Introduction of Cellulolytic Bacterium Bacillus velezensis Z2.6 and Its Cellulase Production Optimization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism, Compost Sample, and Isolation
2.2. Screening of Cellulolytic Bacteria and Maintenance Conditions
2.3. Morphological, Physiological, and Biochemical Analysis and Growth Curve
2.4. 16S rRNA Gene Sequencing and Phylogenetic Analysis
2.5. Enzymatic Assay
2.5.1. Extraction of Crude Enzyme Solution
2.5.2. Crude Cellulase Activity Assay
2.6. Genome Sequencing, Annotation, and Functional Analysis
2.7. Statistical Design for Process Optimization
2.7.1. Complete Randomized Design
2.7.2. Plackett–Burman Design
2.7.3. Path of Steepest Ascent
2.7.4. Response Surface Construction
3. Results and Discussion
3.1. Microorganism Screening and Strain Characteristics
3.2. Identification and Phylogenetic Analysis of Strain Z2.6
3.3. Genome Features and Mining of Potential Cellulases
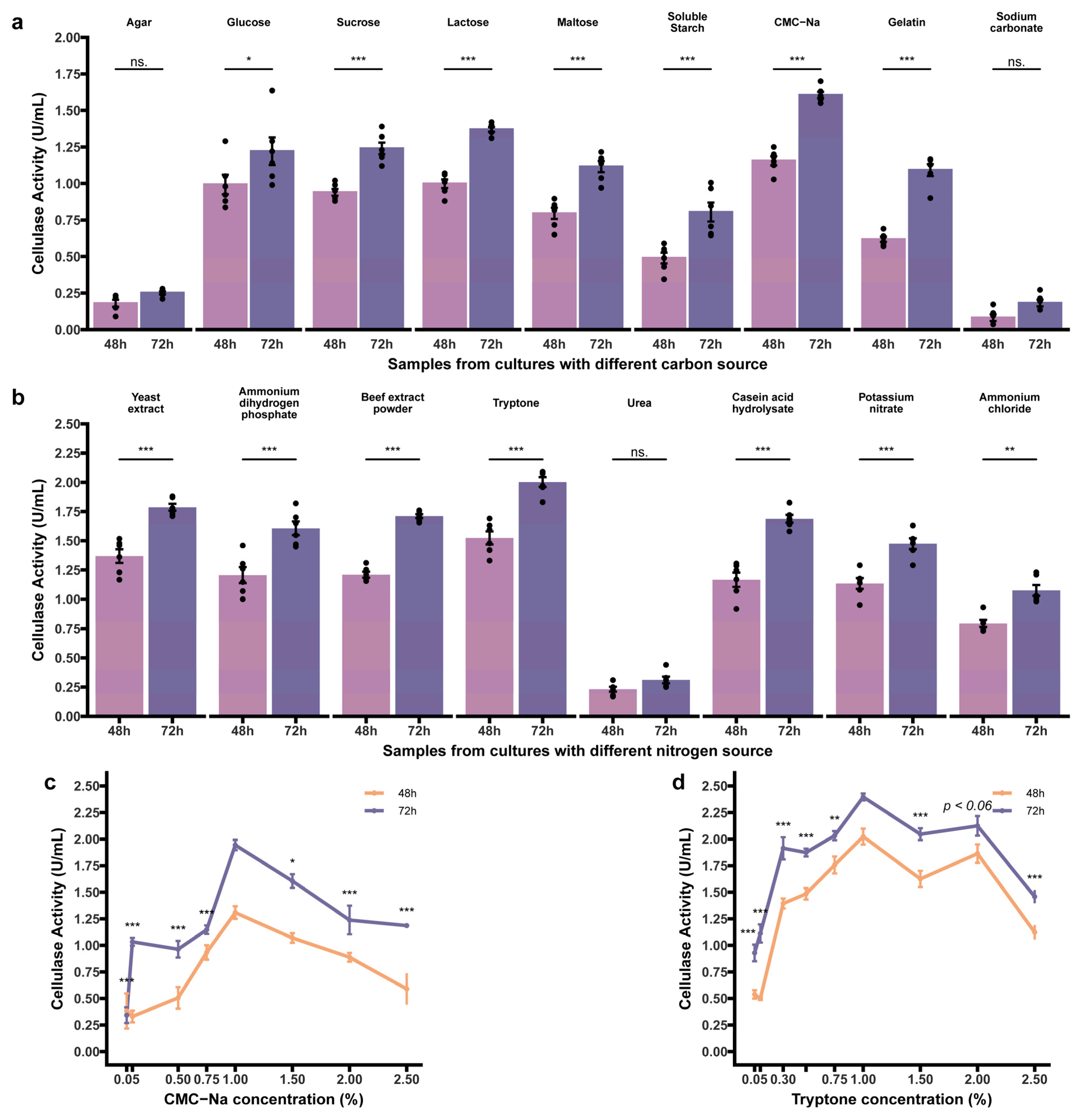
3.4. Process Optimization for Cellulase Production
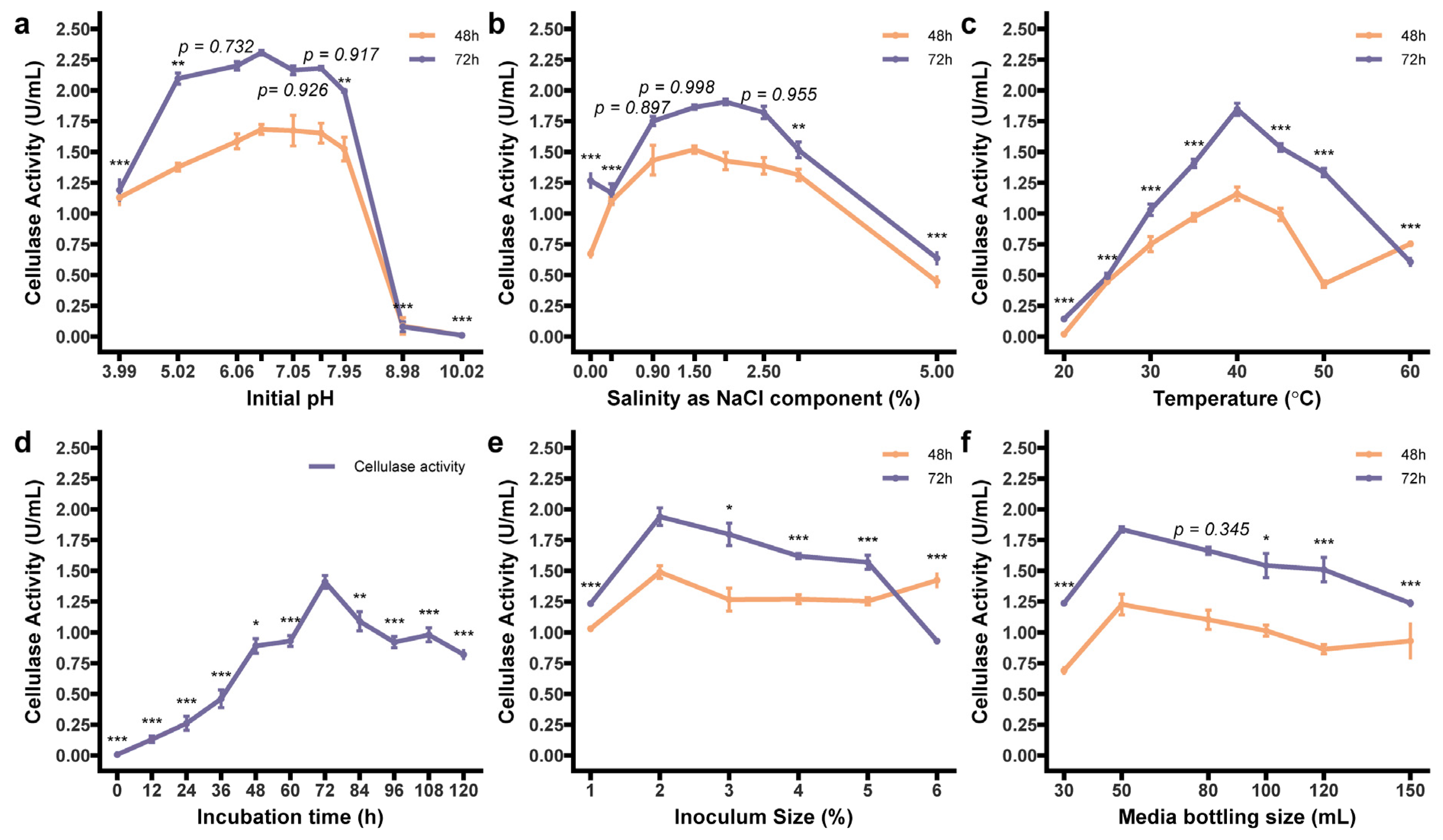
3.4.1. Optimum Enzymatic Condition
3.4.2. Effect of Independent Factors
3.5. Statistical Optimization of Cellulase Production
3.5.1. Independent Factors as Main Effects
3.5.2. Path of Steepest Ascent Design
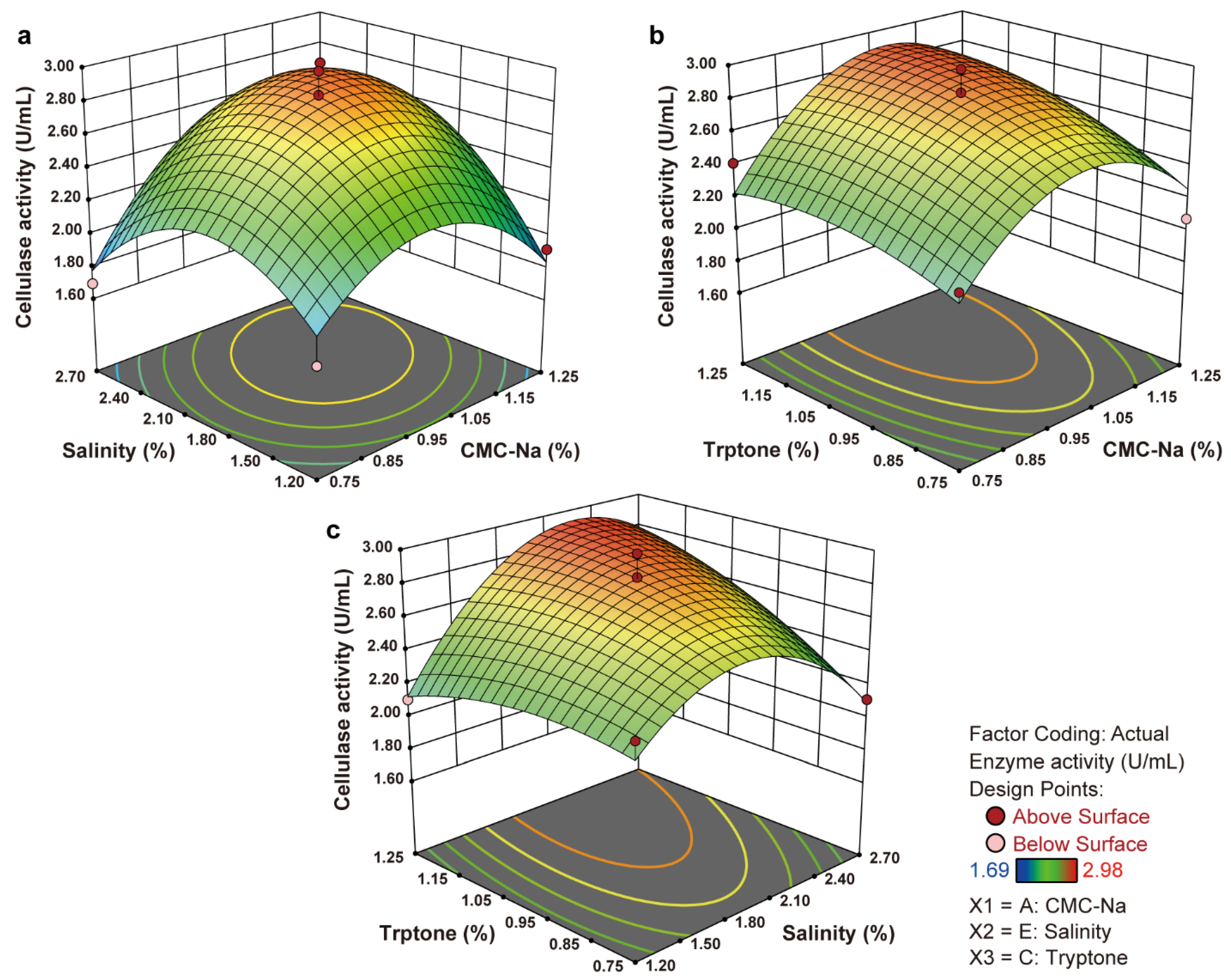
3.5.3. Response Surface Method by Box–Behnken Design
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seddiqi, H.; Oliaei, E.; Honarkar, H.; Jin, J.; Geonzon, L.C.; Bacabac, R.G.; Klein-Nulend, J. Cellulose and its derivatives: Towards biomedical applications. Cellulose 2021, 28, 1893–1931. [Google Scholar] [CrossRef]
- Bhardwaj, N.; Kumar, B.; Agrawal, K.; Verma, P. Current perspective on production and applications of microbial cellulases: A review. Bioresour. Bioprocess. 2021, 8, 95. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-K.; Kim, H.-J.; Lee, J.-W. Rapid Statistical Optimization of Cultural Conditions for Mass Production of Carboxymethylcellulase by a Newly Isolated Marine Bacterium, Bacillus velezensis A-68 from Rice Hulls. J. Life Sci. 2013, 23, 757–769. [Google Scholar] [CrossRef]
- Areeshi, M.Y. Microbial cellulase production using fruit wastes and its applications in biofuels production. Int. J. Food Microbiol. 2022, 378, 109814. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xie, Y.; Gao, X.; Shan, M.; Sun, C.; Niu, Y.D.; Shan, A. Screening of cellulose degradation bacteria from Min pigs and optimization of its cellulase production. Electron. J. Biotechnol. 2020, 48, 29–35. [Google Scholar] [CrossRef]
