Exosomal microRNA/miRNA Dysregulation in Respiratory Diseases: From Mycoplasma-Induced Respiratory Disease to COVID-19 and Beyond
Abstract
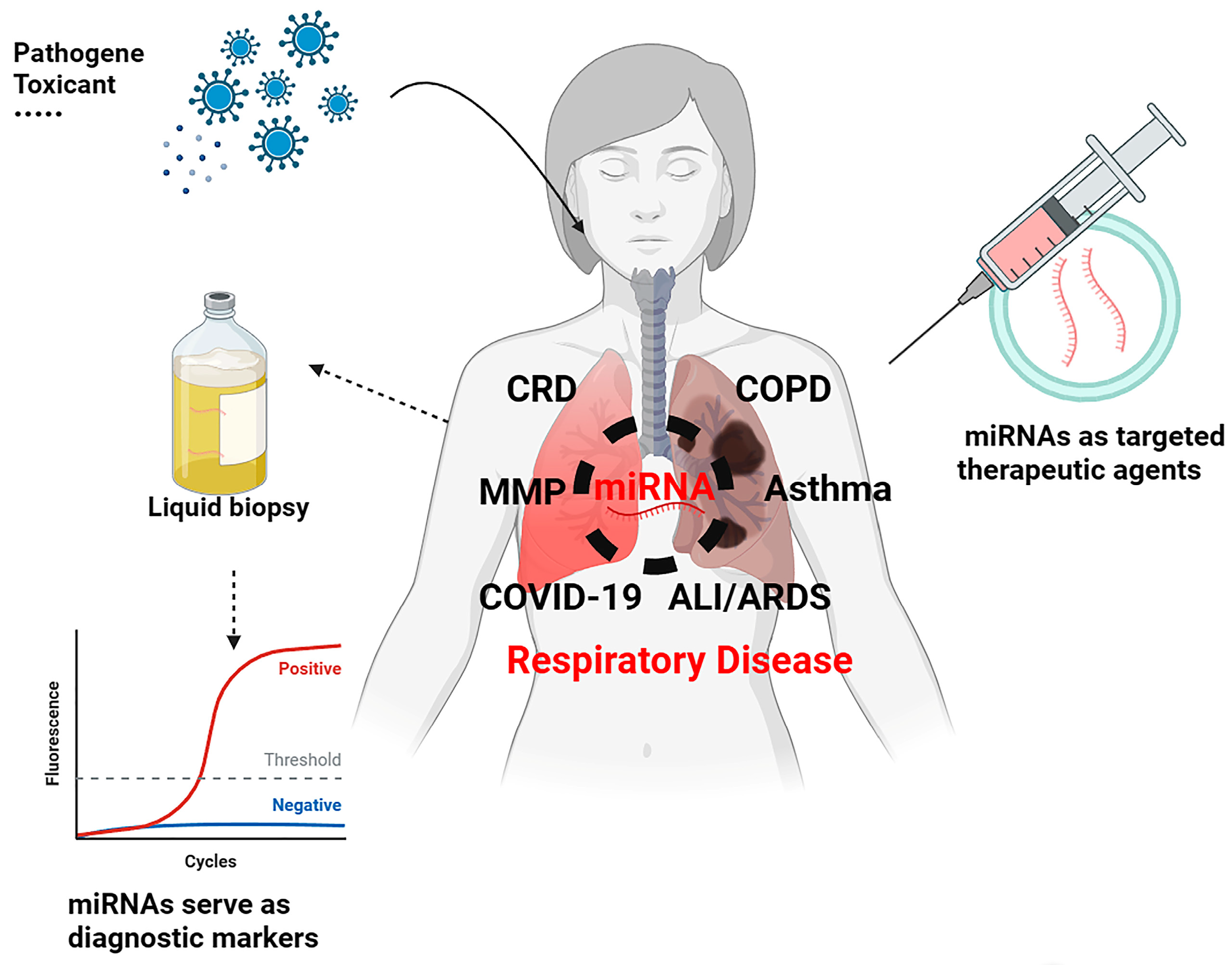
:1. Introduction
2. Pathophysiology Roles of miRNA in Mycoplasma gallisepticum-Induced Respiratory Diseases
2.1. miRNAs in CRD
2.2. Exosomal miRNAs
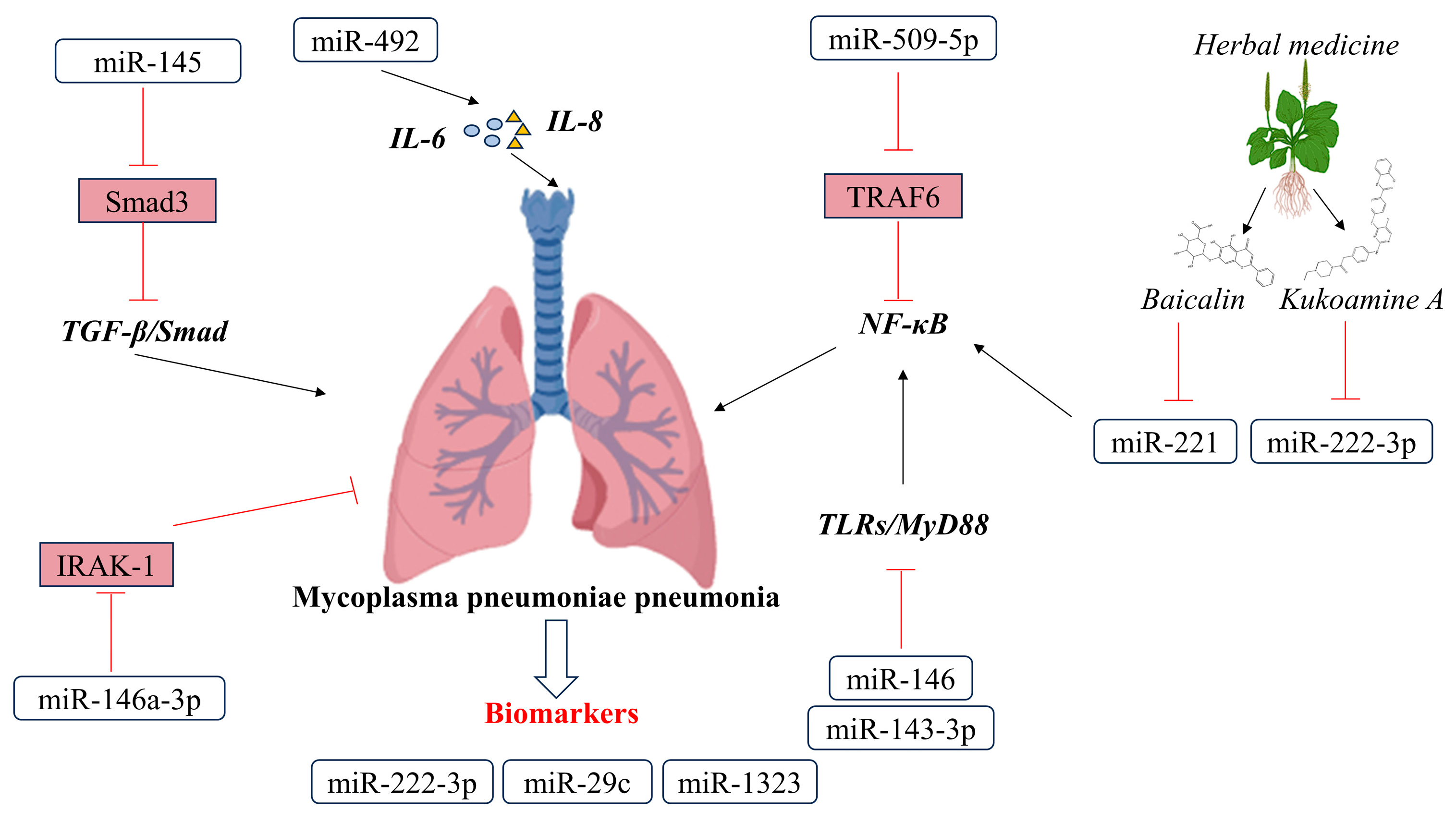
3. Pathophysiology Roles of miRNA in Mycoplasma pneumoniae Pneumonia
3.1. miRNAs in MMP
3.2. Possibilities of miRNA in the Treatment of Mycoplasma pneumoniae Pneumonia
3.3. Possibilities of miRNA in the Diagnosis of Mycoplasma pneumoniae Pneumonia
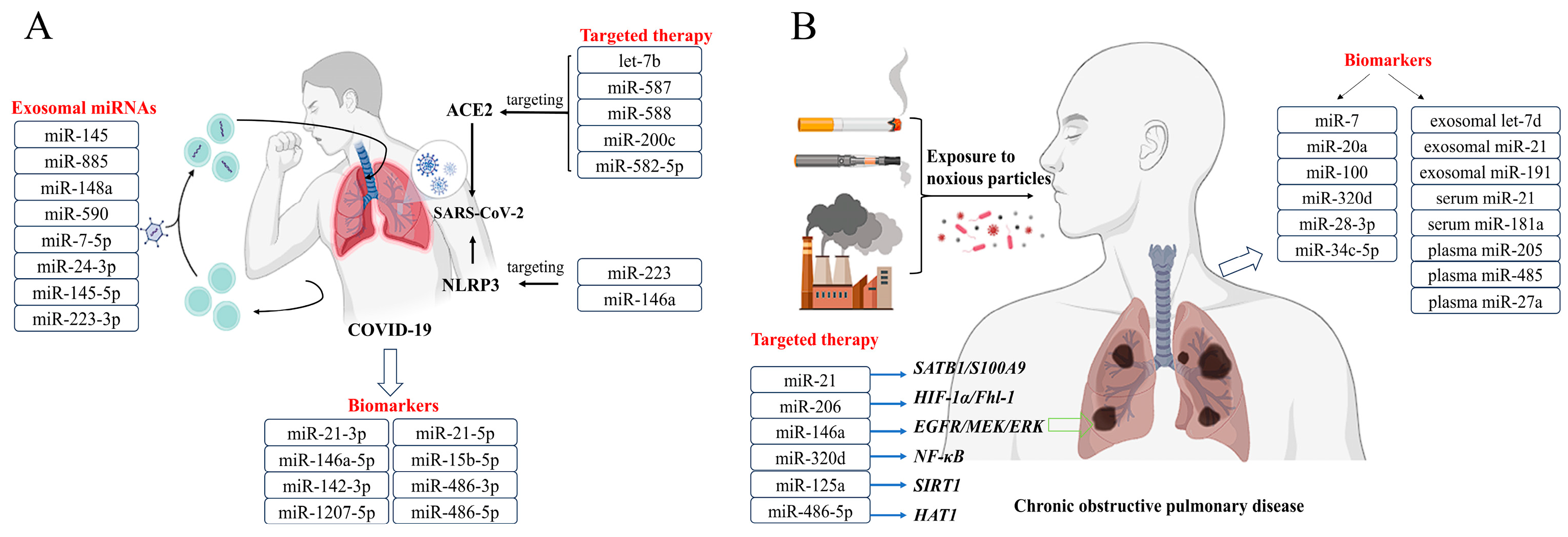
4. Pathophysiology Roles of miRNAs in COVID-19
4.1. miRNAs in COVID-19
4.2. Possibilities of miRNA in the Diagnosis of COVID-19
4.3. Possibilities of miRNA in the Treatment of COVID-19
5. Pathophysiology Roles of miRNAs in COPD
5.1. miRNAs in COPD
5.2. Possibilities of miRNA in the Diagnosis of COPD
5.3. Possibilities of miRNA in the Treatment of COPD
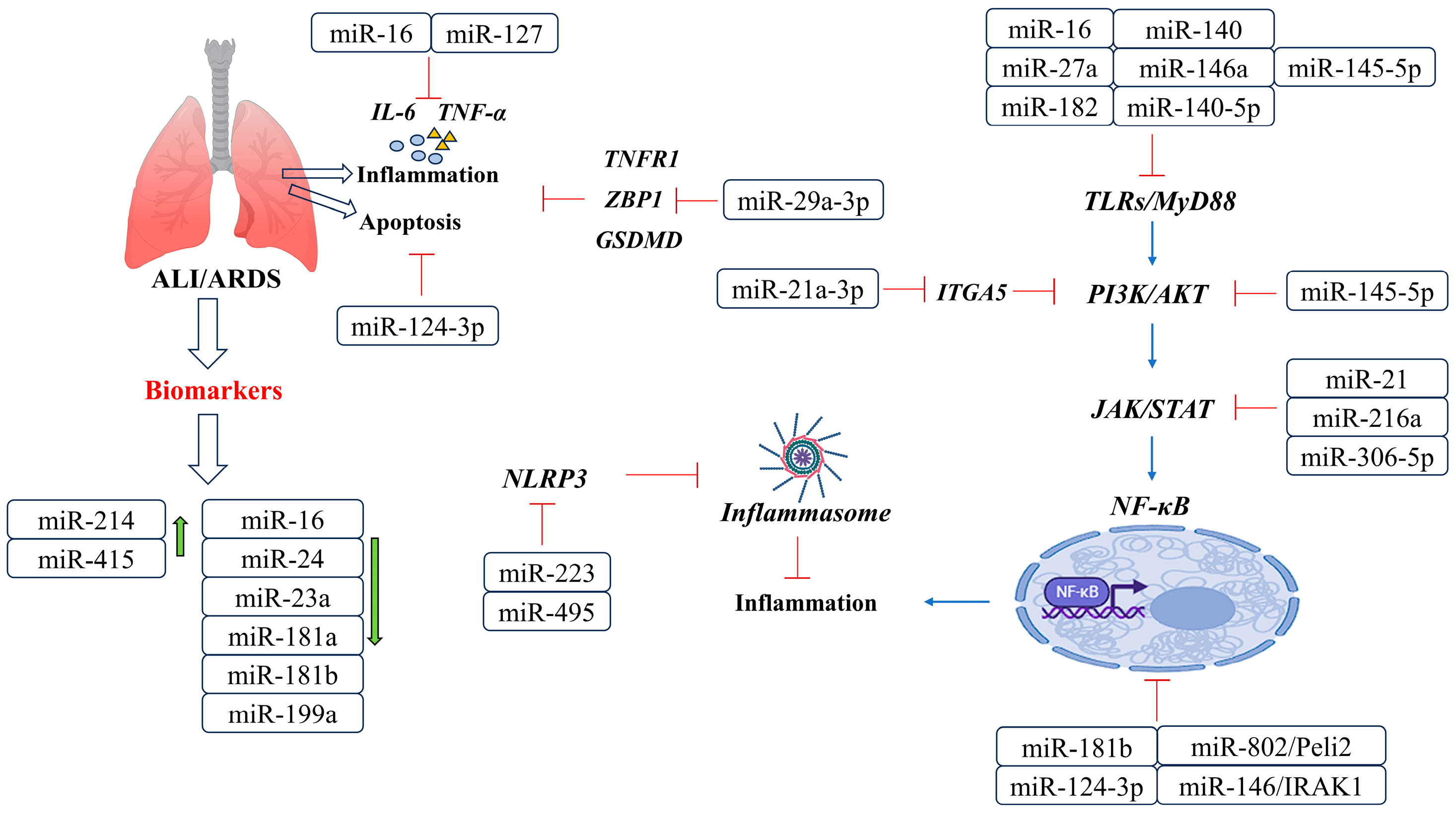
6. Pathophysiology Roles of miRNAs in ALI/ARDS
miRNAs in ALI/ARDS
7. Pathophysiology Roles of miRNAs in Asthma
7.1. miRNAs in Asthma
7.2. Possibilities of miRNA in the Treatment of Asthma
7.3. Possibilities of miRNA in the Diagnosis of Asthma
8. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. TIG 2022, 38, 613–626. [Google Scholar] [CrossRef]
- Chatterjee, B.; Sarkar, M.; Bose, S.; Alam, M.T.; Chaudhary, A.A.; Dixit, A.K.; Tripathi, P.P.; Srivastava, A.K. MicroRNAs: Key modulators of inflammation-associated diseases. Semin. Cell Dev. Biol. 2023. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [PubMed]
- Sun, Z.; Shi, K.; Yang, S.; Liu, J.; Zhou, Q.; Wang, G.; Song, J.; Li, Z.; Zhang, Z.; Yuan, W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol. Cancer 2018, 17, 147. [Google Scholar] [PubMed]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [PubMed]
- Sanz-Rubio, D.; Martin-Burriel, I.; Gil, A.; Cubero, P.; Forner, M.; Khalyfa, A.; Marin, J.M. Stability of Circulating Exosomal miRNAs in Healthy Subjects. Sci. Rep. 2018, 8, 10306. [Google Scholar] [CrossRef]
- Viegi, G.; Maio, S.; Fasola, S.; Baldacci, S. Global Burden of Chronic Respiratory Diseases. J. Aerosol Med. Pulm. Drug Deliv. 2020, 33, 171–177. [Google Scholar] [CrossRef]
- Kleven, S.H. Control of avian mycoplasma infections in commercial poultry. Avian Dis. 2008, 52, 367–374. [Google Scholar] [CrossRef]
- Zhao, Y.; Fu, Y.; Zou, M.; Sun, Y.; Yin, X.; Niu, L.; Gong, Y.; Peng, X. Analysis of deep sequencing exosome-microRNA expression profile derived from CP-II reveals potential role of gga-miRNA-451 in inflammation. J. Cell. Mol. Med. 2020, 24, 6178–6190. [Google Scholar] [CrossRef]
- Zhao, Y.; Hou, Y.; Zhang, K.; Yuan, B.; Peng, X. Identification of differentially expressed miRNAs through high-throughput sequencing in the chicken lung in response to Mycoplasma gallisepticum HS. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 22, 146–156. [Google Scholar] [CrossRef]
- Zhu, M.; Nan, Y.; Zhai, M.; Wang, M.; Shao, Y.; Blair, H.T.; Morris, S.T.; Kenyon, P.R.; Zhao, Z.; Zhang, H. Comparative profiling of the resistance of different genotypes of mannose-binding lectin to Mycoplasma pneumoniae infection in Chinese Merino sheep based on high-throughput sequencing technology. Vet. Immunol. Immunopathol. 2021, 233, 110183. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Pato, A.; Virseda-Berdices, A.; Resino, S.; Ryan, P.; Martínez-González, O.; Pérez-García, F.; Martin-Vicente, M.; Valle-Millares, D.; Brochado-Kith, O.; Blancas, R.; et al. Plasma miRNA profile at COVID-19 onset predicts severity status and mortality. Emerg. Microbes Infect. 2022, 11, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Gao, J.; Sköld, C.M.; Wheelock, Å.M. miRNA-mRNA-protein dysregulated network in COPD in women. Front. Genet. 2022, 13, 1010048. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.; Peffers, M.J.; Ormseth, M.J.; Jurisica, I.; Kapoor, M. The non-coding RNA interactome in joint health and disease. Nat. Rev. Rheumatol. 2021, 17, 692–705. [Google Scholar]
- Kooshkaki, O.; Asghari, A.; Mahdavi, R.; Azarkar, G.; Parsamanesh, N. Potential of MicroRNAs As Biomarkers and Therapeutic Targets in Respiratory Viruses: A Literature Review. DNA Cell Biol. 2022, 41, 544–563. [Google Scholar] [CrossRef]
- Zhao, S.; Wang, H.; Xu, H.; Tan, Y.; Zhang, C.; Zeng, Q.; Liu, L.; Qu, S. Targeting the microRNAs in exosome: A potential therapeutic strategy for alleviation of diabetes-related cardiovascular complication. Pharmacol. Res. 2021, 173, 105868. [Google Scholar] [CrossRef]
- Waites, K.B.; Xiao, L.; Liu, Y.; Balish, M.F.; Atkinson, T.P. Mycoplasma pneumoniae from the Respiratory Tract and Beyond. Clin. Microbiol. Rev. 2017, 30, 747–809. [Google Scholar]
- Wang, Y.; Wang, L.; Luo, R.; Sun, Y.; Zou, M.; Wang, T.; Guo, Q.; Peng, X. Glycyrrhizic Acid against Mycoplasma gallisepticum—Induced Inflammation and Apoptosis Through Suppressing the MAPK Pathway in Chickens. J. Agric. Food Chem. 2022, 70, 1996–2009. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, Y.; Hu, F.; Sun, Y.; Zou, M.; Luo, R.; Peng, X. Chinese herbal formulae defend against Mycoplasma gallisepticum infection. J. Integr. Agric. 2022, 21, 3026–3036. [Google Scholar] [CrossRef]
