CD133 Stimulates Cell Proliferation via the Upregulation of Amphiregulin in Melanoma
Abstract
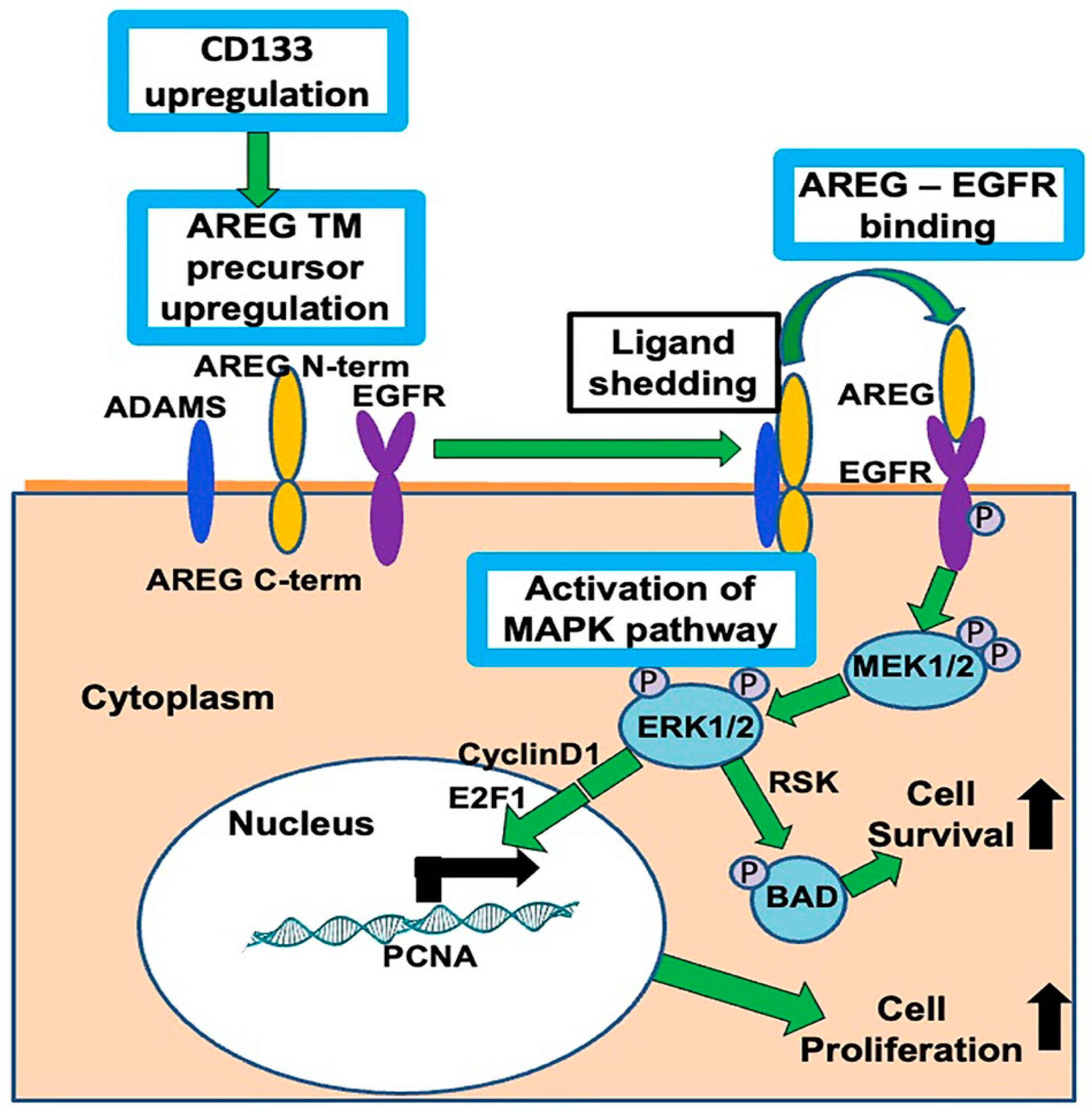
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Generation of Dox-Inducible Cells
2.3. RNA-seq Analysis
2.4. Pathway Analysis
2.5. Quantitative Reverse-Transcription PCR (qRT-PCR)
2.6. Immunoblot Analysis
2.7. Cell Cycle and Cell Growth Analysis
2.8. Bromodeoxyuridine (BrdU) Staining and Fluorescence Microscopy
2.9. Statistical Analysis
3. Results
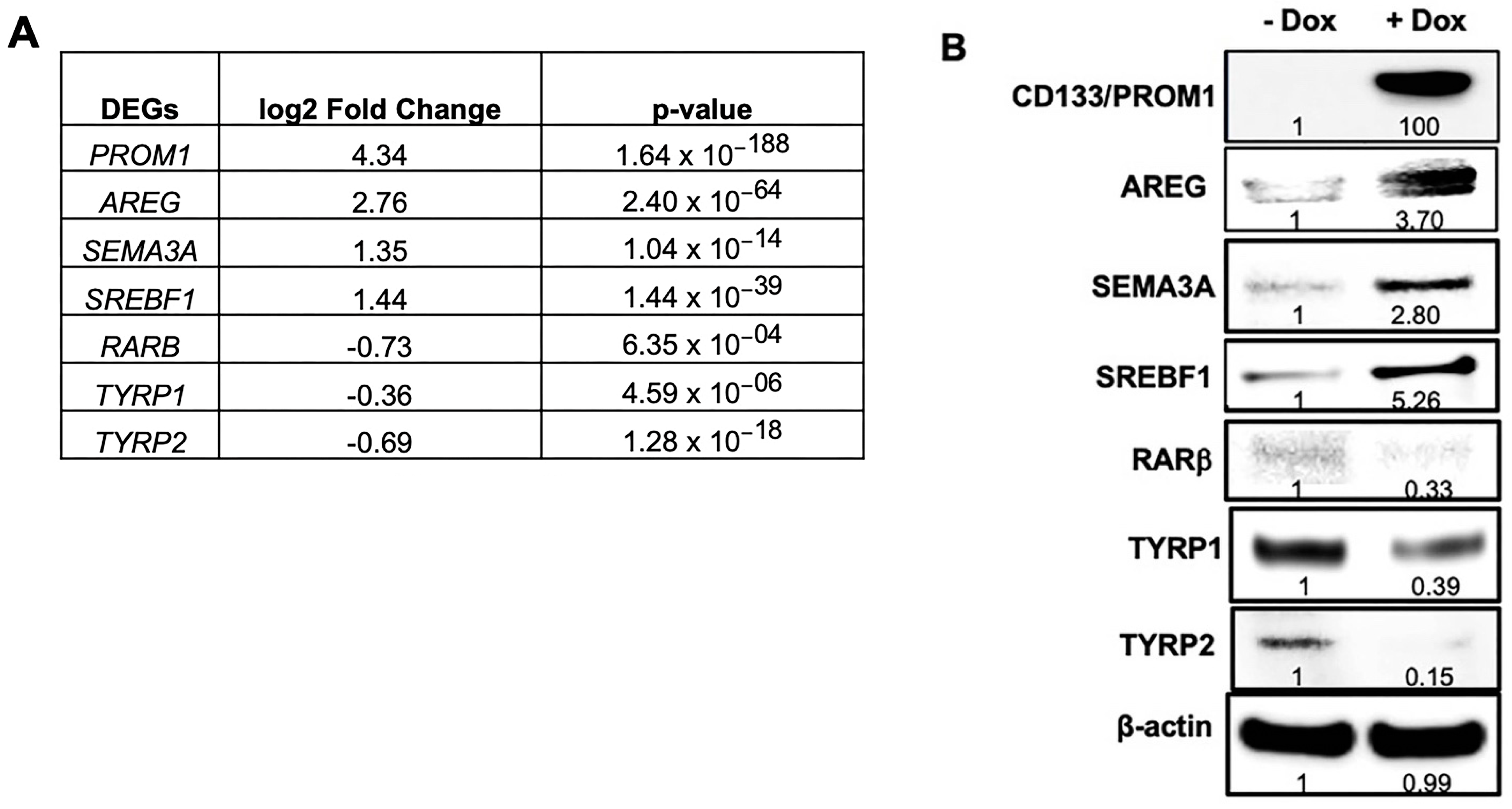
3.1. RNA-seq Analysis Reveal AREG as the Most Differentially Expressed Gene in Patient-Derived BAKP Melanoma Cells after Doxycycline (Dox)-Inducible Expression of CD133
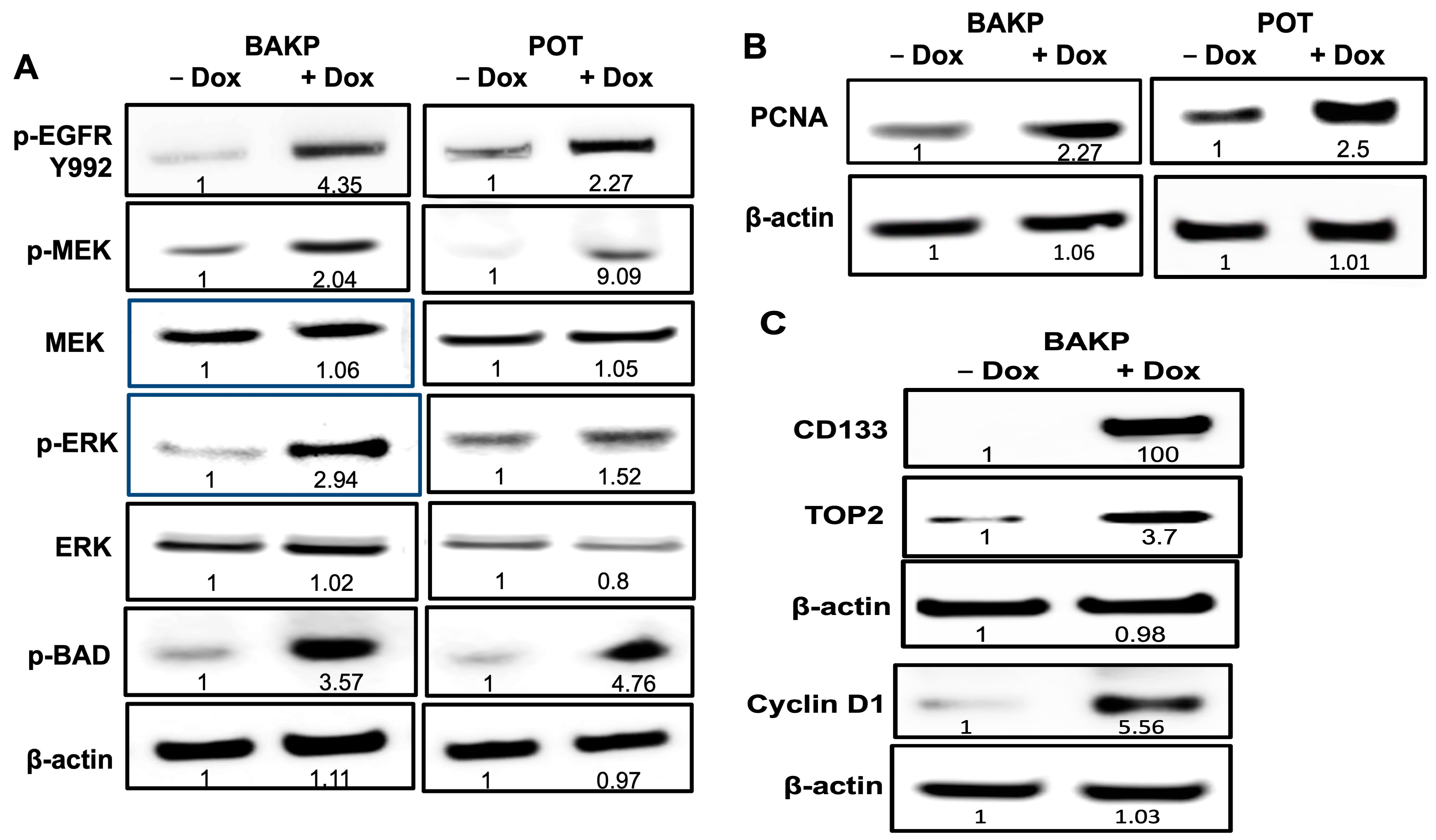
3.2. Validation of Differentially Expressed Genes in RNA-seq by Immunoblot Analysis in BAKP Melanoma Cells after Dox-Inducible Expression of CD133
3.3. Pathway Analysis of Significantly Altered Genes in BAKP Melanoma Cells after Dox-Induced Expression of CD133
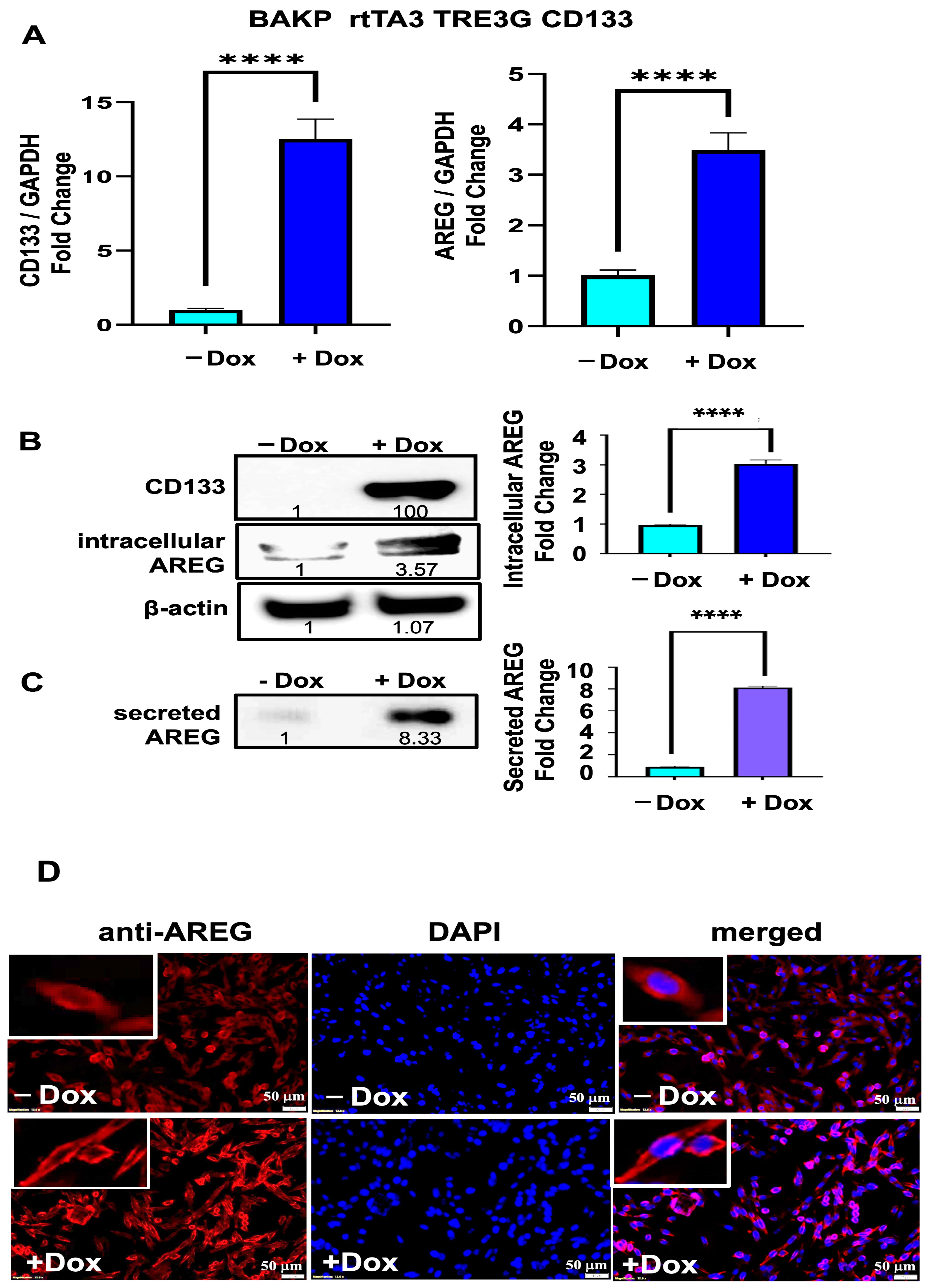
3.4. Both the Intracellular AREG Precursor and Secreted Forms of AREG Are Upregulated in BAKP Melanoma Cells after Dox-Induced CD133 Expression
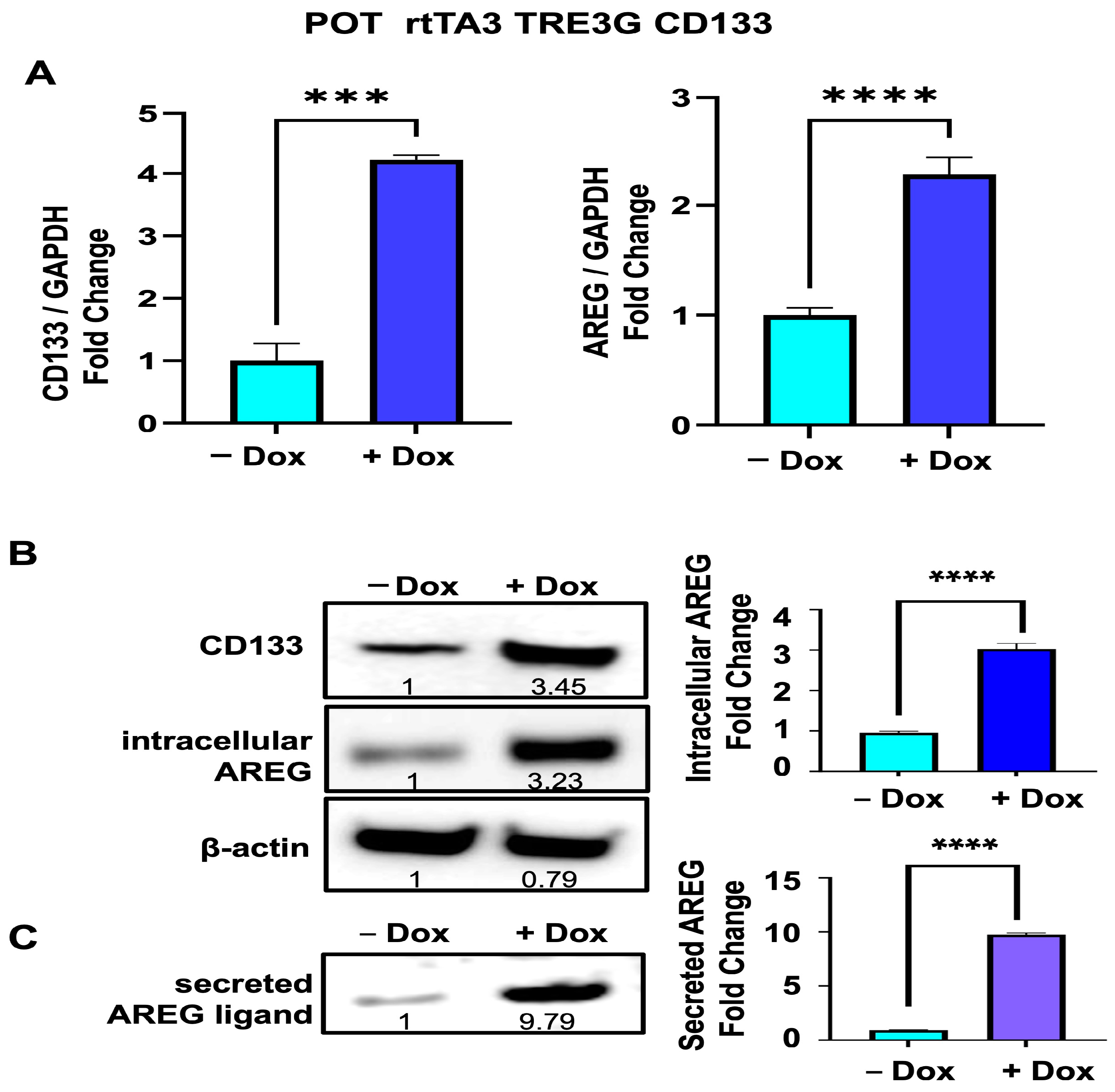
3.5. Upregulation of Intrcellular AREG Precursor and Secreted Forms of AREG Is Reproduced in Another Melanoma Cell Line (POT) after Dox-Inducible Expression of CD133
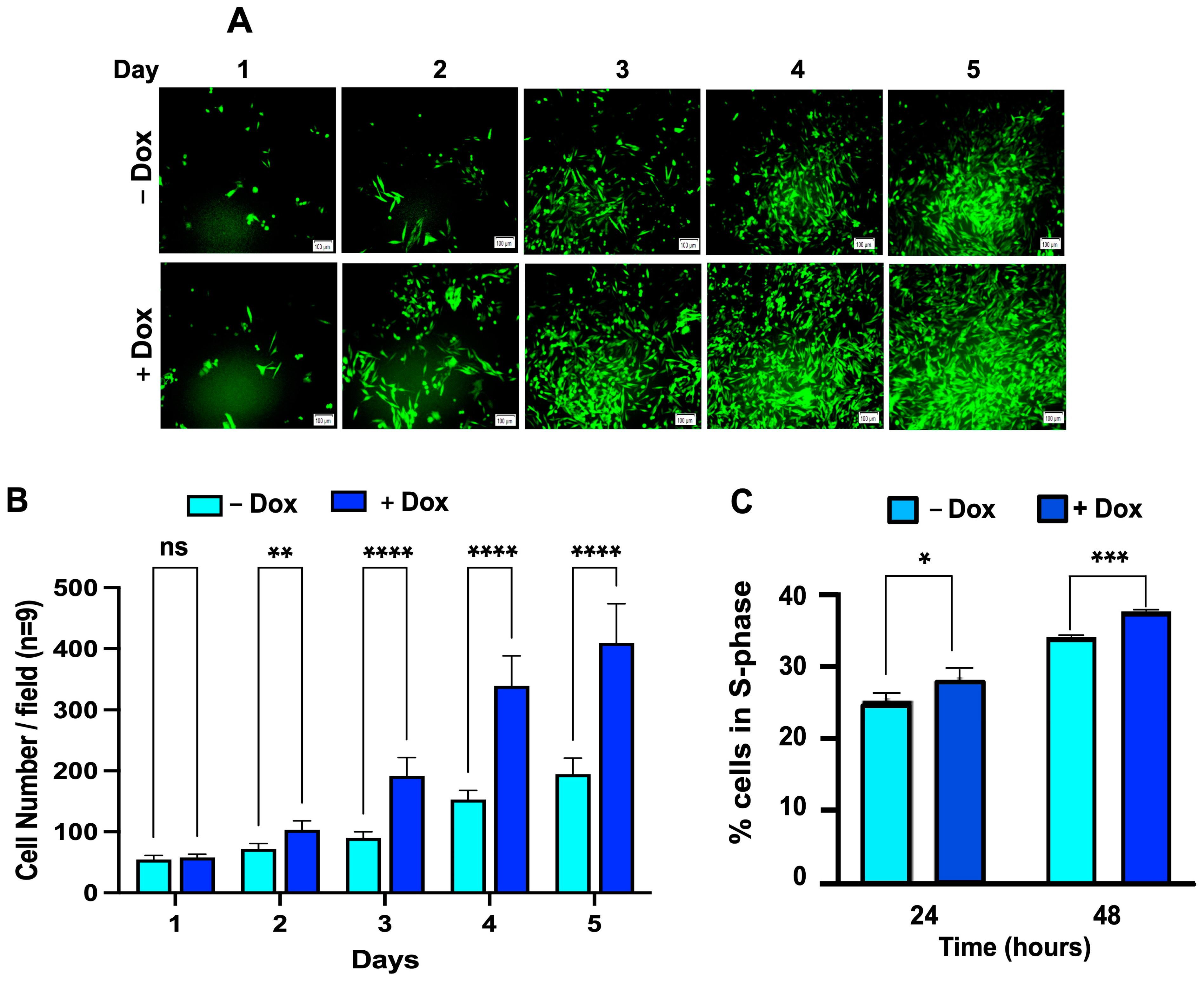
3.6. Dox-Induced CD133 Expression Increases the Percent of Cells in the S-Phase of the Cell Cycle, Leading to Increased Cell Growth in BAKP Melanoma Cells
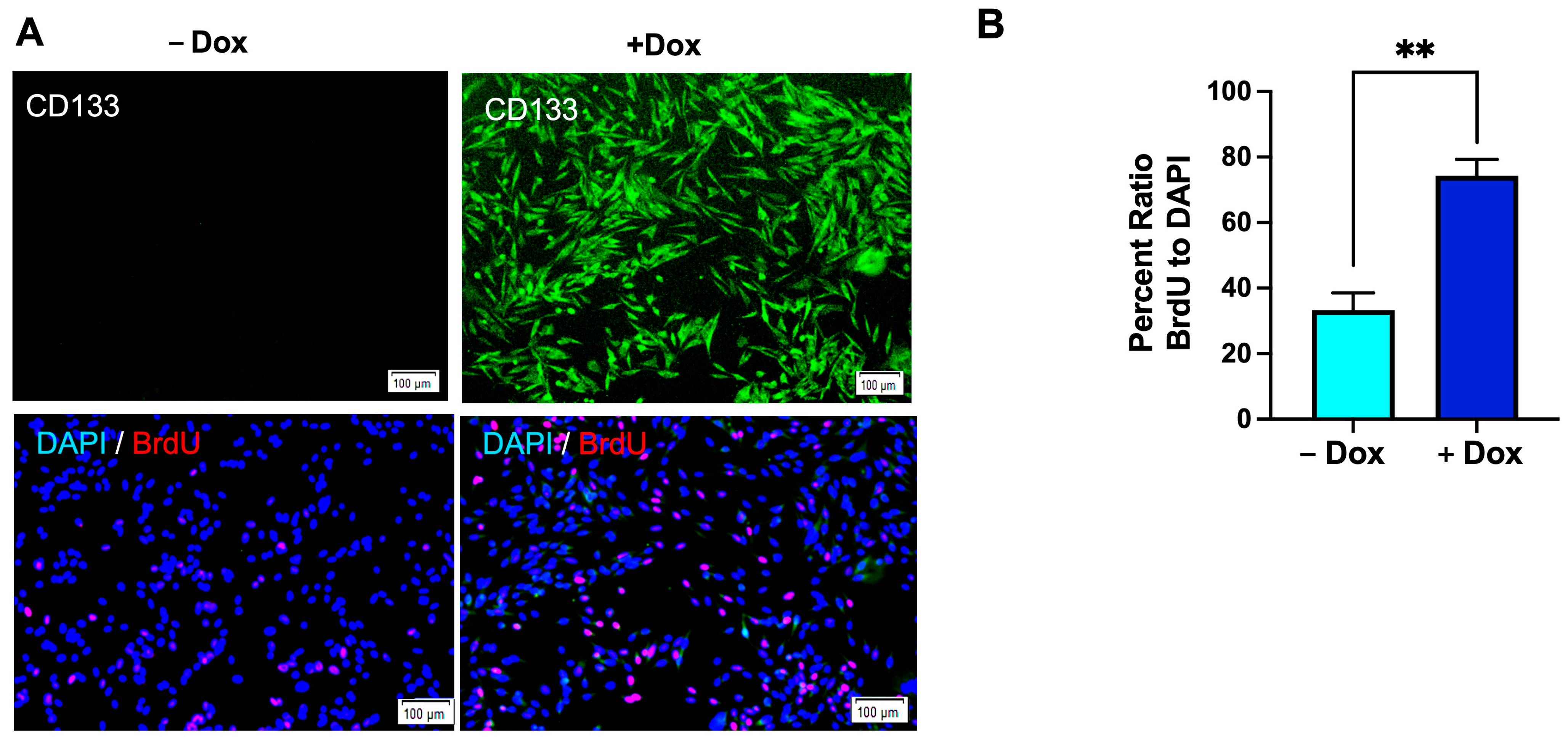
3.7. Enhanced Cell Proliferation in Dox-Induced CD133-Expressing BAKP Cells, as Shown by BrdU Incorporation into Newly Synthesized DNA
3.8. Immunoblot Analysis Reveal Activation of EGFR and the MAPK Pathway in BAKP Cells, as Well as Increased Levels of the Proliferating Cell Nuclear Antigen (PCNA), after Dox-Induction of CD133 Expression
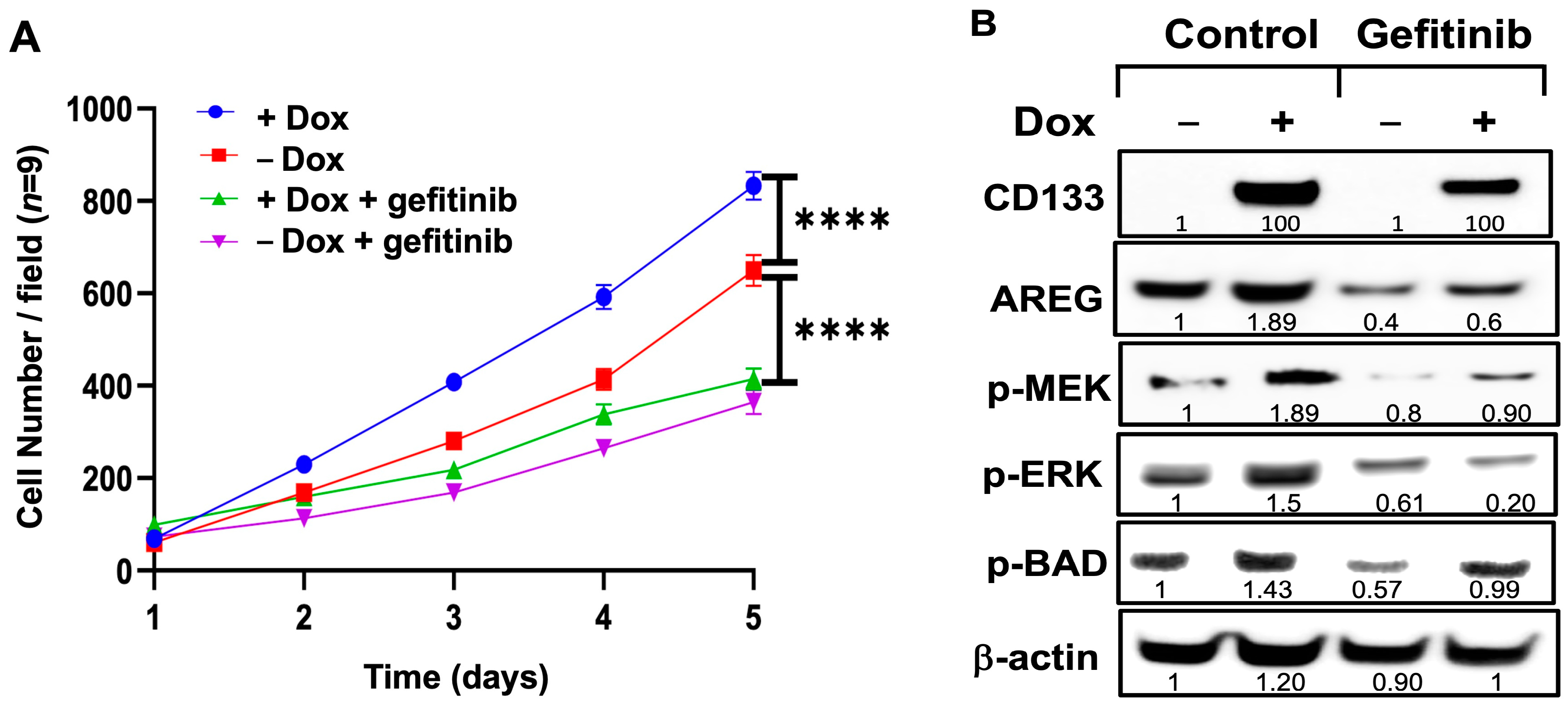
3.9. Exposure of Dox-Induced BAKP Cells to EGFR Inhibitor Gefitinib Reverses CD133-Induced Stimulation of Cell Growth and Activation of the MAPK Pathway
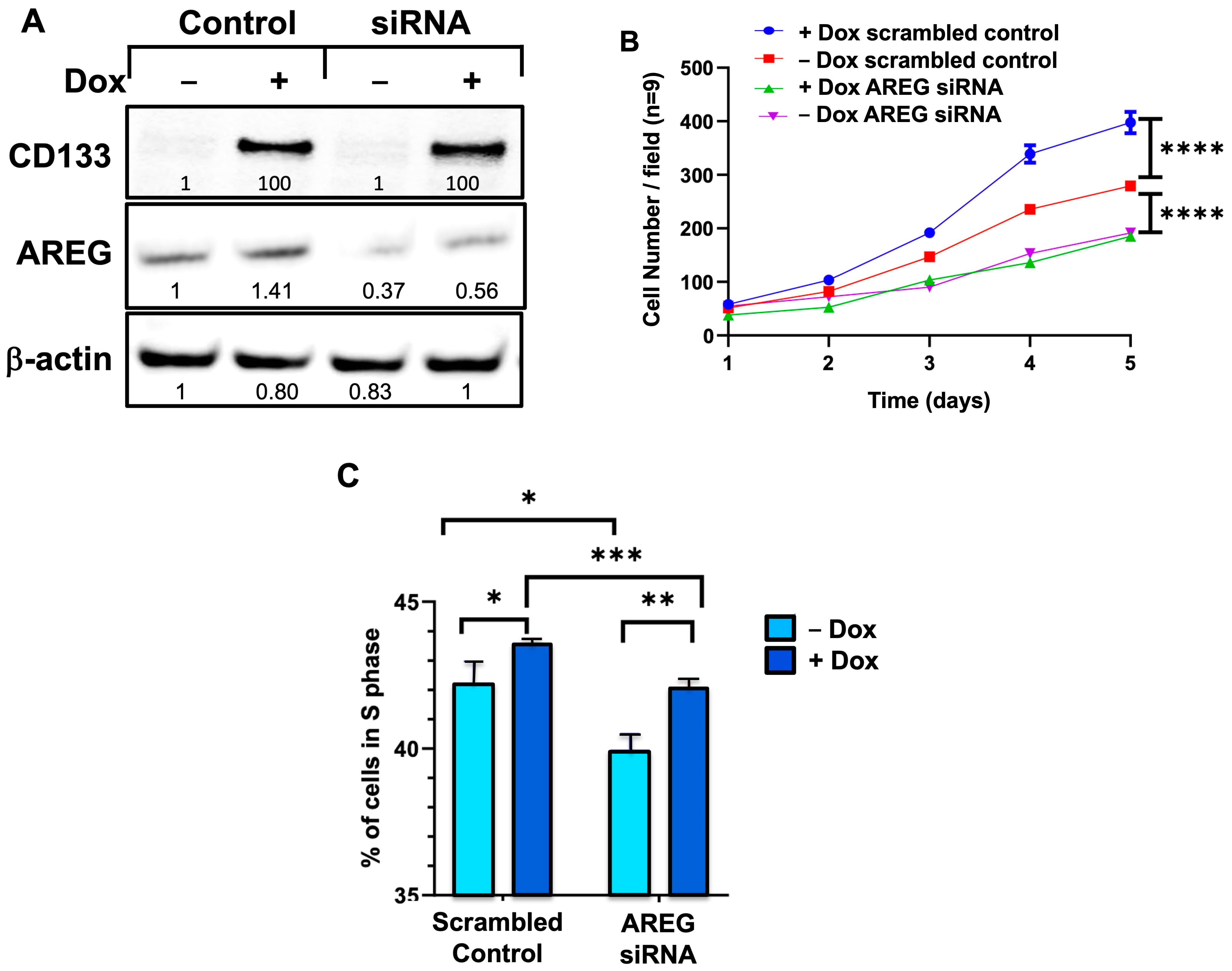