- Horn, S.J.; Vaaje-Kolstad, G.; Westereng, B.; Eijsink, V. Novel enzymes for the degradation of cellulose. Biotechnol. Biofuels 2012, 5, 45. [Google Scholar] [CrossRef]
- Ryckeboer, J.; Mergaert, J.; Vaes, K.; Klammer, S.H.; Clercq, D.D.; Coosemans, J.; Insam, H.; Swings, J. A survey of bacteria and fungi occurring during composting and self-heating processes. Ann. Microbiol. 2003, 53, 349–410. [Google Scholar]
- Han, W.; Clarke, W.; Pratt, S. Composting of waste algae: A review. Waste Manag. 2014, 34, 1148–1155. [Google Scholar] [CrossRef]
- Munir, N.; Malik, K.; Bashir, R.; Altaf, I.; Naz, S. Isolation and Characterization of Polysaccharide-Degrading Microbes from Compost Samples. J. Environ. Sci. Manag. 2021, 24, 10–16. [Google Scholar] [CrossRef]
- Chandna, P.; Nain, L.; Singh, S.; Kuhad, R.C. Assessment of bacterial diversity during composting of agricultural byproducts. BMC Microbiol. 2013, 13, 99. [Google Scholar] [CrossRef]
- Khosravi, F.; Khaleghi, M.; Naghavi, H. Screening and identification of cellulose-degrading bacteria from soil and leaves at Kerman province, Iran. Arch. Microbiol. 2021, 204, 88. [Google Scholar] [CrossRef] [PubMed]
- Maki, M.; Leung, K.T.; Qin, W. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int. J. Biol. Sci. 2009, 5, 500–516. [Google Scholar] [CrossRef] [PubMed]
- Adeniji, A.A.; Loots, D.T.; Babalola, O.O. Bacillus velezensis: Phylogeny, useful applications, and avenues for exploitation. Appl. Microbiol. Biotechnol. 2019, 103, 3669–3682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wei, S.; Liu, Y.; Cheng, C.; Ma, J.; Yue, L.; Gao, Y.; Cheng, Y.; Ren, Y.; Su, S.; et al. Screening and genome-wide analysis of lignocellulose-degrading bacteria from humic soil. Front. Microbiol. 2023, 14, 1167293. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Lee, S.Y.; Weon, H.-Y.; Sang, M.K.; Song, J. Complete genome sequence of Bacillus velezensis M75, a biocontrol agent against fungal plant pathogens, isolated from cotton waste. J. Biotechnol. 2017, 241, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus velezensis FZB42 in 2018: The Gram-Positive Model Strain for Plant Growth Promotion and Biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Qu, Z.; Yu, W.; Zheng, L.; Qiao, H.; Wang, D.; Wei, B.; Zhao, Z. Comparative genomic and transcriptome analysis of Bacillus velezensis CL-4 fermented corn germ meal. AMB