- Wang, Y.; Han, Y.; Wang, L.; Zou, M.; Sun, Y.; Sun, H.; Guo, Q.; Peng, X. Mycoplasma gallisepticum escapes the host immune response via gga-miR-365-3p/SOCS5/STATs axis. Vet. Res. 2022, 53, 103. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Y.; Zou, M.; Wang, T.; Wang, L.; Peng, X. Lnc90386 Sponges miR-33-5p to Mediate Mycoplasma gallisepticum-Induced Inflammation and Apoptosis in Chickens via the JNK Pathway. Front. Immunol. 2022, 13, 887602. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Z.; Hou, Y.; Zhang, K.; Peng, X. gga-miR-99a targets SMARCA5 to regulate Mycoplasma gallisepticum (HS strain) infection by depressing cell proliferation in chicken. Gene 2017, 627, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Fu, Y.; Sun, Y.; Zou, M.; Peng, X. Transcriptional Regulation of gga-miR-451 by AhR:Arnt in Mycoplasma gallisepticum (HS Strain) Infection. Int. J. Mol. Sci. 2019, 20, 3087. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, Y.; Zhao, Y.; Zou, M.; Peng, X. Exosomal miR-181a-5p reduce Mycoplasma gallisepticum (HS strain) infection in chicken by targeting PPM1B and activating the TLR2-mediated MyD88/NF-κB signaling pathway. Mol. Immunol. 2021, 140, 144–157. [Google Scholar] [CrossRef]
- Zou, M.; Fu, Y.; Zhao, Y.; Sun, Y.; Yin, X.; Peng, X. Mycoplasma gallisepticum induced exosomal gga-miR-193a to disturb cell proliferation, apoptosis, and cytokine production by targeting the KRAS/ERK signaling pathway. Int. Immunopharmacol. 2022, 111, 109090. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Bi, D.; Hou, Y.; Zhao, Y.; Sun, J.; Peng, X. Gga-miR-101-3p Plays a Key Role in Mycoplasma gallisepticum (HS Strain) Infection of Chicken. Int. J. Mol. Sci. 2015, 16, 28669–28682. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, Y.; Zou, M.; Deng, G.; Peng, X. gga-miR-142-3p negatively regulates Mycoplasma gallisepticum (HS strain)-induced inflammatory cytokine production via the NF-κB and MAPK signaling by targeting TAB2. Inflamm. Res. 2021, 70, 1217–1231. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, K.; Zou, M.; Sun, Y.; Peng, X. gga-miR-451 Negatively Regulates Mycoplasma gallisepticum (HS Strain)-Induced Inflammatory Cytokine Production via Targeting YWHAZ. Int. J. Mol. Sci. 2018, 19, 1191. [Google Scholar] [CrossRef]
- Zhao, Y.; Zou, M.; Sun, Y.; Zhang, K.; Peng, X. gga-miR-21 modulates Mycoplasma gallisepticum (HS strain)-Induced inflammation via targeting MAP3K1 and activating MAPKs and NF-κB pathways. Vet. Microbiol. 2019, 237, 108407. [Google Scholar] [CrossRef]
- Zhang, K.; Han, Y.; Wang, Z.; Zhao, Y.; Fu, Y.; Peng, X. gga-miR-146c Activates TLR6/MyD88/NF-κB Pathway through Targeting MMP16 to Prevent Mycoplasma Gallisepticum (HS Strain) Infection in Chickens. Cells 2019, 8, 501. [Google Scholar] [CrossRef]
- Yuan, B.; Zou, M.; Zhao, Y.; Zhang, K.; Sun, Y.; Peng, X. Up-Regulation of miR-130b-3p Activates the PTEN/PI3K/AKT/NF-κB Pathway to Defense against Mycoplasma gallisepticum (HS Strain) Infection of Chicken. Int. J. Mol. Sci. 2018, 19, 2172. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Zhao, Y.; Wang, Z.; Hou, Y.; Bi, D.; Sun, J.; Peng, X. Chicken gga-miR-19a Targets ZMYND11 and Plays an Important Role in Host Defense against Mycoplasma gallisepticum (HS Strain) Infection. Front. Cell. Infect. Microbiol. 2016, 6, 102. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Wang, Y.; Sun, Y.; Han, Y.; Sun, H.; Zou, M.; Luo, R.; Peng, X. Down-regulated gga-miR-223 inhibits proliferation and induces apoptosis of MG-infected DF-1 cells by targeting FOXO3. Microb. Pathog. 2021, 155, 104927. [Google Scholar] [CrossRef]
- Wang, Y.; Tong, D.; Sun, Y.; Sun, H.; Liu, F.; Zou, M.; Luo, R.; Peng, X. DF-1 cells prevent MG-HS infection through gga-miR-24-3p/RAP1B mediated decreased proliferation and increased apoptosis. Res. Vet. Sci. 2021, 141, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Han, Y.; Zhao, Y.; Sun, Y.; Zou, M.; Fu, Y.; Peng, X. Upregulated gga-miR-16-5p Inhibits the Proliferation Cycle and Promotes the Apoptosis of MG-Infected DF-1 Cells by Repressing PIK3R1-Mediated the PI3K/Akt/NF-κB Pathway to Exert Anti-Inflammatory Effect. Int. J. Mol. Sci. 2019, 20, 1036. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, H.; Zhao, W.; Wang, T.; Zou, M.; Han, Y.; Sun, Y.; Peng, X. Low let-7d microRNA levels in chick embryos enhance innate immunity against Mycoplasma gallisepticum by suppressing the mitogen-activated protein kinase pathway. Vet. Res. 2023, 54, 50. [Google Scholar] [CrossRef]
- Pigati, L.; Yaddanapudi, S.C.; Iyengar, R.; Kim, D.J.; Hearn, S.A.; Danforth, D.; Hastings, M.L.; Duelli, D.M. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS ONE 2010, 5, e13515. [Google Scholar]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; Simpson, D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012, 13, 357. [Google Scholar] [CrossRef]
- Mansel, J.K.; Rosenow, E.C., 3rd; Smith, T.F.; Martin, J.W., Jr. Mycoplasma pneumoniae pneumonia. Chest 1989, 95, 639–646. [Google Scholar] [CrossRef]
- Tsai, T.A.; Tsai, C.K.; Kuo, K.C.; Yu, H.R. Rational stepwise approach for Mycoplasma pneumoniae pneumonia in children. J. Microbiol. Immunol. Infect. 2021, 54, 557–565. [Google Scholar] [CrossRef]
- Meyer Sauteur, P.M.; Goetschel, P.; Lautenschlager, S. Mycoplasma pneumoniae and mucositis—Part of the Stevens-Johnson syndrome spectrum. J. Der Dtsch. Dermatol. Ges. J. Ger. Soc. Dermatol. JDDG 2012, 10, 740–746. [Google Scholar]