3.10. siRNA Knockdown of AREG Reverses the CD133-Induced Stimulation of Cell Proliferation in BAKP Melanoma Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040. JAMA Dermatol. 2022, 158, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Dahlman, K.B.; Xia, J.; Hutchinson, K.; Ng, C.; Hucks, D.; Jia, P.; Atefi, M.; Su, Z.; Branch, S.; Lyle, P.L.; et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov. 2012, 2, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Padua, R.A.; Barrass, N.; Currie, G.A. A novel transforming gene in a human malignant melanoma cell line. Nature 1984, 311, 671–673. [Google Scholar] [CrossRef] [PubMed]
- Fedorenko, I.V.; Gibney, G.T.; Smalley, K.S. NRAS mutant melanoma: Biological behavior and future strategies for therapeutic management. Oncogene 2013, 32, 3009–3018. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Jakob, J.A.; Bassett, R.L., Jr.; Ng, C.S.; Curry, J.L.; Joseph, R.W.; Alvarado, G.C.; Rohlfs, M.L.; Richard, J.; Gershenwald, J.E.; Kim, K.B.; et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer 2012, 118, 4014–4023. [Google Scholar] [CrossRef] [PubMed]
- Christofi, T.; Baritaki, S.; Falzone, L.; Libra, M.; Zaravinos, A. Current Perspectives in Cancer Immunotherapy. Cancers 2019, 11, 1472. [Google Scholar] [CrossRef] [PubMed]
- Li, Z. CD133: A stem cell biomarker and beyond. Experimental. Hematol. Oncol. 2013, 2, 17. [Google Scholar] [CrossRef]
- Brescia, P.; Ortensi, B.; Fornasari, L.; Levi, D.; Broggi, G.; Pelicci, G. CD133 is essential for glioblastoma stem cell maintenance. Stem Cells 2013, 31, 857–869. [Google Scholar] [CrossRef]
- Ikram, D.; Masadah, R.; Nelwan, B.J.; Zainuddin, A.A.; Ghaznawie, M.; Wahid, S. CD133 Act as an Essential Marker in Ovarian Carcinogenesis. Asian Pac. J. Cancer Prev. 2021, 22, 3525–3531. [Google Scholar] [CrossRef] [PubMed]
- Ma, S. Biology and clinical implications of CD133(+) liver cancer stem cells. Exp. Cell Res. 2013, 319, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Aljitawi, O.; Van Veldhuizen, P. Prostate Cancer Stem Cells: The Role of CD133. Cancers 2022, 14, 5448. [Google Scholar] [CrossRef] [PubMed]