Express 2023, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Djelid, H.; Flahaut, S.; Vander Wauven, C.; Oudjama, Y.; Hiligsmann, S.; Cornu, B.; Cherfia, R.; Gares, M.; Kacem Chaouche, N. Production of a halotolerant endo-1,4-β-glucanase by a newly isolated Bacillus velezensis H1 on olive mill wastes without pretreatment: Purification and characterization of the enzyme. Arch. Microbiol. 2022, 204, 681. [Google Scholar] [CrossRef] [PubMed]
- Khalid, A.; Ye, M.; Wei, C.; Dai, B.; Yang, R.; Huang, S.; Wang, Z. Production of β-glucanase and protease from Bacillus velezensis strain isolated from the manure of piglets. Prep. Biochem. Biotechnol. 2020, 51, 497–510. [Google Scholar] [CrossRef]
- Hungate, R.E. Studies on Cellulose Fermentation: I. The Culture and Physiology of an Anaerobic Cellulose-digesting Bacterium. J. Bacteriol. 1944, 48, 499–513. [Google Scholar] [CrossRef]
- Teather, R.M.; Wood, P.J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 1982, 43, 777–780. [Google Scholar] [CrossRef] [PubMed]
- Samira, M.S.; Mohammad, R.A.; Gholamreza, G. Carboxymethyl-cellulase and Filter-paperase Activity of New Strains Isolated from Persian Gulf. Microbiology 2011, 1, 8–16. [Google Scholar] [CrossRef]
- Dong, X.; Cai, M. Common Bacterial System Identification Manual; Science Press: Beijing, China, 2001. [Google Scholar]
- López, S.; Prieto, M.; Dijkstra, J.; Dhanoa, M.S.; France, J. Statistical evaluation of mathematical models for microbial growth. Int. J. Food Microbiol. 2004, 96, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S.; Battistuzzi, F.U. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Ghose, T.K. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Makarova, K.S.; Wolf, Y.I.; Koonin, E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015, 43, D261–D269. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef] [PubMed]
- Plackett, R.L.; Burman, J.P. The Dedign if Optimum Multifactorial Experiments. Biometrika 1946, 33, 305–325. [Google Scholar] [CrossRef]
- Jiang, D.; Zhen, F.; Chin, S.-x.; Tian, X.-f.; Su, T.-c. Biohydrogen Production from Hydrolysates of Selected Tropical Biomass Wastes with Clostridium Butyricum. Sci. Rep. 2016, 6, 27205. [Google Scholar] [CrossRef]
- Box, G.E.P.; Behnken, D.W. Some New Three Level Designs for the Study of Quantitative Variables. Technometrics 1960, 2, 455–475. [Google Scholar] [CrossRef]
