Detection of Glutamate Decarboxylase Antibodies and Simultaneous Multi-Molecular Translocation Exploration by Glass Nanopores
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical and Reagents
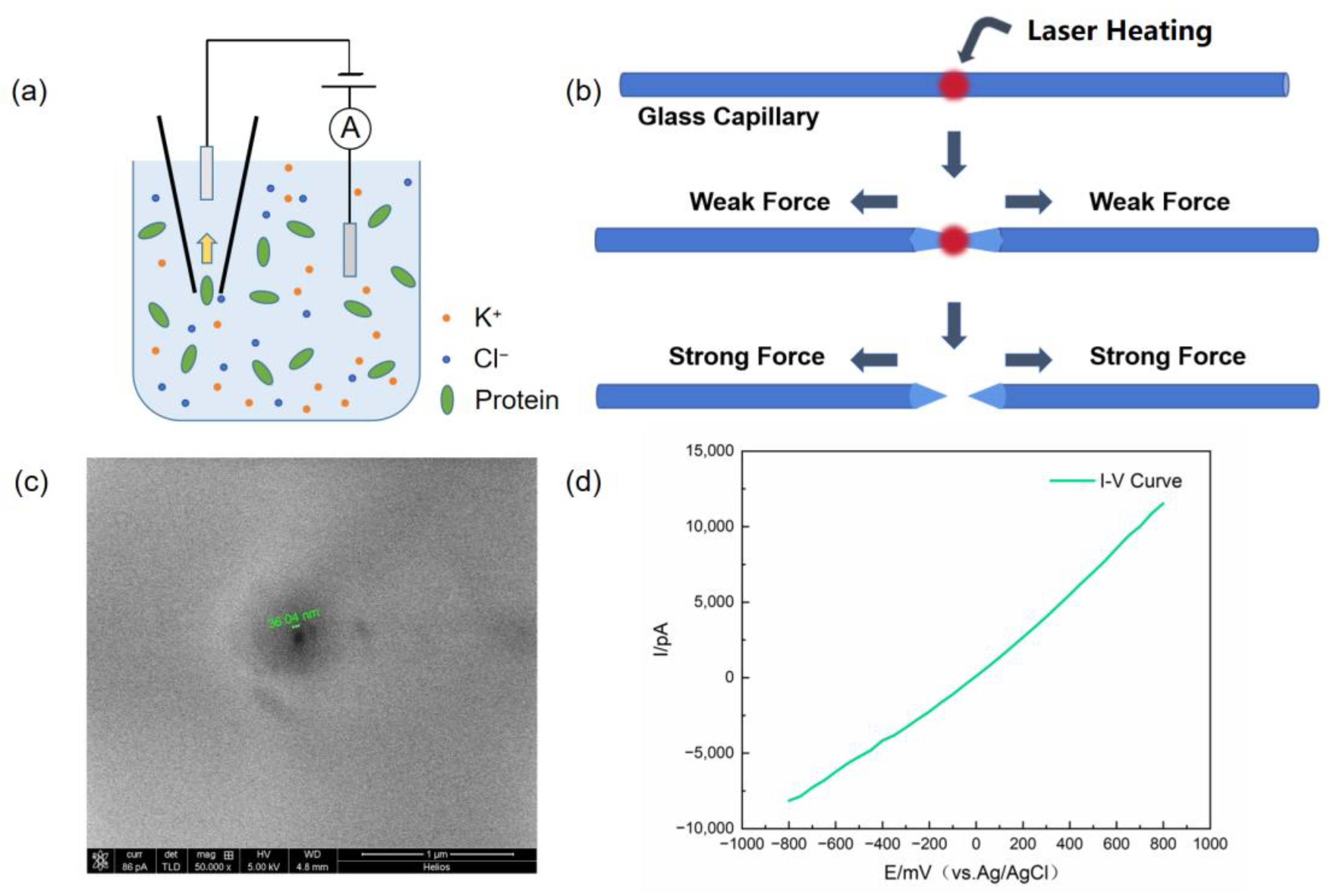
2.2. Device Construction
2.3. Fabrication of Glass Nanopores and Characterization
2.4. Protein Translocation
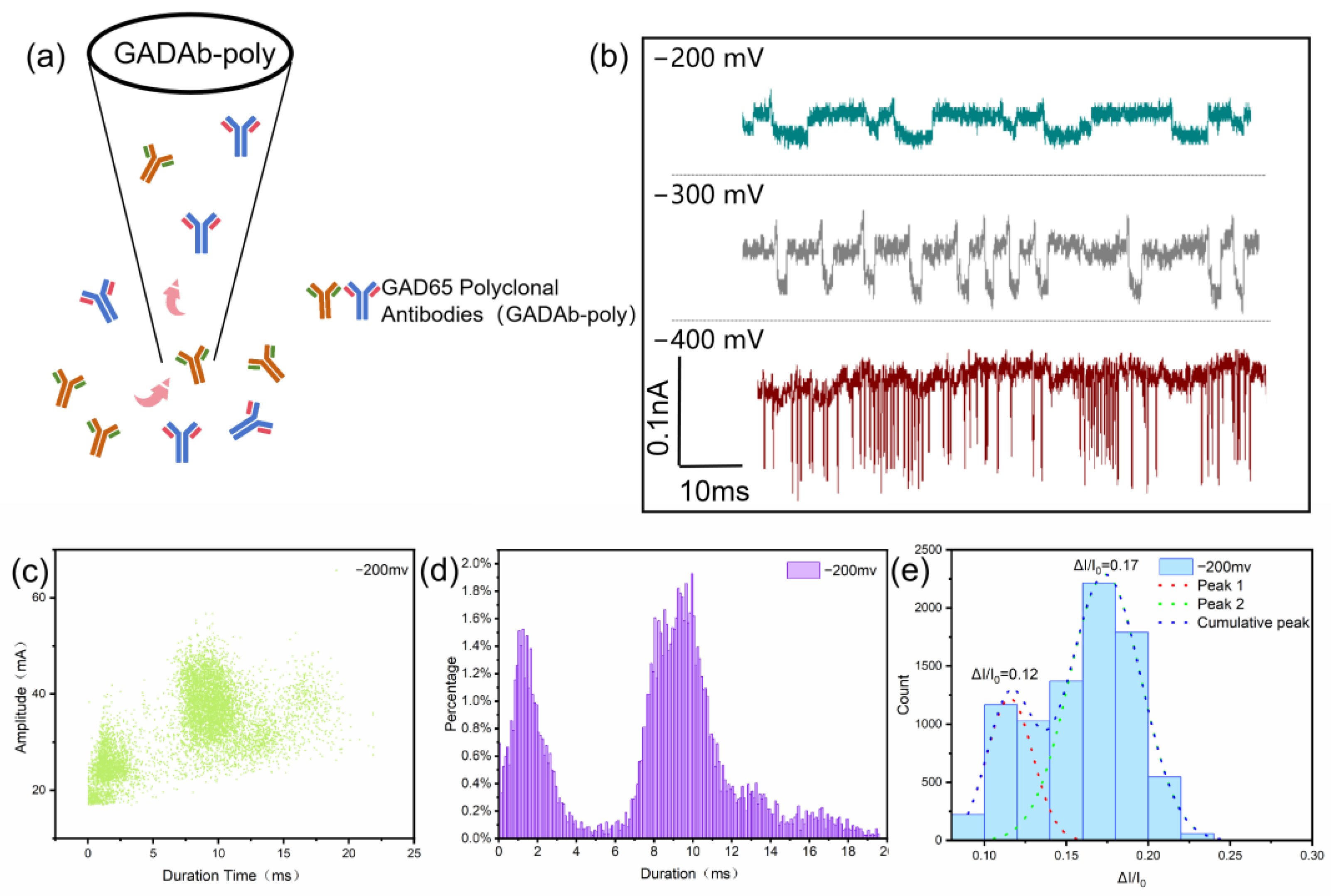
2.5. Detection of Immune Complex
2.6. Statistical Analysis Methods
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Diabetes Association. 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2021. Diabetes Care 2020, 44 (Suppl. S1), S15–S33. [Google Scholar] [CrossRef] [PubMed]
- Singla, R.; Homko, C.; Schey, R.; Parkman, H.P. Diabetes-Related Autoantibodies in Diabetic Gastroparesis. Dig. Dis. Sci. 2015, 60, 1733–1737. [Google Scholar] [CrossRef] [PubMed]
- Towns, R.; Pietropaolo, M. GAD-65 Autoantibodies and their role as biomarkers of type 1 diabetes and latent autoimmune diabetes in adults (LADA). Drugs Future 2011, 36, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, M.; Achenbach, P.; Powell, M.; Coles, R.; Chlebowska, M.; Carr, L.; Furmaniak, J.; Scholz, M.; Bonifacio, E.; Ziegler, A.-G.; et al. 3 Screen islet cell autoantibody ELISA: A sensitive and specific ELISA for the combined measurement of autoantibodies to GAD65, to IA-2 and to ZnT8. Clin. Chim. Acta 2016, 462, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Huang, J.; Hu, J. Improved diagnosis of type-1 diabetes mellitus using multiplexed autoantibodies ELISA array. Anal. Biochem. 2022, 649, 114722. [Google Scholar] [CrossRef] [PubMed]
- Murayama, H.; Matsuura, N.; Kawamura, T.; Maruyama, T.; Kikuchi, N.; Kobayashi, T.; Nishibe, F.; Nagata, A. A sensitive radioimmunoassay of insulin autoantibody: Reduction of non-specific binding of [125I]insulin. J. Autoimmun. 2006, 26, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Ankelo, M.; Westerlund, A.; Blomberg, K.; Knip, M.; Ilonen, J.; Hinkkanen, A.E. Time-Resolved Immunofluorometric Dual-Label Assay for Simultaneous Detection of Autoantibodies to GAD65 and IA-2 in Children with Type 1 Diabetes. Clin. Chem. 2007, 53, 472–479. [Google Scholar] [CrossRef]