- Nomura, A.; Banerjee, S.; Chugh, R.; Dudeja, V.; Yamamoto, M.; Vickers, S.M.; Saluja, A.K. CD133 initiates tumors, induces epithelial-mesenchymal transition and increases metastasis in pancreatic cancer. Oncotarget 2015, 6, 8313–8322. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Sheng, W.Q.; Du, X. CD133: A cancer stem cells marker, is used in colorectal cancers. World J. Gastroenterol. 2013, 19, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.K.; Manglik, V.; O’Connell, M.; Weeraratna, A.; McCarron, E.C.; Broussard, J.N.; Divito, K.A.; Simbulan-Rosenthal, C.M.; Rosenthal, D.S.; Zapas, J.L. Clonal dominance of CD133+ subset population as risk factor in tumor progression and disease recurrence of human cutaneous melanoma. Int. J. Oncol. 2012, 41, 1570–1576. [Google Scholar] [CrossRef] [PubMed]
- Simbulan-Rosenthal, C.M.; Dougherty, R.; Vakili, S.; Ferraro, A.M.; Kuo, L.W.; Alobaidi, R.; Aljehane, L.; Gaur, A.; Sykora, P.; Glasgow, E.; et al. CRISPR-Cas9 Knockdown and Induced Expression of CD133 Reveal Essential Roles in Melanoma Invasion and Metastasis. Cancers 2019, 11, 1490. [Google Scholar] [CrossRef] [PubMed]
- Simbulan-Rosenthal, C.M.; Gaur, A.; Zhou, H.; AbdusSamad, M.; Qin, Q.; Dougherty, R.; Aljehane, L.; Kuo, L.W.; Vakili, S.; Karna, K.; et al. CD133 Is Associated with Increased Melanoma Cell Survival after Multikinase Inhibition. J. Oncol. 2019, 2019, 6486173. [Google Scholar] [CrossRef] [PubMed]
- Simbulan-Rosenthal, C.M.; Haribabu, Y.; Vakili, S.; Kuo, L.W.; Clark, H.; Dougherty, R.; Alobaidi, R.; Carney, B.; Sykora, P.; Rosenthal, D.S. Employing CRISPR-Cas9 to Generate CD133 Synthetic Lethal Melanoma Stem Cells. Int. J. Mol. Sci. 2022, 23, 2333. [Google Scholar] [CrossRef]
- Das, A.T.; Tenenbaum, L.; Berkhout, B. Tet-On Systems For Doxycycline-inducible Gene Expression. Curr. Gene Ther. 2016, 16, 156–167. [Google Scholar] [CrossRef]
- Das, A.T.; Zhou, X.; Metz, S.W.; Vink, M.A.; Berkhout, B. Selecting the optimal Tet-On system for doxycycline-inducible gene expression in transiently transfected and stably transduced mammalian cells. Biotechnol. J. 2016, 11, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Cawthorne, C.; Swindell, R.; Stratford, I.J.; Dive, C.; Welman, A. Comparison of doxycycline delivery methods for Tet-inducible gene expression in a subcutaneous xenograft model. J. Biomol. Tech. 2007, 18, 120–123. [Google Scholar] [PubMed]