- Chantarasiri, A. Novel Halotolerant Cellulolytic Bacillus methylotrophicus RYC01101 Isolated from Ruminant Feces in Thailand and its Application for Bioethanol Production. KMUTNB Int. J. Appl. Sci. Technol. 2014, 7, 63–68. [Google Scholar] [CrossRef]
- Bergey, D.H.; Holt, J.G. Bergey’s Manual of Determinative Bacteriology; Williams & Wilkins: Philadelphia, PA, USA, 1994. [Google Scholar]
- Dunlap, C.A.; Kim, S.J.; Kwon, S.W.; Rooney, A.P. Phylogenomic analysis shows that Bacillus amyloliquefaciens subsp. plantarum is a later heterotypic synonym of Bacillus methylotrophicus. Int. J. Syst. Evol. Microbiol. 2015, 65, 2104–2109. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-García, C.; Béjar, V.; Martínez-Checa, F.; Llamas, I.; Quesada, E. Bacillus velezensis sp. nov., a surfactant-producing bacterium isolated from the river Vélez in Málaga, southern Spain. Int. J. Syst. Evol. Microbiol. 2005, 55, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.-Y.; Liu, Y.; Miao, L.-L.; Li, E.-W.; Sun, G.-x.; Liu, Y.; Liu, Z.-P. Characterization and mechanism of anti-Aeromonas salmonicida activity of a marine probiotic strain, Bacillus velezensis V4. Appl. Microbiol. Biotechnol. 2017, 101, 3759–3768. [Google Scholar] [CrossRef] [PubMed]
- Thurlow, C.M.; Williams, M.A.; Carrias, A.; Ran, C.; Newman, M.; Tweedie, J.; Allison, E.; Jescovitch, L.N.; Wilson, A.E.; Terhune, J.S.; et al. Bacillus velezensis AP193 exerts probiotic effects in channel catfish (Ictalurus punctatus) and reduces aquaculture pond eutrophication. Aquaculture 2019, 503, 347–356. [Google Scholar] [CrossRef]
- Lee, H.-H.; Park, J.; Lim, J.Y.; Kim, H.; Choi, G.J.; Kim, J.-C.; Seo, Y.-S. Complete genome sequence of Bacillus velezensis G341, a strain with a broad inhibitory spectrum against plant pathogens. J. Biotechnol. 2015, 211, 97–98. [Google Scholar] [CrossRef]
- Shu, X.; Wang, Y.; Zhou, Q.; Li, M.; Hu, H.; Ma, Y.; Chen, X.; Ni, J.; Zhao, W.; Huang, S.; et al. Biological Degradation of Aflatoxin B1 by Cell-Free Extracts of Bacillus velezensis DY3108 with Broad pH Stability and Excellent Thermostability. Toxins 2018, 10, 330. [Google Scholar] [CrossRef] [PubMed]
- Jani, M.; Azad, R.K. Discovery of mosaic genomic islands in Pseudomonas spp. Arch. Microbiol. 2021, 203, 2735–2742. [Google Scholar] [CrossRef]
- Tan, M.C.Y.; Zakaria, M.R.; Liew, K.J.; Chong, C.S. Draft genome sequence of Hahella sp. CR1 and its ability in producing cellulases for saccharifying agricultural biomass. Arch. Microbiol. 2023, 205, 278. [Google Scholar] [CrossRef]