- Zhu, M.; Cao, S.; Zheng, W.; Zhai, M.; Wang, M.; Blair, H.T.; Morris, S.T.; Zhang, H.; Zhao, Z. miR-509-5p anti-infection response for mycoplasma pneumonia in sheep by targeting NF-κB pathway. Vet. Immunol. Immunopathol. 2021, 238, 110275. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Sun, Q.; Zheng, Y.; Xu, J.; Wang, Y. The immunogenic involvement of miRNA-492 in mycoplasma pneumoniae infection in pediatric patients. J. Pediatr. 2023, 99, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Wu, Y.X.; Sun, J.X.; Wang, F.C.; Cui, Z.Q.; Xu, X.H. The role of miR-145 in promoting the fibrosis of pulmonary fibroblasts. J. Biol. Regul. Homeost. Agents 2019, 33, 1337–1345. [Google Scholar] [PubMed]
- Li, H.N.; Zhao, X.; Zha, Y.J.; Du, F.; Liu, J.; Sun, L. miR-146a-5p suppresses ATP-binding cassette subfamily G member 1 dysregulation in patients with refractory Mycoplasma pneumoniae via interleukin 1 receptor-associated kinase 1 downregulation. Int. J. Mol. Med. 2019, 44, 2003–2014. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Shi, Y.; Wang, S.; Xu, Y.; Li, H.; Liu, D. miR-143-3p impacts on pulmonary inflammatory factors and cell apoptosis in mice with mycoplasmal pneumonia by regulating TLR4/MyD88/NF-κB pathway. Biosci. Rep. 2020, 40, BSR20193419. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.; Wang, J.; Cheng, Q.; Shang, Y.; Wang, G. Baicalin relieves Mycoplasma pneumoniae infection-induced lung injury through regulating microRNA-221 to inhibit the TLR4/NF-κB signaling pathway. Mol. Med. Rep. 2021, 24, 571. [Google Scholar] [CrossRef]
- Chu, C.; Lei, X.; Li, Y.; Luo, Y.; Ding, Y.; Zhou, W.; Ji, W. High expression of miR-222-3p in children with Mycoplasma pneumoniae pneumonia. Ital. J. Pediatr. 2019, 45, 163. [Google Scholar] [CrossRef]
- Liu, X.X.; Wang, M.J.; Kan, Q.N.; Li, C.; Xiao, Z.; Jiang, Y.H.; Li, W.; Li, X.; Jiang, Z.Y. Kukoamine A Improves Mycoplasma pneumoniae Pneumonia by Regulating miR-222-3p/Superoxide Dismutase 2. BioMed Res. Int. 2022, 2022, 2064013. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, X.; Liu, X. Long non-coding RNA GAS5 protects against Mycoplasma pneumoniae pneumonia by regulating the microRNA-222-3p/TIMP3 axis. Mol. Med. Rep. 2021, 23, 380. [Google Scholar] [CrossRef]
- Li, Q.L.; Wu, Y.Y.; Sun, H.M.; Gu, W.J.; Zhang, X.X.; Wang, M.J.; Yan, Y.D.; Hao, C.L.; Ji, W.; Chen, Z.R. The role of miR-29c/B7-H3/Th17 axis in children with Mycoplasma pneumoniae pneumonia. Ital. J. Pediatr. 2019, 45, 61. [Google Scholar] [CrossRef]
- Yin, L.; Ma, Y.; Wang, W.; Zhu, Y. The critical function of miR-1323/Il6 axis in children with Mycoplasma pneumoniae pneumonia. J. Pediatr. 2021, 97, 552–558. [Google Scholar] [CrossRef]
- Ramasamy, S.; Subbian, S. Critical Determinants of Cytokine Storm and Type I Interferon Response in COVID-19 Pathogenesis. Clin. Microbiol. Rev. 2021, 34, e00299-20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Amahong, K.; Sun, X.; Lian, X.; Liu, J.; Sun, H.; Lou, Y.; Zhu, F.; Qiu, Y. The miRNA: A small but powerful RNA for COVID-19. Brief. Bioinform. 2021, 22, 1137–1149. [Google Scholar] [PubMed]
- Bertolazzi, G.; Cipollina, C.; Benos, P.V.; Tumminello, M.; Coronnello, C. miR-1207-5p Can Contribute to Dysregulation of Inflammatory Response in COVID-19 via Targeting SARS-CoV-2 RNA. Front. Cell. Infect. Microbiol. 2020, 10, 586592. [Google Scholar] [CrossRef]
- Nersisyan, S.; Engibaryan, N.; Gorbonos, A.; Kirdey, K.; Makhonin, A.; Tonevitsky, A. Potential role of cellular miRNAs in coronavirus-host interplay. PeerJ 2020, 8, e9994. [Google Scholar] [CrossRef]
- Tang, H.; Gao, Y.; Li, Z.; Miao, Y.; Huang, Z.; Liu, X.; Xie, L.; Li, H.; Wen, W.; Zheng, Y.; et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin. Transl. Med. 2020, 10, e200. [Google Scholar] [CrossRef]
- Jafarinejad-Farsangi, S.; Jazi, M.M.; Rostamzadeh, F.; Hadizadeh, M. High affinity of host human microRNAs to SARS-CoV-2 genome: An in silico analysis. Non-Coding RNA Res. 2020, 5, 222–231. [Google Scholar] [CrossRef]
- Sabbatinelli, J.; Giuliani, A.; Matacchione, G.; Latini, S.; Laprovitera, N.; Pomponio, G.; Ferrarini, A.; Svegliati Baroni, S.; Pavani, M.; Moretti, M.; et al. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech. Ageing Dev. 2021, 193, 111413. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; Seeliger, B.; Derda, A.A.; Xiao, K.; Gietz, A.; Scherf, K.; Sonnenschein, K.; Pink, I.; Hoeper, M.M.; Welte, T.; et al. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur. J. Heart Fail. 2021, 23, 468–475. [Google Scholar] [CrossRef]
- Lu, D.; Chatterjee, S.; Xiao, K.; Riedel, I.; Wang, Y.; Foo, R.; Bär, C.; Thum, T. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J. Mol. Cell. Cardiol. 2020, 148, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Latini, A.; Vancheri, C.; Amati, F.; Morini, E.; Grelli, S.; Matteucci, C.; Petrone, V.; Colona, V.L.; Murdocca, M.; Andreoni, M.; et al. Expression analysis of miRNA hsa-let7b-5p in naso-oropharyngeal swabs of COVID-19 patients supports its role in regulating ACE2 and DPP4 receptors. J. Cell. Mol. Med. 2022, 26, 4940–4948. [Google Scholar] [CrossRef] [PubMed]
- Houshmandfar, S.; Saeedi-Boroujeni, A.; Rashno, M.; Khodadadi, A.; Mahmoudian-Sani, M.R. miRNA-223 as a regulator of inflammation and NLRP3 inflammasome, the main fragments in the puzzle of immunopathogenesis of different inflammatory diseases and COVID-19. Naunyn-Schmiedeb. Arch. Pharmacol. 2021, 394, 2187–2195. [Google Scholar]
- Lu, F.; Chen, H.; Hong, Y.; Lin, Y.; Liu, L.; Wei, N.; Wu, Q.; Liao, S.; Yang, S.; He, J.; et al. A gain-of-function NLRP3 3’-UTR polymorphism causes miR-146a-mediated suppression of NLRP3 expression and confers protection against sepsis progression. Sci. Rep. 2021, 11, 13300. [Google Scholar] [CrossRef]