- Simbulan-Rosenthal, C.M.; Rosenthal, D.S.; Hilz, H.; Hickey, R.; Malkas, L.; Applegren, N.; Wu, Y.; Bers, G.; Smulson, M.E. The expression of poly(ADP-ribose) polymerase during differentiation-linked DNA replication reveals that it is a component of the multiprotein DNA replication complex. Biochemistry 1996, 35, 11622–11633. [Google Scholar] [CrossRef] [PubMed]
- Hung, Y.H.; Hou, Y.C.; Hsu, S.H.; Wang, L.Y.; Tsai, Y.L.; Shan, Y.S.; Su, Y.Y.; Hung, W.C.; Chen, L.T. Pancreatic cancer cell-derived semaphorin 3A promotes neuron recruitment to accelerate tumor growth and dissemination. Am. J. Cancer Res. 2023, 13, 3417–3432. [Google Scholar] [PubMed]
- Yang, H.; Zhou, Y.; Wang, L.; Lv, M.; Sun, J.; Luo, Z.; He, J. Sema3A alleviates the malignant behaviors of gastric cancer cells by inhibiting NRP-1. Curr. Mol. Med. 2023, 24, 931–939. [Google Scholar] [CrossRef]
- Shubhra, Q.T.H. RARRES2’s impact on lipid metabolism in triple-negative breast cancer: A pathway to brain metastasis. Mil. Med. Res. 2023, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Bell, E.H.; Mischel, P.; Chakravarti, A. Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr. Pharm. Des. 2014, 20, 2619–2626. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Yu, X.; Peng, C.; Liu, N.; Chen, W.; Xu, H.; Wei, H.; Fang, K.; Dong, Z.; Fu, C.; et al. Activation of SREBP-1c alters lipogenesis and promotes tumor growth and metastasis in gastric cancer. Biomed. Pharmacother. 2020, 128, 110274. [Google Scholar] [CrossRef] [PubMed]
- Han, S.Y.; Han, H.B.; Tian, X.Y.; Sun, H.; Xue, D.; Zhao, C.; Jiang, S.T.; He, X.R.; Zheng, W.X.; Wang, J.; et al. MicroRNA-33a-3p suppresses cell migration and invasion by directly targeting PBX3 in human hepatocellular carcinoma. Oncotarget 2016, 7, 42461–42473. [Google Scholar] [CrossRef]
- Berasain, C.; Avila, M.A. Amphiregulin. Semin. Cell Dev. Biol. 2014, 28, 31–41. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef] [PubMed]