- Nair, A.S.; Al-Battashi, H.; Al-Akzawi, A.; Annamalai, N.; Gujarathi, A.; Al-Bahry, S.; Dhillon, G.S.; Sivakumar, N. Waste office paper: A potential feedstock for cellulase production by a novel strain Bacillus velezensis ASN1. Waste Manag. 2018, 79, 491–500. [Google Scholar] [CrossRef]
- Liu, H.; Zeng, L.; Jin, Y.; Nie, K.; Deng, L.; Wang, F. Effect of Different Carbon Sources on Cellulase Production by Marine Strain Microbulbifer hydrolyticus IRE-31-192. Appl. Biochem. Biotechnol. 2019, 188, 741–749. [Google Scholar] [CrossRef]
- Ma, L.; Aizhan, R.; Wang, X.; Yi, Y.; Shan, Y.; Liu, B.; Zhou, Y.; Lü, X. Cloning and characterization of low-temperature adapted GH5-CBM3 endo-cellulase from Bacillus subtilis 1AJ3 and their application in the saccharification of switchgrass and coffee grounds. AMB Express 2020, 10, 42. [Google Scholar] [CrossRef]
- Ramana, K.V.; Tomar, A.; Singh, L. Effect of various carbon and nitrogen sources on cellulose synthesis by Acetobacter xylinum. World J. Microbiol. Biotechnol. 2000, 16, 245–248. [Google Scholar] [CrossRef]
- Wu, Y.-F.; Zheng, A.-R.; Bai, Y.-Y. Effects of colloid sources and concentrations of colloidal organic carbon on the growth of colloids’ own nature bacteria community. J. Oceanogr. Taiwan Strait 2009, 28, 343–348. [Google Scholar]
- Deka, D.; Bhargavi, P.; Sharma, A.; Goyal, D.; Jawed, M.; Goyal, A. Enhancement of Cellulase Activity from a New Strain of Bacillus subtilis by Medium Optimization and Analysis with Various Cellulosic Substrates. Enzym. Res. 2011, 2011, 151656. [Google Scholar] [CrossRef]
- Vijayaraghavan, P.; Prakash Vincent, S.G.; Dhillon, G.S. Solid-substrate bioprocessing of cow dung for the production of carboxymethyl cellulase by Bacillus halodurans IND18. Waste Manag. 2016, 48, 513–520. [Google Scholar] [CrossRef]
- Kazemi, A.; Rasoul-Amini, S.; Shahbazi, M.; Safari, A.; Ghasemi, Y. Isolation, identification, and media optimization of high-level cellulase production by Bacillus sp. BCCS A3, in a fermentation system using response surface methodology. Prep. Biochem. Biotechnol. 2014, 44, 107–118. [Google Scholar] [CrossRef]
- Singh, J.; Batra, N.; Sobti, R.C. A highly thermostable, alkaline CMCase produced by a newly isolated Bacillus sp VG1. World J. Microbiol. Biotechnol. 2001, 17, 761–765. [Google Scholar] [CrossRef]
- Ray, A.K.; Bairagi, A.; Sarkar Ghosh, K.; Sen, S.K. Optimization of fermentation conditions for cellulase production by Bacillus subtilis CY5 and Bacillus circulans TP3 isolated from fish gut. Acta Ichthyol. Et Piscat. 2007, 37, 47–53. [Google Scholar] [CrossRef]
