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boorboori, M.R.; Li, Z.; Yan, X.; Dan, M.; Zhang, Z.; Lin, W.; Fang, C. Comparison of Silicon-Evoked Responses on Arsenic Stress between Different Dular Rice Genotypes. Plants 2021, 10, 2210. https://doi.org/10.3390/plants10102210
Boorboori MR, Li Z, Yan X, Dan M, Zhang Z, Lin W, Fang C. Comparison of Silicon-Evoked Responses on Arsenic Stress between Different Dular Rice Genotypes. Plants. 2021; 10(10):2210. https://doi.org/10.3390/plants10102210
Chicago/Turabian StyleBoorboori, Mohammad Reza, Zhou Li, Xue Yan, Mu Dan, Zhixing Zhang, Wenxiong Lin, and Changxun Fang. 2021. "Comparison of Silicon-Evoked Responses on Arsenic Stress between Different Dular Rice Genotypes" Plants 10, no. 10: 2210. https://doi.org/10.3390/plants10102210
APA StyleBoorboori, M. R., Li, Z., Yan, X., Dan, M., Zhang, Z., Lin, W., & Fang, C. (2021). Comparison of Silicon-Evoked Responses on Arsenic Stress between Different Dular Rice Genotypes. Plants, 10(10), 2210. https://doi.org/10.3390/plants10102210