- Castillo, J.; Erroba, E.; Perugorria, M.J.; Santamaria, M.; Lee, D.C.; Prieto, J.; Avila, M.A.; Berasain, C. Amphiregulin contributes to the transformed phenotype of human hepatocellular carcinoma cells. Cancer Res. 2006, 66, 6129–6138. [Google Scholar] [CrossRef] [PubMed]
- Kelman, Z. PCNA: Structure, functions and interactions. Oncogene 1997, 14, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Essers, J.; Theil, A.F.; Baldeyron, C.; van Cappellen, W.A.; Houtsmuller, A.B.; Kanaar, R.; Vermeulen, W. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell. Biol. 2005, 25, 9350–9359. [Google Scholar] [CrossRef] [PubMed]
- Vecchiato, A.; Rossi, C.R.; Montesco, M.C.; Frizzera, E.; Seno, A.; Piccoli, A.; Martello, T.; Ninfo, V.; Lise, M. Proliferating cell nuclear antigen (PCNA) and recurrence in patients with cutaneous melanoma. Melanoma Res. 1994, 4, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Heck, M.M.; Earnshaw, W.C. Topoisomerase II: A specific marker for cell proliferation. J. Cell Biol. 1986, 103, 2569–2581. [Google Scholar] [CrossRef] [PubMed]
- Riverso, M.; Montagnani, V.; Stecca, B. KLF4 is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes melanoma cell growth. Oncogene 2017, 36, 3322–3333. [Google Scholar] [CrossRef] [PubMed]
- Ishida, S.; Huang, E.; Zuzan, H.; Spang, R.; Leone, G.; West, M.; Nevins, J.R. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 2001, 21, 4684–4699. [Google Scholar] [CrossRef]
- Tsoi, J.; Robert, L.; Paraiso, K.; Galvan, C.; Sheu, K.M.; Lay, J.; Wong, D.J.L.; Atefi, M.; Shirazi, R.; Wang, X.; et al. Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell 2018, 33, 890–904.e895. [Google Scholar] [CrossRef]
- Simbulan-Rosenthal, C.M.; Islam, N.; Alobaidi, R.; Martin, A.; Moreno, A.; Sykora, P.; Rosenthal, D.S. Roles of AKT1, 2, and 3 in Melanoma Survial; Department of Biochemstry and Molecular & Cellular Biology, Georgetown University: Washington, DC, USA, 2024; manuscript in preparation. [Google Scholar]
- Dong, J.; Opresko, L.K.; Dempsey, P.J.; Lauffenburger, D.A.; Coffey, R.J.; Wiley, H.S. Metalloprotease-mediated ligand release regulates autocrine signaling through the epidermal growth factor receptor. Proc. Natl. Acad. Sci. USA 1999, 96, 6235–6240. [Google Scholar] [CrossRef]