- Gambardella, J.; Kansakar, U.; Sardu, C.; Messina, V.; Jankauskas, S.S.; Marfella, R.; Maggi, P.; Wang, X.; Mone, P.; Paolisso, G.; et al. Exosomal miR-145 and miR-885 Regulate Thrombosis in COVID-19. J. Pharmacol. Exp. Ther. 2023, 384, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Tiwari, A.; McGeachie, M.J. Recent miRNA Research in Asthma. Curr. Allergy Asthma Rep. 2022, 22, 231–258. [Google Scholar]
- Wang, Y.; Zhu, X.; Jiang, X.M.; Guo, J.; Fu, Z.; Zhou, Z.; Yang, P.; Guo, H.; Guo, X.; Liang, G.; et al. Decreased inhibition of exosomal miRNAs on SARS-CoV-2 replication underlies poor outcomes in elderly people and diabetic patients. Signal Transduct. Target. Ther. 2021, 6, 300. [Google Scholar]
- Kim, W.R.; Park, E.G.; Kang, K.W.; Lee, S.M.; Kim, B.; Kim, H.S. Expression Analyses of MicroRNAs in Hamster Lung Tissues Infected by SARS-CoV-2. Mol. Cells 2020, 43, 953–963. [Google Scholar] [CrossRef]
- Agustí, A.; Vogelmeier, C.; Faner, R. COPD 2020: Changes and challenges. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 319, L879–L883. [Google Scholar] [CrossRef]
- Serban, K.A.; Rezania, S.; Petrusca, D.N.; Poirier, C.; Cao, D.; Justice, M.J.; Patel, M.; Tsvetkova, I.; Kamocki, K.; Mikosz, A.; et al. Structural and functional characterization of endothelial microparticles released by cigarette smoke. Sci. Rep. 2016, 6, 31596. [Google Scholar]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Wu, M.; Lin, H.; Liu, C.; Yang, H.; Zhan, J.; Sun, S. An increased ratio of serum miR-21 to miR-181a levels is associated with the early pathogenic process of chronic obstructive pulmonary disease in asymptomatic heavy smokers. Mol. Biosyst. 2014, 10, 1072–1081. [Google Scholar] [PubMed]
- Butt, Y.; Kurdowska, A.; Allen, T.C. Acute Lung Injury: A Clinical and Molecular Review. Arch. Pathol. Lab. Med. 2016, 140, 345–350. [Google Scholar] [PubMed]
- Cai, Z.G.; Zhang, S.M.; Zhang, Y.; Zhou, Y.Y.; Wu, H.B.; Xu, X.P. MicroRNAs are dynamically regulated and play an important role in LPS-induced lung injury. Can. J. Physiol. Pharmacol. 2012, 90, 37–43. [Google Scholar] [CrossRef]
- Vencken, S.F.; Greene, C.M.; McKiernan, P.J. Non-coding RNA as lung disease biomarkers. Thorax 2015, 70, 501–503. [Google Scholar] [CrossRef]
- Soccio, P.; Moriondo, G.; Lacedonia, D.; Tondo, P.; Quarato, C.M.I.; Foschino Barbaro, M.P.; Scioscia, G. EVs-miRNA: The New Molecular Markers for Chronic Respiratory Diseases. Life 2022, 12, 1544. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, X.; Lin, F.; Li, W.; Zhao, Y.; Zhu, F.; Yang, H.; Rao, M.; Li, Y.; Liang, H.; et al. MiR-29a-3p Improves Acute Lung Injury by Reducing Alveolar Epithelial Cell PANoptosis. Aging Dis. 2022, 13, 899–909. [Google Scholar] [CrossRef]
- Li, W.; Qiu, X.; Liu, J.; Han, Y.; Wei, D.; Ji, G.; Jiang, H. miR-27a protects against acute lung injury in LPS-treated mice by inhibiting NF-κB-mediated inflammatory response. Int. J. Clin. Exp. Pathol. 2018, 11, 2980–2989. [Google Scholar]
- Ju, M.; Liu, B.; He, H.; Gu, Z.; Liu, Y.; Su, Y.; Zhu, D.; Cang, J.; Luo, Z. MicroRNA-27a alleviates LPS-induced acute lung injury in mice via inhibiting inflammation and apoptosis through modulating TLR4/MyD88/NF-κB pathway. Cell Cycle 2018, 17, 2001–2018. [Google Scholar] [CrossRef]
- Faiz, A.; Steiling, K.; Roffel, M.P.; Postma, D.S.; Spira, A.; Lenburg, M.E.; Borggrewe, M.; Eijgenraam, T.R.; Jonker, M.R.; Koppelman, G.H.; et al. Effect of long-term corticosteroid treatment on microRNA and gene-expression profiles in COPD. Eur. Respir. J. 2019, 53, 1801202. [Google Scholar] [CrossRef]
- Xu, H.; Ling, M.; Xue, J.; Dai, X.; Sun, Q.; Chen, C.; Liu, Y.; Zhou, L.; Liu, J.; Luo, F.; et al. Exosomal microRNA-21 derived from bronchial epithelial cells is involved in aberrant epithelium-fibroblast cross-talk in COPD induced by cigarette smoking. Theranostics 2018, 8, 5419–5433. [Google Scholar] [CrossRef] [PubMed]
- Akbas, F.; Coskunpinar, E.; Aynaci, E.; Oltulu, Y.M.; Yildiz, P. Analysis of serum micro-RNAs as potential biomarker in chronic obstructive pulmonary disease. Exp. Lung Res. 2012, 38, 286–294. [Google Scholar] [CrossRef] [PubMed]
- O’Farrell, H.E.; Bowman, R.V.; Fong, K.M.; Yang, I.A. Plasma Extracellular Vesicle miRNAs Can Identify Lung Cancer, Current Smoking Status, and Stable COPD. Int. J. Mol. Sci. 2021, 22, 5803. [Google Scholar] [CrossRef]
- Li, N.; Ouyang, B.S.; Liu, L.; Lin, C.S.; Xing, D.D.; Liu, J. Dexmedetomidine protected COPD-induced lung injury by regulating miRNA-146a. Bratisl. Lek. Listy 2016, 117, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Novák, J.; Kružliak, P.; Bienertová-Vašků, J.; Slabý, O.; Novák, M. MicroRNA-206: A promising theranostic marker. Theranostics 2014, 4, 119–133. [Google Scholar] [CrossRef]
- Yang, J.; Chen, Y.; Jiang, K.; Zhao, G.; Guo, S.; Liu, J.; Yang, Y.; Deng, G. MicroRNA-182 supplies negative feedback regulation to ameliorate lipopolysaccharide-induced ALI in mice by targeting TLR4. J. Cell. Physiol. 2020, 235, 5925–5937. [Google Scholar] [CrossRef]
- Zeng, Z.; Gong, H.; Li, Y.; Jie, K.; Ding, C.; Shao, Q.; Liu, F.; Zhan, Y.; Nie, C.; Zhu, W.; et al. Upregulation of miR-146a contributes to the suppression of inflammatory responses in LPS-induced acute lung injury. Exp. Lung Res. 2013, 39, 275–282. [Google Scholar] [CrossRef]