- Sethi, S.; Datta, A.; Gupta, B.L.; Gupta, S. Optimization of cellulase production from bacteria isolated from soil. ISRN Biotechnol. 2013, 2013, 985685. [Google Scholar] [CrossRef]
- Meng, F.; Ma, L.; Ji, S.; Yang, W.; Cao, B. Isolation and characterization of Bacillus subtilis strain BY-3, a thermophilic and efficient cellulase-producing bacterium on untreated plant biomass. Lett. Appl. Microbiol. 2014, 59, 306–312. [Google Scholar] [CrossRef]
- Thakkar, A.; Saraf, M. Application of Statistically Based Experimental Designs to Optimize Cellulase Production and Identification of Gene. Nat. Prod. Bioprospecting 2014, 4, 341–351. [Google Scholar] [CrossRef]
- Sherief, A.A.; El-Tanash, A.B.; Atia, N. Cellulase Production by Aspergillus fumigatus Grown on Mixed Substrate of Rice Straw and Wheat Bran. Res. J. Microbiol. 2010, 5, 199–211. [Google Scholar] [CrossRef]
- Singh, K.; Richa, K.; Bose, H.; Karthik, L.; Kumar, G.; Bhaskara Rao, K.V. Statistical media optimization and cellulase production from marine Bacillus VITRKHB. 3 Biotech 2014, 4, 591–598. [Google Scholar] [CrossRef]
- Jiang, L.; Long, C.; Wu, X.; Xu, H.; Shao, Z.; Long, M. Optimization of thermophilic fermentative hydrogen production by the newly isolated Caloranaerobacter azorensis H53214 from deep-sea hydrothermal vent environment. Int. J. Hydrogen Energy 2014, 39, 14154–14160. [Google Scholar] [CrossRef]
- Thite, V.S.; Nerurkar, A.S.; Baxi, N.N. Optimization of concurrent production of xylanolytic and pectinolytic enzymes by Bacillus safensis M35 and Bacillus altitudinis J208 using agro-industrial biomass through Response Surface Methodology. Sci. Rep. 2020, 10, 3824. [Google Scholar] [CrossRef]
- Peixoto, S.B.; Cladera-Olivera, F.; Daroit, D.J.; Brandelli, A. Cellulase-producing Bacillus strains isolated from the intestine of Amazon basin fish. Aquac. Res. 2011, 42, 887–891. [Google Scholar] [CrossRef]
- Özbek Yazıcı, S.; Özmen, I. Optimization for coproduction of protease and cellulase from Bacillus subtilis M-11 by the Box–Behnken design and their detergent compatibility. Braz. J. Chem. Eng. 2020, 37, 49–59. [Google Scholar] [CrossRef]
- Premalatha, N.; Gopal, N.O.; Jose, P.A.; Anandham, R.; Kwon, S.-W. Optimization of cellulase production by Enhydrobacter sp. ACCA2 and its application in biomass saccharification. Front. Microbiol. 2015, 6, 1046. [Google Scholar] [CrossRef]