- Gschwind, A.; Hart, S.; Fischer, O.M.; Ullrich, A. TACE cleavage of proamphiregulin regulates GPCR-induced proliferation and motility of cancer cells. EMBO J. 2003, 22, 2411–2421. [Google Scholar] [CrossRef]
- Busser, B.; Sancey, L.; Brambilla, E.; Coll, J.L.; Hurbin, A. The multiple roles of amphiregulin in human cancer. Biochim. Biophys. Acta 2011, 1816, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Baillo, A.; Giroux, C.; Ethier, S.P. Knock-down of amphiregulin inhibits cellular invasion in inflammatory breast cancer. J. Cell. Physiol. 2011, 226, 2691–2701. [Google Scholar] [CrossRef]
- Willmarth, N.E.; Baillo, A.; Dziubinski, M.L.; Wilson, K.; Riese, D.J., 2nd; Ethier, S.P. Altered EGFR localization and degradation in human breast cancer cells with an amphiregulin/EGFR autocrine loop. Cell Signal. 2009, 21, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Willmarth, N.E.; Ethier, S.P. Autocrine and juxtacrine effects of amphiregulin on the proliferative, invasive, and migratory properties of normal and neoplastic human mammary epithelial cells. J. Biol. Chem. 2006, 281, 37728–37737. [Google Scholar] [CrossRef]
- Damstrup, L.; Kuwada, S.K.; Dempsey, P.J.; Brown, C.L.; Hawkey, C.J.; Poulsen, H.S.; Wiley, H.S.; Coffey, R.J. Amphiregulin acts as an autocrine growth factor in two human polarizing colon cancer lines that exhibit domain selective EGF receptor mitogenesis. Br. J. Cancer 1999, 80, 1012–1019. [Google Scholar] [CrossRef]
- Ruck, A.; Paulie, S. EGF, TGF alpha, AR and HB-EGF are autocrine growth factors for human bladder carcinoma cell lines. Anticancer Res. 1998, 18, 1447–1452. [Google Scholar] [PubMed]
- Ichikawa, W.; Terashima, M.; Ochiai, A.; Kitada, K.; Kurahashi, I.; Sakuramoto, S.; Katai, H.; Sano, T.; Imamura, H.; Sasako, M. Impact of insulin-like growth factor-1 receptor and amphiregulin expression on survival in patients with stage II/III gastric cancer enrolled in the Adjuvant Chemotherapy Trial of S-1 for Gastric Cancer. Gastric. Cancer 2017, 20, 263–273. [Google Scholar] [CrossRef]
- Jing, C.; Jin, Y.H.; You, Z.; Qiong, Q.; Jun, Z. Prognostic value of amphiregulin and epiregulin mRNA expression in metastatic colorectal cancer patients. Oncotarget 2016, 7, 55890–55899. [Google Scholar] [CrossRef]