- He, R.; Li, Y.; Zhou, L.; Su, X.; Li, Y.; Pan, P.; Hu, C. miR-146b overexpression ameliorates lipopolysaccharide-induced acute lung injury in vivo and in vitro. J. Cell. Biochem. 2019, 120, 2929–2939. [Google Scholar] [CrossRef]
- Liang, Y.; Xie, J.; Che, D.; Zhang, C.; Lin, Y.; Feng, L.; Chen, J.; Chen, J.; Chen, L.; Wu, Z. MiR-124-3p helps to protect against acute respiratory distress syndrome by targeting p65. Biosci. Rep. 2020, 40, BSR20192132. [Google Scholar] [CrossRef]
- Yang, H.; Lu, Z.; Huo, C.; Chen, Y.; Cao, H.; Xie, P.; Zhou, H.; Liu, D.; Liu, J.; Yu, L. Liang-Ge-San, a Classic Traditional Chinese Medicine Formula, Attenuates Lipopolysaccharide-Induced Acute Lung Injury Through Up-Regulating miR-21. Front. Pharmacol. 2019, 10, 1332. [Google Scholar] [CrossRef]
- Kong, F.; Sun, Y.; Song, W.; Zhou, Y.; Zhu, S. MiR-216a alleviates LPS-induced acute lung injury via regulating JAK2/STAT3 and NF-κB signaling. Hum. Cell 2020, 33, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Chen, Y.L. The Functional Mechanisms of miR-30b-5p in Acute Lung Injury in Children. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 40–51. [Google Scholar] [CrossRef]
- Xie, T.; Liang, J.; Liu, N.; Wang, Q.; Li, Y.; Noble, P.W.; Jiang, D. MicroRNA-127 inhibits lung inflammation by targeting IgG Fcγ receptor I. J. Immunol. 2012, 188, 2437–2444. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhou, F. Inhibition of microRNA-92a ameliorates lipopolysaccharide-induced endothelial barrier dysfunction by targeting ITGA5 through the PI3K/Akt signaling pathway in human pulmonary microvascular endothelial cells. Int. Immunopharmacol. 2020, 78, 106060. [Google Scholar] [PubMed]
- Zhou, Y.; Yang, Y.; Liang, T.; Hu, Y.; Tang, H.; Song, D.; Fang, H. The regulatory effect of microRNA-21a-3p on the promotion of telocyte angiogenesis mediated by PI3K (p110α)/AKT/mTOR in LPS induced mice ARDS. J. Transl. Med. 2019, 17, 427. [Google Scholar]
- Ying, Y.; Mao, Y.; Yao, M. NLRP3 Inflammasome Activation by MicroRNA-495 Promoter Methylation May Contribute to the Progression of Acute Lung Injury. Mol. Ther. Nucleic Acids 2019, 18, 801–814. [Google Scholar] [CrossRef]
- Humphries, F.; Bergin, R.; Jackson, R.; Delagic, N.; Wang, B.; Yang, S.; Dubois, A.V.; Ingram, R.J.; Moynagh, P.N. The E3 ubiquitin ligase Pellino2 mediates priming of the NLRP3 inflammasome. Nat. Commun. 2018, 9, 1560. [Google Scholar]
- Sun, X.; Icli, B.; Wara, A.K.; Belkin, N.; He, S.; Kobzik, L.; Hunninghake, G.M.; Vera, M.P.; Blackwell, T.S.; Baron, R.M.; et al. MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J. Clin. Investig. 2012, 122, 1973–1990. [Google Scholar] [CrossRef]
- Papi, A.; Brightling, C.; Pedersen, S.E.; Reddel, H.K. Asthma. Lancet 2018, 391, 783–800. [Google Scholar]
- Szymczak, I.; Wieczfinska, J.; Pawliczak, R. Molecular Background of miRNA Role in Asthma and COPD: An Updated Insight. BioMed Res. Int. 2016, 2016, 7802521. [Google Scholar] [CrossRef]
- Zhong, J.; Liu, M.; Chen, S.; Liu, S.; Li, F.; Li, C. Study of the Regulatory Mechanism of miR-26a-5p in Allergic Asthma. Cells 2022, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Alharris, E.; Alghetaa, H.; Seth, R.; Chatterjee, S.; Singh, N.P.; Nagarkatti, M.; Nagarkatti, P. Resveratrol Attenuates Allergic Asthma and Associated Inflammation in the Lungs Through Regulation of miRNA-34a That Targets FoxP3 in Mice. Front. Immunol. 2018, 9, 2992. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, F.; Yu, X.; Wang, B.; Yang, Y.; Zhou, X.; Cheng, R.; Xia, S.; Zhou, X. miR-16 inhibits NLRP3 inflammasome activation by directly targeting TLR4 in acute lung injury. Biomed. Pharmacother. 2019, 112, 108664. [Google Scholar] [CrossRef]
- Zhang, K.; Feng, Y.; Liang, Y.; Wu, W.; Chang, C.; Chen, D.; Chen, S.; Gao, J.; Chen, G.; Yi, L.; et al. Epithelial miR-206 targets CD39/extracellular ATP to upregulate airway IL-25 and TSLP in type 2-high asthma. JCI Insight 2021, 6, e148103. [Google Scholar] [PubMed]
- Lu, T.X.; Munitz, A.; Rothenberg, M.E. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 2009, 182, 4994–5002. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Qi, N.; Zhou, Q. LncRNA H19 Inhibits Proliferation and Migration of Airway Smooth Muscle Cells Induced by PDGF-BB Through miR-21/PTEN/Akt Axis. J. Asthma Allergy 2021, 14, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, J.; Gao, P.; Wang, Q.; Zhang, J. miR-155: A Novel Target in Allergic Asthma. Int. J. Mol. Sci. 2016, 17, 1773. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, E.; Li, X.; Zhang, M.; Tang, Z.; He, L.; Lv, K. miR-155 contributes to Df1-induced asthma by increasing the proliferative response of Th cells via CTLA-4 downregulation. Cell. Immunol. 2017, 314, 1–9. [Google Scholar] [CrossRef]
- Malmhäll, C.; Alawieh, S.; Lu, Y.; Sjöstrand, M.; Bossios, A.; Eldh, M.; Rådinger, M. MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. J. Allergy Clin. Immunol. 2014, 133, 1429–1438.e7. [Google Scholar] [CrossRef]
- Kumar, M.; Ahmad, T.; Sharma, A.; Mabalirajan, U.; Kulshreshtha, A.; Agrawal, A.; Ghosh, B. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J. Allergy Clin. Immunol. 2011, 128, 1077–1085.e10. [Google Scholar] [CrossRef]
- ElKashef, S.; Ahmad, S.E.; Soliman, Y.M.A.; Mostafa, M.S. Role of microRNA-21 and microRNA-155 as biomarkers for bronchial asthma. Innate Immun. 2021, 27, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Panganiban, R.P.; Pinkerton, M.H.; Maru, S.Y.; Jefferson, S.J.; Roff, A.N.; Ishmael, F.T. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am. J. Clin. Exp. Immunol. 2012, 1, 154–165. [Google Scholar] [PubMed]