- Singh, S.; Dikshit, P.K.; Moholkar, V.S.; Goyal, A. Purification and characterization of acidic cellulase from Bacillus amyloliquefaciens SS35 for hydrolyzingParthenium hysterophorusbiomass. Environ. Prog. Sustain. Energy 2015, 34, 810–818. [Google Scholar] [CrossRef]
- Shajahan, S.; Moorthy, I.G.; Sivakumar, N.; Selvakumar, G. Statistical modeling and optimization of cellulase production by Bacillus licheniformis NCIM 5556 isolated from the hot spring, Maharashtra, India. J. King Saud Univ.-Sci. 2017, 29, 302–310. [Google Scholar] [CrossRef]


| Characteristic | Results 1 | Characteristic | Results 1 |
|---|---|---|---|
| Lactose | + | Catalase | + |
| Sucrose | + | Urea | − |
| Glycerol | + | Lysine | − |
| Mannose | + | Arginine | − |
| Sorbitol | − | Ornithine | − |
| Salicin | − | Indole | − |
| Dulcitol | − | VP test | − |
| Esculin | + | Methyl red test | − |
| Raffinose | − | 6% NaCl | + |
| ONPG | + | 8% NaCl | + |
| Gelatin | + | 10% NaCl | − |
| Propionate | − | pH 6.0 | + |
| Malonate | − | pH 5.0 | + |
| Simon’s citrate salt | − | Growth at 10 °C | + |
| Amylolysis | + | Growth at 70 °C | − |
| Locus Accession Number | CAZy Family | Annotation (EC Codes) | CBM Family 1 | Signal Peptide | Most Identical Sequence to B. velezensis (% Identity) | Most Identical Sequence to Non-bacillus [Species] (% Identity) |
|---|---|---|---|---|---|---|
| V7S33_01155 | GH1 | Alpha-N-arabinofuranosidase (EC 3.2.1.55) | − 2 | − | WP_326142774.1 99.80% | P45797.1 [Paenibacillus polymyxa] 75.46% |
| V7S33_04525 | GH1 | 6-phospho-beta-glucosidase (EC 3.2.1.86) | − | − | WP_129091804.1 99.39% | Q9KBR4.1 [Halalkalibacterium halodurans C-125] 73.03% |
| V7S33_05150 | GH5 subfamily 2 | Endoglucanase (EC 3.2.1.4) | CBM3 | − | WP_032875077.1 100% | Q46829.2 [Escherichia coli K-12] 65.40% |
| V7S33_13785 | GH1 | Beta-glucosidase (EC 3.2.1.21) | − | − | WP_285980062.1 99.57% | Q47096.1 [Pectobacterium carotovorum subsp. carotovorum] 53.50% |
| V7S33_13965 | GH16 subfamily 21 | Beta-glucanase (EC 3.2.1.73) | − | Yes | WP_308826282.1 99.59% | P26208.1 [Acetivibrio thermocellus ATCC 27405] 38.48% |
| Run Order | Variables 1 | Response Cellulase Activity | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | ||
| 1 | 0.5 | 1 | 45 | 6.5 | 0 | 2 | 50 | 72 | 1.177 ± 0.08 |
| 2 | 0.5 | 0.5 | 40 | 6 | 0 | 2 | 50 | 48 | 0.756 ± 0.07 |
| 3 | 0.5 | 0.5 | 45 | 6 | 1.5 | 4 | 50 | 72 | 1.229 ± 0.08 |
| 4 | 1 | 1 | 40 | 6 | 0 | 4 | 50 | 72 | 2.167 ± 0.11 |