- Haghgoo, S.M.; Khosravi, A.; Mortaz, E.; Pourabdollah-Toutkaboni, M.; Seifi, S.; Sabour, S.; Allameh, A. Prognostic value of rare and complex mutations in EGFR and serum levels of soluble EGFR and its ligands in non-small cell lung carcinoma patients. Clin. Biochem. 2017, 50, 293–300. [Google Scholar] [CrossRef]
- Kim, J.W.; Kim, D.K.; Min, A.; Lee, K.H.; Nam, H.J.; Kim, J.H.; Kim, J.S.; Kim, T.Y.; Im, S.A.; Park, I.A. Amphiregulin confers trastuzumab resistance via AKT and ERK activation in HER2-positive breast cancer. J. Cancer Res. Clin. Oncol. 2016, 142, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Seefried, F.; Haller, L.; Fukuda, S.; Thongmao, A.; Schneider, N.; Utikal, J.; Higashiyama, S.; Bosserhoff, A.K.; Kuphal, S. Nuclear AREG affects a low-proliferative phenotype and contributes to drug resistance of melanoma. Int. J. Cancer 2022, 151, 2244–2264. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.J.; Pan, W.W.; Liu, S.B.; Shen, Z.F.; Xu, Y.; Hu, L.L. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020, 19, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, S.; Lindzen, M.; Lauriola, M.; Shirazi, N.; Sinha, S.; Abdul-Hai, A.; Levanon, K.; Korach, J.; Barshack, I.; Cohen, Y.; et al. An antibody to amphiregulin, an abundant growth factor in patients’ fluids, inhibits ovarian tumors. Oncogene 2016, 35, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.R.; Saeki, T.; Gordon, A.W.; Shoyab, M.; Salomon, D.S.; Stromberg, K. Autocrine action of amphiregulin in a colon carcinoma cell line and immunocytochemical localization of amphiregulin in human colon. J. Cell Biol. 1992, 118, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Reiss, L.K.; Fragoulis, A.; Siegl, S.; Platen, C.; Kan, Y.W.; Nautiyal, J.; Parker, M.; Pufe, T.; Uhlig, U.; Martin, C.; et al. Interplay between nuclear factor erythroid 2-related factor 2 and amphiregulin during mechanical ventilation. Am. J. Respir Cell Mol. Biol. 2014, 51, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Panupinthu, N.; Yu, S.; Zhang, D.; Zhang, F.; Gagea, M.; Lu, Y.; Grandis, J.R.; Dunn, S.E.; Lee, H.Y.; Mills, G.B. Self-reinforcing loop of amphiregulin and Y-box binding protein-1 contributes to poor outcomes in ovarian cancer. Oncogene 2014, 33, 2846–2856. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, Y.; Anderson, E.M.; Birmingham, A.; Reynolds, A.; Karpilow, J.; Robinson, K.; Leake, D.; Marshall, W.S.; Khvorova, A. Off-target effects by siRNA can induce toxic phenotype. RNA 2006, 12, 1188–1196. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.; Thoma, C.; Goodall, R.J.; Lyons, T.J.; Gaitskell, K.; Wiggans, A.J.; Bryant, A. Epidermal growth factor receptor blockers for the treatment of ovarian cancer. Cochrane Database Syst. Rev. 2018, 10, CD007927. [Google Scholar] [CrossRef]
- Richards, F.M.; Tape, C.J.; Jodrell, D.I.; Murphy, G. Anti-tumour effects of a specific anti-ADAM17 antibody in an ovarian cancer model in vivo. PLoS ONE 2012, 7, e40597. [Google Scholar] [CrossRef]
- Sobocan, M.; Bracic, S.; Knez, J.; Takac, I.; Haybaeck, J. The Communication Between the PI3K/AKT/mTOR Pathway and Y-box Binding Protein-1 in Gynecological Cancer. Cancers 2020, 12, 205. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simbulan-Rosenthal, C.M.; Islam, N.; Haribabu, Y.; Alobaidi, R.; Shalamzari, A.; Graham, G.; Kuo, L.-W.; Sykora, P.; Rosenthal, D.S. CD133 Stimulates Cell Proliferation via the Upregulation of Amphiregulin in Melanoma. Cells 2024, 13, 777. https://doi.org/10.3390/cells13090777
Simbulan-Rosenthal CM, Islam N, Haribabu Y, Alobaidi R, Shalamzari A, Graham G, Kuo L-W, Sykora P, Rosenthal DS. CD133 Stimulates Cell Proliferation via the Upregulation of Amphiregulin in Melanoma. Cells. 2024; 13(9):777. https://doi.org/10.3390/cells13090777
Chicago/Turabian StyleSimbulan-Rosenthal, Cynthia M, Nusrat Islam, Yogameenakshi Haribabu, Ryyan Alobaidi, Azadeh Shalamzari, Garrett Graham, Li-Wei Kuo, Peter Sykora, and Dean S Rosenthal. 2024. "CD133 Stimulates Cell Proliferation via the Upregulation of Amphiregulin in Melanoma" Cells 13, no. 9: 777. https://doi.org/10.3390/cells13090777
APA StyleSimbulan-Rosenthal, C. M., Islam, N., Haribabu, Y., Alobaidi, R., Shalamzari, A., Graham, G., Kuo, L.-W., Sykora, P., & Rosenthal, D. S. (2024). CD133 Stimulates Cell Proliferation via the Upregulation of Amphiregulin in Melanoma. Cells, 13(9), 777. https://doi.org/10.3390/cells13090777