- Zhong, Z.Y.; Peng, N.; Qing, Y.; Shan, J.L.; Li, M.X.; Guan, W.; Dai, N.; Gu, X.Q.; Wang, D. An electrochemical immunosensor for simultaneous multiplexed detection of neuron-specific enolase and pro-gastrin-releasing peptide using liposomes as enhancer. Electrochim. Acta 2011, 56, 5624–5629. [Google Scholar] [CrossRef]
- Parker, D.M.; Winkenbach, L.P.; Parker, A.; Boyson, S.; Nishimura, E.O. Improved Methods for Single-Molecule Fluorescence In Situ Hybridization and Immunofluorescence in Caenorhabditis elegans Embryos. Curr. Protoc. 2021, 1, e299. [Google Scholar] [CrossRef]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef]
- He, Y.; Tsutsui, M.; Zhou, Y.; Miao, X.-S. Solid-state nanopore systems: From materials to applications. NPG Asia Mater. 2021, 13, 48. [Google Scholar] [CrossRef]
- Xue, L.; Yamazaki, H.; Ren, R.; Wanunu, M.; Ivanov, A.P.; Edel, J.B. Solid-state nanopore sensors. Nat. Rev. Mater. 2020, 5, 931–951. [Google Scholar] [CrossRef]
- Coulter, W.H. Means for Counting Particles Suspended in a Fluid. US2656508A, 20 October 1953. [Google Scholar]
- Ying, Y.L.; Cao, C.; Long, Y.T. Single molecule analysis by biological nanopore sensors. Analyst 2014, 139, 3826–3835. [Google Scholar] [CrossRef] [PubMed]
- Arjmandi, N.; Van Roy, W.; Lagae, L.; Borghs, G. Measuring the Electric Charge and Zeta Potential of Nanometer-Sized Objects Using Pyramidal-Shaped Nanopores. Anal. Chem. 2012, 84, 8490–8496. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, M.; Yoshida, T.; Yokota, K.; Yasaki, H.; Yasui, T.; Arima, A.; Tonomura, W.; Nagashima, K.; Yanagida, T.; Kaji, N.; et al. Discriminating single-bacterial shape using low-aspect-ratio pores. Sci. Rep. 2017, 7, 17371. [Google Scholar] [CrossRef] [PubMed]
- Houghtaling, J.; Ying, C.; Eggenberger, O.M.; Fennouri, A.; Nandivada, S.; Acharjee, M.; Li, J.; Hall, A.R.; Mayer, M. Estimation of Shape, Volume, and Dipole Moment of Individual Proteins Freely Transiting a Synthetic Nanopore. ACS Nano 2019, 13, 5231–5242. [Google Scholar] [CrossRef] [PubMed]