| 5 | 1 | 0.5 | 45 | 6.5 | 1.5 | 2 | 50 | 48 | 1.694 ± 0.06 |
| 6 | 1 | 1 | 45 | 6 | 0 | 2 | 80 | 48 | 1.947 ± 0.04 |
| 7 | 0.5 | 0.5 | 40 | 6.5 | 0 | 4 | 80 | 48 | 0.535 ± 0.06 |
| 8 | 0.5 | 1 | 40 | 6.5 | 1.5 | 2 | 80 | 72 | 2.003 ± 0.04 |
| 9 | 0.5 | 1 | 45 | 6 | 1.5 | 4 | 80 | 48 | 1.876 ± 0.10 |
| 10 | 1 | 0.5 | 45 | 6.5 | 0 | 4 | 80 | 72 | 1.976 ± 0.06 |
| 11 | 1 | 1 | 40 | 6.5 | 1.5 | 4 | 50 | 48 | 2.642 ± 0.05 |
| 12 | 1 | 0.5 | 40 | 6 | 1.5 | 2 | 80 | 72 | 2.499 ± 0.09 |
| Run | CMC-Na (%) | Salinity (%) | Tryptone (%) | Cellulase Activity (U/mL) |
|---|---|---|---|---|
| 1 | 0.50 | 1.05 | 0.50 | 0.891 ± 0.03 |
| 2 | 0.75 | 1.50 | 0.75 | 1.678 ± 0.08 |
| 3 | 1.00 | 1.95 | 1.00 | 2.764 ± 0.04 |
| 4 | 1.25 | 2.40 | 1.25 | 2.509 ± 0.05 |
| 5 | 1.50 | 2.85 | 1.50 | 1.988 ± 0.11 |
| Run Order | Variable | Response: Enzyme Activity (U/mL) | ||
|---|---|---|---|---|
| Factor 1: CMC-Na (%) | Factor 2: Salinity (%) | Factor 3: Tryptone (%) | ||
| 1 | 1 | 0.65 | 1 | 2.979 ± 0.10 |
| 2 | 1 | 0.9 | 1.25 | 2.685 ± 0.11 |
| 3 | 1 | 0.4 | 1.25 | 2.106 ± 0.09 |
| 4 | 1.25 | 0.65 | 0.75 | 2.076 ± 0.06 |
| 5 | 1.25 | 0.65 | 1.25 | 2.621 ± 0.14 |
| 6 | 1 | 0.65 | 1 | 2.814 ± 0.08 |
| 7 | 1 | 0.9 | 0.75 | 2.114 ± 0.03 |
| 8 | 0.75 | 0.65 | 0.75 | 2.162 ± 0.07 |
| 9 | 1.25 | 0.9 | 1 | 2.671 ± 0.05 |
| 10 | 1 | 0.65 | 1 | 2.705 ± 0.07 |
| 11 | 1 | 0.4 | 0.75 | 2.377 ± 0.04 |
| 12 | 0.75 | 0.9 | 1 | 1.695 ± 0.05 |
| 13 | 1 | 0.65 | 1 | 2.819 ± 0.05 |
| 14 | 1 | 0.65 | 1 | 2.840 ± 0.09 |
| 15 | 0.75 | 0.65 | 1.25 | 2.413 ± 0.10 |
| 16 | 0.75 | 0.4 | 1 | 1.787 ± 0.02 |
| 17 | 1.25 | 0.4 | 1 | 1.916 ± 0.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Z.; Wang, Y.; You, Y.; Yang, N.; Lu, S.; Xue, J.; Xing, X.; Sha, S.; Zhao, L. Introduction of Cellulolytic Bacterium Bacillus velezensis Z2.6 and Its Cellulase Production Optimization. Microorganisms 2024, 12, 979. https://doi.org/10.3390/microorganisms12050979
Cai Z, Wang Y, You Y, Yang N, Lu S, Xue J, Xing X, Sha S, Zhao L. Introduction of Cellulolytic Bacterium Bacillus velezensis Z2.6 and Its Cellulase Production Optimization. Microorganisms. 2024; 12(5):979. https://doi.org/10.3390/microorganisms12050979
Chicago/Turabian StyleCai, Zhi, Yi Wang, Yang You, Nan Yang, Shanshan Lu, Jianheng Xue, Xiang Xing, Sha Sha, and Lihua Zhao. 2024. "Introduction of Cellulolytic Bacterium Bacillus velezensis Z2.6 and Its Cellulase Production Optimization" Microorganisms 12, no. 5: 979. https://doi.org/10.3390/microorganisms12050979