- Levänen, B.; Bhakta, N.R.; Torregrosa Paredes, P.; Barbeau, R.; Hiltbrunner, S.; Pollack, J.L.; Sköld, C.M.; Svartengren, M.; Grunewald, J.; Gabrielsson, S.; et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J. Allergy Clin. Immunol. 2013, 131, 894–903. [Google Scholar]
- Kastelowitz, N.; Yin, H. Exosomes and microvesicles: Identification and targeting by particle size and lipid chemical probes. ChemBioChem 2014, 15, 923–928. [Google Scholar] [PubMed]
- Pizzirani, C.; Ferrari, D.; Chiozzi, P.; Adinolfi, E.; Sandonà, D.; Savaglio, E.; Di Virgilio, F. Stimulation of P2 receptors causes release of IL-1beta-loaded microvesicles from human dendritic cells. Blood 2007, 109, 3856–3864. [Google Scholar] [CrossRef] [PubMed]
- Bianco, F.; Pravettoni, E.; Colombo, A.; Schenk, U.; Möller, T.; Matteoli, M.; Verderio, C. Astrocyte-derived ATP induces vesicle shedding and IL-1 beta release from microglia. J. Immunol. 2005, 174, 7268–7277. [Google Scholar] [CrossRef]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowska-Wieczorek, A.; Ratajczak, M.Z. Membrane-derived microvesicles: Important and underappreciated mediators of cell-to-cell communication. Leukemia 2006, 20, 1487–1495. [Google Scholar]
- Crunkhorn, S. miR-21 antagomir reverses COPD pathology. Nat. Rev. Drug Discov. 2022, 21, 20. [Google Scholar]
- Zheng, W.; Zhao, J.; Tao, Y.; Guo, M.; Ya, Z.; Chen, C.; Qin, N.; Zheng, J.; Luo, J.; Xu, L. MicroRNA-21: A promising biomarker for the prognosis and diagnosis of non-small cell lung cancer. Oncol. Lett. 2018, 16, 2777–2782. [Google Scholar]
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011, 8, 706–713. [Google Scholar] [CrossRef]
- Osada, H.; Takahashi, T. let-7 and miR-17-92: Small-sized major players in lung cancer development. Cancer Sci. 2011, 102, 9–17. [Google Scholar] [PubMed]
- Wang, X.; Lu, X.; Ma, C.; Ma, L.; Han, S. Combination of TLR agonist and miR146a mimics attenuates ovalbumin-induced asthma. Mol. Med. 2020, 26, 65. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Lee, H.Y.; Choi, J.Y.; Hur, J.; Kim, I.K.; Kim, Y.K.; Kang, J.Y.; Lee, S.Y. Inhibition of MicroRNA-21 by an antagomir ameliorates allergic inflammation in a mouse model of asthma. Exp. Lung Res. 2017, 43, 109–119. [Google Scholar] [CrossRef]
- Gil-Martínez, M.; Rodrigo-Muñoz, J.M.; Sastre, B.; Cañas, J.A.; García-Latorre, R.; Redondo, N.; de la Fuente, L.; Mínguez, P.; Mahíllo-Fernández, I.; Sastre, J.; et al. Serum microRNAs Catalog Asthma Patients by Phenotype. J. Investig. Allergol. Clin. Immunol. 2022, 32, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Jude, J.A.; Dileepan, M.; Subramanian, S.; Solway, J.; Panettieri, R.A.; Walseth, T.F., Jr.; Kannan, M.S. miR-140-3p regulation of TNF-α-induced CD38 expression in human airway smooth muscle cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2012, 303, L460–L468. [Google Scholar] [CrossRef]
- Roffel, M.P.; Bracke, K.R.; Heijink, I.H.; Maes, T. miR-223: A Key Regulator in the Innate Immune Response in Asthma and COPD. Front. Med. 2020, 7, 196. [Google Scholar]
- Ding, Y.; Hou, Y.; Liu, Y.; Xie, X.; Cui, Y.; Nie, H. Prospects for miR-21 as a Target in the Treatment of Lung Diseases. Curr. Pharm. Des. 2021, 27, 415–422. [Google Scholar]
- Kupczyk, M.; Kuna, P. MicroRNAs—New biomarkers of respiratory tract diseases. Pneumonol. I Alergol. Pol. 2014, 82, 183–190. [Google Scholar]
- Wang, H.; Zhu, Y.; Zhao, M.; Wu, C.; Zhang, P.; Tang, L.; Zhang, H.; Chen, X.; Yang, Y.; Liu, G. miRNA-29c suppresses lung cancer cell adhesion to extracellular matrix and metastasis by targeting integrin β1 and matrix metalloproteinase2 (MMP2). PLoS ONE 2013, 8, e70192. [Google Scholar]
- Roa, W.H.; Kim, J.O.; Razzak, R.; Du, H.; Guo, L.; Singh, R.; Gazala, S.; Ghosh, S.; Wong, E.; Joy, A.A.; et al. Sputum microRNA profiling: A novel approach for the early detection of non-small cell lung cancer. Clin. Investig. Med. Med. Clin. Et Exp. 2012, 35, E271. [Google Scholar] [CrossRef]
- Molina-Pinelo, S.; Pastor, M.D.; Suarez, R.; Romero-Romero, B.; González De la Peña, M.; Salinas, A.; García-Carbonero, R.; De Miguel, M.J.; Rodríguez-Panadero, F.; Carnero, A.; et al. MicroRNA clusters: Dysregulation in lung adenocarcinoma and COPD. Eur. Respir. J. 2014, 43, 1740–1749. [Google Scholar] [CrossRef] [PubMed]
- Mirra, D.; Esposito, R.; Spaziano, G.; La Torre, C.; Vocca, C.; Tallarico, M.; Cione, E.; Gallelli, L.; D’Agostino, B. Lung microRNAs Expression in Lung Cancer and COPD: A Preliminary Study. Biomedicines 2023, 11, 736. [Google Scholar]
- Tong, L.; Shen, S.; Huang, Q.; Fu, J.; Wang, T.; Pan, L.; Zhang, P.; Chen, G.; Huang, T.; Li, K.; et al. Proteasome-dependent degradation of Smad7 is critical for lung cancer metastasis. Cell Death Differ. 2020, 27, 1795–1806. [Google Scholar] [CrossRef]
- Wang, H.; Zhan, Y.; Jin, J.; Zhang, C.; Li, W. MicroRNA-15b promotes proliferation and invasion of non-small cell lung carcinoma cells by directly targeting TIMP2. Oncol. Rep. 2017, 37, 3305–3312. [Google Scholar] [CrossRef]
- Duan, F.G.; Wang, M.F.; Cao, Y.B.; Dan, L.; Li, R.Z.; Fan, X.X.; Khan, I.; Lai, H.L.; Zhang, Y.Z.; Hsiao, W.W.; et al. MicroRNA-421 confers paclitaxel resistance by binding to the KEAP1 3’UTR and predicts poor survival in non-small cell lung cancer. Cell Death Dis. 2019, 10, 821. [Google Scholar] [PubMed]