- Stoddart, D.; Heron, A.J.; Klingelhoefer, J.; Mikhailova, E.; Maglia, G.; Bayley, H. Nucleobase Recognition in ssDNA at the Central Constriction of the α-Hemolysin Pore. Nano Lett. 2010, 10, 3633–3637. [Google Scholar] [CrossRef] [PubMed]
- Hiratani, M.; Kawano, R. DNA Logic Operation with Nanopore Decoding To Recognize MicroRNA Patterns in Small Cell Lung Cancer. Anal. Chem. 2018, 90, 8531–8537. [Google Scholar] [CrossRef] [PubMed]
- Merstorf, C.; Cressiot, B.; Pastoriza-Gallego, M.; Oukhaled, A.; Betton, J.M.; Auvray, L.; Pelta, J. Wild Type, Mutant Protein Unfolding and Phase Transition Detected by Single-Nanopore Recording. ACS Chem. Biol. 2012, 7, 652–658. [Google Scholar] [CrossRef]
- Ying, Y.L.; Yu, R.J.; Hu, Y.X.; Gao, R.; Long, Y.T. Single antibody-antigen interactions monitored via transient ionic current recording using nanopore sensors. Chem. Commun. 2017, 53, 8620–8623. [Google Scholar] [CrossRef]
- Lin, K.B.; Li, Z.W.; Tao, Y.; Li, K.; Yang, H.J.; Ma, J.; Li, T.; Sha, J.J.; Chen, Y.F. Surface Charge Density Inside a Silicon Nitride Nanopore. Langmuir 2021, 37, 10521–10528. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, B.M.; Dorvel, B.; Yemenicioglu, S.; Watkins, N.; Petrov, I.; Bashir, R. Highly Sensitive, Mechanically Stable Nanopore Sensors for DNA Analysis. Adv. Mater. 2009, 21, 2771–2776. [Google Scholar] [CrossRef] [PubMed]
- Steinbock, L.J.; Krishnan, S.; Bulushev, R.D.; Borgeaud, S.; Blokesch, M.; Feletti, L.; Radenovic, A. Probing the size of proteins with glass nanopores. Nanoscale 2014, 6, 14380–14387. [Google Scholar] [CrossRef] [PubMed]
- Bandara, Y.M.N.D.Y.; Freedman, K.J. Enhanced Signal to Noise Ratio Enables High Bandwidth Nanopore Recordings and Molecular Weight Profiling of Proteins. ACS Nano 2022, 16, 14111–14120. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhao, C.; Zhao, P.; Chen, F.; Qiao, D.; Feng, J. MoS2 nanopore identifies single amino acids with sub-1 Dalton resolution. Nat. Commun. 2023, 14, 2895. [Google Scholar] [CrossRef]
- Plesa, C.; Dekker, C. Data analysis methods for solid-state nanopores. Nanotechnology 2015, 26, 084003. [Google Scholar] [CrossRef]