- Bozgeyik, I. Therapeutic potential of miRNAs targeting SARS-CoV-2 host cell receptor ACE2. Meta Gene 2021, 27, 100831. [Google Scholar] [CrossRef]
- Yang, T.; Li, H.; Chen, T.; Ren, H.; Shi, P.; Chen, M. LncRNA MALAT1 Depressed Chemo-Sensitivity of NSCLC Cells through Directly Functioning on miR-197-3p/p120 Catenin Axis. Mol. Cells 2019, 42, 270–283. [Google Scholar]
- De Smet, E.G.; Van Eeckhoutte, H.P.; Avila Cobos, F.; Blomme, E.; Verhamme, F.M.; Provoost, S.; Verleden, S.E.; Venken, K.; Maes, T.; Joos, G.F.; et al. The role of miR-155 in cigarette smoke-induced pulmonary inflammation and COPD. Mucosal Immunol. 2020, 13, 423–436. [Google Scholar]
- Xie, Y.; Lv, Y.; Zhang, Y.; Liang, Z.; Han, L.; Xie, Y. LATS2 promotes apoptosis in non-small cell lung cancer A549 cells via triggering Mff-dependent mitochondrial fission and activating the JNK signaling pathway. Biomed. Pharmacother. 2019, 109, 679–689. [Google Scholar] [CrossRef]
- Jiang, K.; Yang, J.; Guo, S.; Zhao, G.; Wu, H.; Deng, G. Peripheral Circulating Exosome-Mediated Delivery of miR-155 as a Novel Mechanism for Acute Lung Inflammation. Mol. Ther. J. Am. Soc. Gene Ther. 2019, 27, 1758–1771. [Google Scholar] [CrossRef]
- Wang, D.; Wang, X.; Song, Y.; Si, M.; Sun, Y.; Liu, X.; Cui, S.; Qu, X.; Yu, X. Exosomal miR-146a-5p and miR-155-5p promote CXCL12/CXCR7-induced metastasis of colorectal cancer by crosstalk with cancer-associated fibroblasts. Cell Death Dis. 2022, 13, 380. [Google Scholar] [CrossRef] [PubMed]
- Kassif-Lerner, R.; Zloto, K.; Rubin, N.; Asraf, K.; Doolman, R.; Paret, G.; Nevo-Caspi, Y. miR-155: A Potential Biomarker for Predicting Mortality in COVID-19 Patients. J. Pers. Med. 2022, 12, 324. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Xue, J.; Ruan, M.; Lu, J.; Xu, Q.; Shi, Y.; Yu, F. Expression of Serum miR-155 in Children with Mycoplasma pneumoniae Pneumonia and Its Role in Immunity to Mycoplasma pneumoniae. Infect. Drug Resist. 2021, 14, 1273–1281. [Google Scholar] [CrossRef]
- Wang, Z.F.; Yang, Y.M.; Fan, H. Diagnostic value of miR-155 for acute lung injury/acute respiratory distress syndrome in patients with sepsis. J. Int. Med. Res. 2020, 48, 300060520943070. [Google Scholar] [CrossRef]
- Li, C.; Hu, X.; Li, L.; Li, J.H. Differential microRNA expression in the peripheral blood from human patients with COVID-19. J. Clin. Lab. Anal. 2020, 34, e23590. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Banerjea, A.C. SARS-CoV-2 Spike Targets USP33-IRF9 Axis via Exosomal miR-148a to Activate Human Microglia. Front. Immunol. 2021, 12, 656700. [Google Scholar] [PubMed]
- Graff, J.W.; Powers, L.S.; Dickson, A.M.; Kim, J.; Reisetter, A.C.; Hassan, I.H.; Kremens, K.; Gross, T.J.; Wilson, M.E.; Monick, M.M. Cigarette smoking decreases global microRNA expression in human alveolar macrophages. PLoS ONE 2012, 7, e44066. [Google Scholar]
- Christenson, S.A.; Brandsma, C.A.; Campbell, J.D.; Knight, D.A.; Pechkovsky, D.V.; Hogg, J.C.; Timens, W.; Postma, D.S.; Lenburg, M.; Spira, A. miR-638 regulates gene expression networks associated with emphysematous lung destruction. Genome Med. 2013, 5, 114. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, S.; Bogaard, H.J.; Gomez-Arroyo, J.; Alhussaini, A.; Kraskauskas, D.; Cool, C.D.; Voelkel, N.F. MicroRNA-199a-5p is associated with hypoxia-inducible factor-1α expression in lungs from patients with COPD. Chest 2012, 142, 663–672. [Google Scholar] [CrossRef]
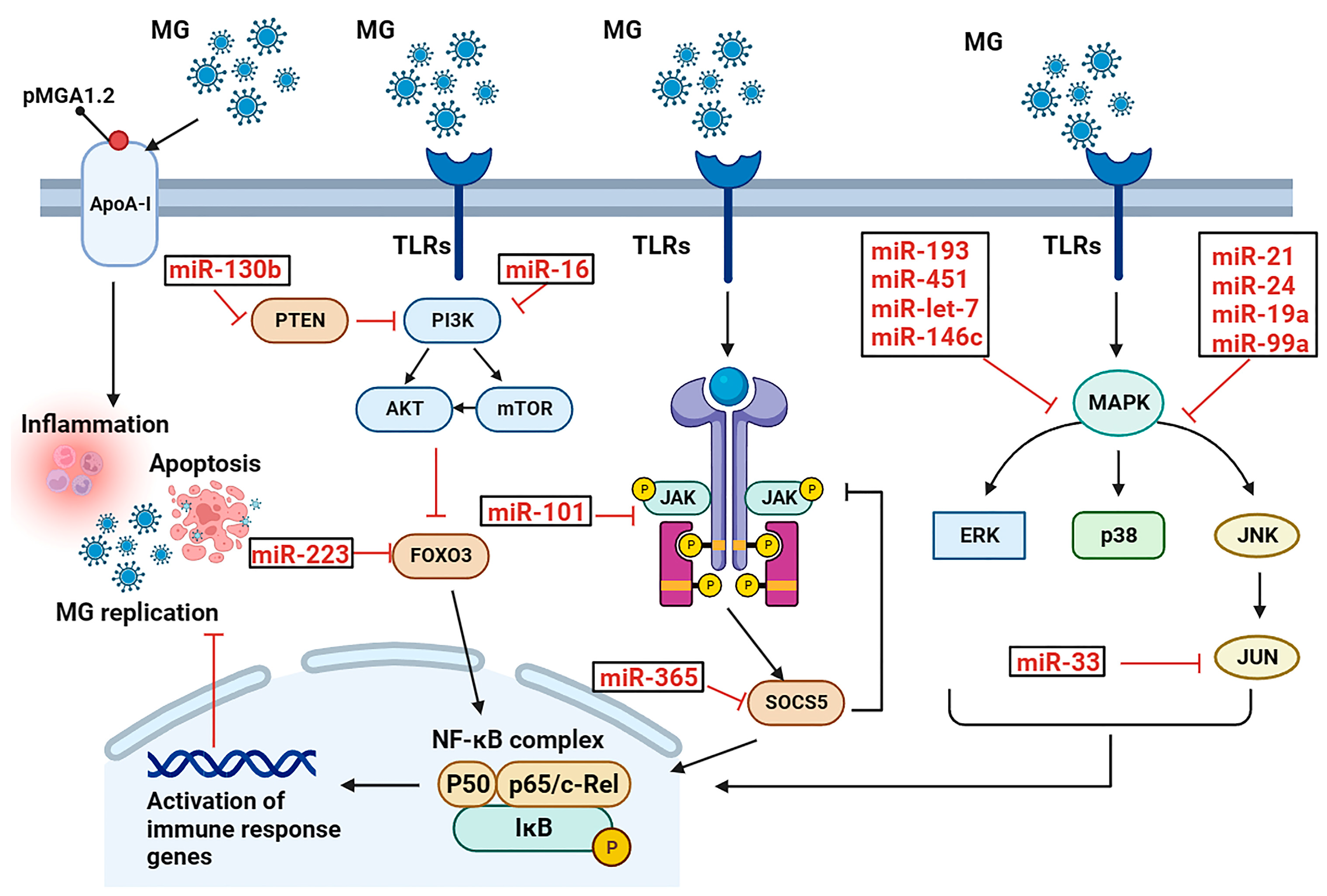
| miRNA | Target | Function | Reference |
|---|---|---|---|
| miR-451 | YWHAZ | inhibits cell cycle progression and cell proliferation and promotes cell apoptosis | [28] |
| miR-193a | KRAS | inhibits cell proliferation, promotes apoptosis, and increases inflammation | [25] |
| miR-181a | PPM1B | activates the TLR2-mediated MyD88/NF-κB pathways to promote inflammation | [24] |
| miR-19a | ZMYND11 | activates the NF-κB signaling pathway to defend against MG infection | [32] |
| miR-99a | SMARCA5 | represses the proliferation of MG-infected DF-1 cells by inhibiting cell cycle | [22] |
| miR-101 | EZH2 | inhibits proliferation of MG-infected DF-1 cells by inhibiting cell cycle | [26] |
| miR-146c | MMP16 | activates the TLR6/MyD88/NF-κB pathway to defend against MG infection | [30] |
| miR-16 | PIK3R1 | inhibits PI3K/Akt/NF-κB pathway to exert an anti-inflammatory effect | [35] |
| miR-130b | PTEN | activates the PI3K/AKT/NF-κB pathway | [31] |
| miR-21 | MAP3K1 | promotes inflammation and cell proliferation to defend against MG infection | [29] |
| miR-142 | TAB2 | facilitates cell proliferation by inhibiting cell apoptosis to defend against MG infection | [27] |
| miR-223 | FOXO3 | decreases proliferation and cycle progression, and increases apoptosis to promote MG infection | [33] |
| miR-24 | RAP1B | decreases proliferation but increases apoptosis | [34] |
| let-7 | MPK1 | an inhibitor of MAPK pathway to effectively mitigate MG adhesion | [36] |
| miR-365 | SOCS5 | a key factor for MG evasion of host immunity | [20] |
| miR-33 | JNK1 | lnc90386 sponges miR-33-5p to defend against MG infection | [21] |
| miRNAs | Function | References |
|---|---|---|
| miR-1207 | may contribute to uncontrolled inflammation in most severe COVID-19 cases | [55] |
| miR-21 | has the largest probability of binding the human coronavirus RNAs | [56] |
| miR-146a miR-21 miR-142 miR-15b | as potential contributors to the disease pathogenesis, possibly serving as biomarkers of severe COVID-19 | [57,58,59,60] |