- Cai, H.; Wang, Y.; Yu, Y.; Mirkin, M.V.; Bhakta, S.; Bishop, G.W.; Joshi, A.A.; Rusling, J.F. Resistive-Pulse Measurements with Nanopipettes: Detection of Vascular Endothelial Growth Factor C (VEGF-C) Using Antibody-Decorated Nanoparticles. Anal. Chem. 2015, 87, 6403–6410. [Google Scholar] [CrossRef] [PubMed]
- Kovarik, M.L.; Zhou, K.M.; Jacobson, S.C. Effect of Conical Nanopore Diameter on Ion Current Rectification. J. Phys. Chem. B 2009, 113, 15960–15966. [Google Scholar] [CrossRef] [PubMed]
- Woermann, D. Electrochemical transport properties of a cone-shaped nanopore: High and low electrical conductivity states depending on the sign of an applied electrical potential difference. Phys. Chem. Chem. Phys. 2003, 5, 1853–1858. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Y.; Guo, W.; Ji, H.; Xue, J.; Ouyang, Q. Asymmetric properties of ion transport in a charged conical nanopore. Phys. Rev. E 2007, 75, 6. [Google Scholar] [CrossRef]
- Siwy, Z.; Fulinski, A. Fabrication of a synthetic nanopore ion pump. Phys. Rev. Lett. 2002, 89, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tang, H.; Yang, C.; Li, Y. Selective Single Molecule Nanopore Sensing of microRNA Using PNA Functionalized Magnetic Core-shell Fe3O4-Au Nanoparticles. Anal. Chem. 2019, 91, 7965–7970. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Qi, G.; Zhou, Y.; Zhang, Y.; Wang, B.; Hu, P.; Jin, Y. Single-cell ATP detection and content analyses in electrostimulus-induced apoptosis using functionalized glass nanopipettes. Chem. Commun. 2020, 56, 1561–1564. [Google Scholar] [CrossRef] [PubMed]
- Fenalti, G.; Law, R.H.P.; Buckle, A.M.; Langendorf, C.; Tuck, K.; Rosado, C.J.; Faux, N.G.; Mahmood, K.; Hampe, C.S.; Banga, J.P.; et al. GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop. Nat. Struct. Mol. Biol. 2007, 14, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Sha, J.; Hasan, T.; Milana, S.; Bertulli, C.; Bell, N.A.W.; Privitera, G.; Ni, Z.; Chen, Y.; Bonaccorso, F.; Ferrari, A.C.; et al. Nanotubes Complexed with DNA and Proteins for Resistive-Pulse Sensing. ACS Nano 2013, 7, 8857–8869. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lin, Y.; Long, Y.-T.; Minteer, S.D.; Ying, Y.