| miR-126 | involved in vascular endothelial function | [35] |
| miR-208a | associated with myocardial injury in COVID-19 patients | [60] |
| miR-200c let-7b | involved in the regulation of ACE2 expression | [61,62] |
| miR-223 miR-146a | enhance the antiviral immune response | [63,64] |
| miR-145 miR-885 | may contribute to the thrombotic complications observed in COVID-19 patients | [65] |
| miR-148a miR-590 | significantly upregulated in the exosomes of patients with neurological manifestations | [66] |
| miR-7 miR-24 miR-145 miR-223 | are associated with high mortality rates of COVID-19 in the elderly | [67] |
| miR-588 miR-587 miR-582 | enhance lung pathogenesis and injury | [68] |
| miRNAs | Function | References |
|---|---|---|
| miR-320d | suppresses inflammatory cytokine production by regulating NF-κB activity | [70] |
| miR-21 | regulates the HIF-1α signaling pathway, which is responsible for myofibroblast differentiation, to treat COPD | [71] |
| let-7d miR-191 | affect the clearance of apoptotic cells by specialized macrophages and may contribute to the pathogenesis of COPD | [72] |
| miR-206 miR-146a | have potential value for the diagnosis and treatment of COPD | [73,74] |
| miR-21 miR-181a | their ratio is suggested as a potential biomarker in predicting the development of COPD in asymptomatic smokers | [75] |
| miR-100 miR-20a miR-34c-5p miR-28-3p miR-7 | potential biomarkers for COPD and may have a biological function in the pathogenesis of COPD | [76] |
| miR-452 | increases the expression of MMP12 and causes emphysema | [77] |
| miR-638 | positively correlated with emphysema severity | [78] |
| miR-199a-5p | decreases hypoxia inducible factor 1α (HIF-1α) expression | [79] |
| miRNAs | Function | References |
|---|---|---|
| miR-214 miR-415 miR-16 miR-23a miR-24 miR-181 miR-181b miR-199a | a group of miRNAs that were differentially expressed in ALI mice; miR-16 may play a role in alleviating in ALI by inhibiting LPS-induced IL-6 and TNF-a | [86] |
| miR-29a | may play a role in regulating the inflammatory response in ALI by targeting TNFR1; reduces alveolar epithelial cell PANoptosis in the ALI mouse model | [87] |
| miR-27a miR-16 miR-182 miR-145-5p miR-140 miR-140-5p miR-146a | reduce the release of pro-inflammatory cytokines and downstream TLR4/MyD88/NF-κB signaling pathways, ultimately suppressing the inflammatory response | [88,89,90,91,92] |
| miR-146b | reduces lung inflammation and increases lung permeability by targeting IRAK1 to inhibit NF-κB signaling | [93] |
| miR-124-3p | promotes macrophage apoptosis and plays a protective role in ARDS by targeting p65 | [94] |
| miR-21 | inhibits the JAK2/STAT3 signaling pathway, thereby reducing the infiltration of inflammatory cells in the lung tissue of ALI/ARDS | [95] |
| miR-216a | inhibits the JAK2/STAT3 signaling pathway, inhibiting cell apoptosis, autophagy, and the release of inflammatory factors | [96] |
| miR-30b-5p | negatively regulates the JAK2/STAT3 pathway | [97] |
| miR-127 | suppresses lung inflammation by targeting macrophage CD64 | [98] |
| miR-92a | inhibits PI3K/AKT pathway, improves endothelial cell barrier function, and protects alveolar vascular endothelial cells | [99] |
| miR-21a-3p | regulates the PI3K (p110α)/Akt/mTOR pathway and promotes lung tissue repair and angiogenesis | [100] |
| miR-802 | improves lung injury induced by LPS | [101] |
| miR-495 miR-223 | inhibits NLRP3 activation, leading to reduced inflammation and improved ALI/ARDS | [102] |
| miRNAs | Function | References |
|---|---|---|
| miR-34a miR-206 | modulate airway inflammation by regulating the expression of cytokines | [107,109] |
| miR-26 | regulates airway remodeling by regulating the expression of collagen genes | [108] |
| miR-21 | promotes airway remodeling by regulating the expression of matrix metalloproteinases | [110] |
| miR-155 | promotes Th cell proliferation through downregulation of CTLA-4, thereby participating in the development of allergic asthma | [111] |
| miR-146a | reduces airway inflammation | [112] |
| let-7 | reduces airway inflammation | [113] |
| miR-1248 miR-155 miR-26a miR-376a | potential biomarkers for asthma | [114,115,116] |
| let-7a miRNA-658 miRNA-24 miRNA-26a miRNA-99a miRNA-200c miRNA-1268 | potential biomarkers and strongly correlated with forced expiratory volume in 1 s (FEV1) within asthmatic patients | [117] |
| miR-140 | appears to play an influential role in airway smooth muscle cell hyperplasia | [118] |
| let-7 families miRNA-200 families | dysregulated in the exosomes isolated from BAL fluid of asthmatic patients, potential biomarkers for asthma | [119,120,121] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zou, M.; Zhao, Y.; Kabir, M.A.; Peng, X. Exosomal microRNA/miRNA Dysregulation in Respiratory Diseases: From Mycoplasma-Induced Respiratory Disease to COVID-19 and Beyond. Cells 2023, 12, 2421. https://doi.org/10.3390/cells12192421
Wang Y, Zou M, Zhao Y, Kabir MA, Peng X. Exosomal microRNA/miRNA Dysregulation in Respiratory Diseases: From Mycoplasma-Induced Respiratory Disease to COVID-19 and Beyond. Cells. 2023; 12(19):2421. https://doi.org/10.3390/cells12192421
Chicago/Turabian StyleWang, Yingjie, Mengyun Zou, Yabo Zhao, Md. Ahsanul Kabir, and Xiuli Peng. 2023. "Exosomal microRNA/miRNA Dysregulation in Respiratory Diseases: From Mycoplasma-Induced Respiratory Disease to COVID-19 and Beyond" Cells 12, no. 19: 2421. https://doi.org/10.3390/cells12192421