-L. Nanopore-based measurement of the interaction of P450cam monooxygenase and putidaredoxin at the single-molecule level. Faraday Discuss. 2022, 233, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Sha, J.; Si, W.; Xu, B.; Zhang, S.; Li, K.; Lin, K.; Shi, H.; Chen, Y. Identification of Spherical and Nonspherical Proteins by a Solid-State Nanopore. Anal. Chem. 2018, 90, 13826–13831. [Google Scholar] [CrossRef] [PubMed]
- Lipman, N.S.; Jackson, L.R.; Trudel, L.J.; Weis-Garcia, F. Monoclonal Versus Polyclonal Antibodies: Distinguishing Characteristics, Applications, and Information Resources. ILAR J. 2005, 46, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Fu, F.; Zhou, Z.Z.; Sun, Q.; Xu, B.; Sha, J. Label-free Detection of PD-1 Antibody and Antigen Immunoreaction Using Nano-Sensors. Acta Chim. 2019, 77, 287–292. [Google Scholar] [CrossRef]



| Heat | Filament | Velocity | Delay | Pull |
|---|---|---|---|---|
| 760 | 4 | 29 | 140 | 168 |
| Grouping of Events | Analysis Type | −300 mV | −400 mV | −500 mV | −600 mV |
|---|---|---|---|---|---|
| 0–60% | Duration Time | 0.052 ms | 0.056 ms | 0.053 ms | 0.056 ms |
| ΔI/I0 | 0.051 | 0.055 | 0.060 | 0.060 | |
| 60–100% | Duration Time | 0.12 ms | 0.28 ms | 0.66 ms | 0.29 ms |
| ΔI/I0 | 0.081 | 0.089 | 0.10 | 0.090 | |
| 0–100% | Duration Time | 0.053 ms | 0.077 ms | 0.087 ms | 0.086 ms |
| ΔI/I0 | 0.056 | 0.065 | 0.070 | 0.066 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, C.; Bai, Y.; Chen, J.; Lu, J.; Bi, Y.; Li, J. Detection of Glutamate Decarboxylase Antibodies and Simultaneous Multi-Molecular Translocation Exploration by Glass Nanopores. Biosensors 2024, 14, 255. https://doi.org/10.3390/bios14050255
Tao C, Bai Y, Chen J, Lu J, Bi Y, Li J. Detection of Glutamate Decarboxylase Antibodies and Simultaneous Multi-Molecular Translocation Exploration by Glass Nanopores. Biosensors. 2024; 14(5):255. https://doi.org/10.3390/bios14050255
Chicago/Turabian StyleTao, Chongxin, Yun Bai, Jiang Chen, Jing Lu, Yan Bi, and Jian Li. 2024. "Detection of Glutamate Decarboxylase Antibodies and Simultaneous Multi-Molecular Translocation Exploration by Glass Nanopores" Biosensors 14, no. 5: 255. https://doi.org/10.3390/bios14050255





