Investigating the Inhibitory Factors of Sucrose Hydrolysis in Sugar Beet Molasses with Yeast and Invertase
Abstract
:1. Introduction
2. Results and Discussion
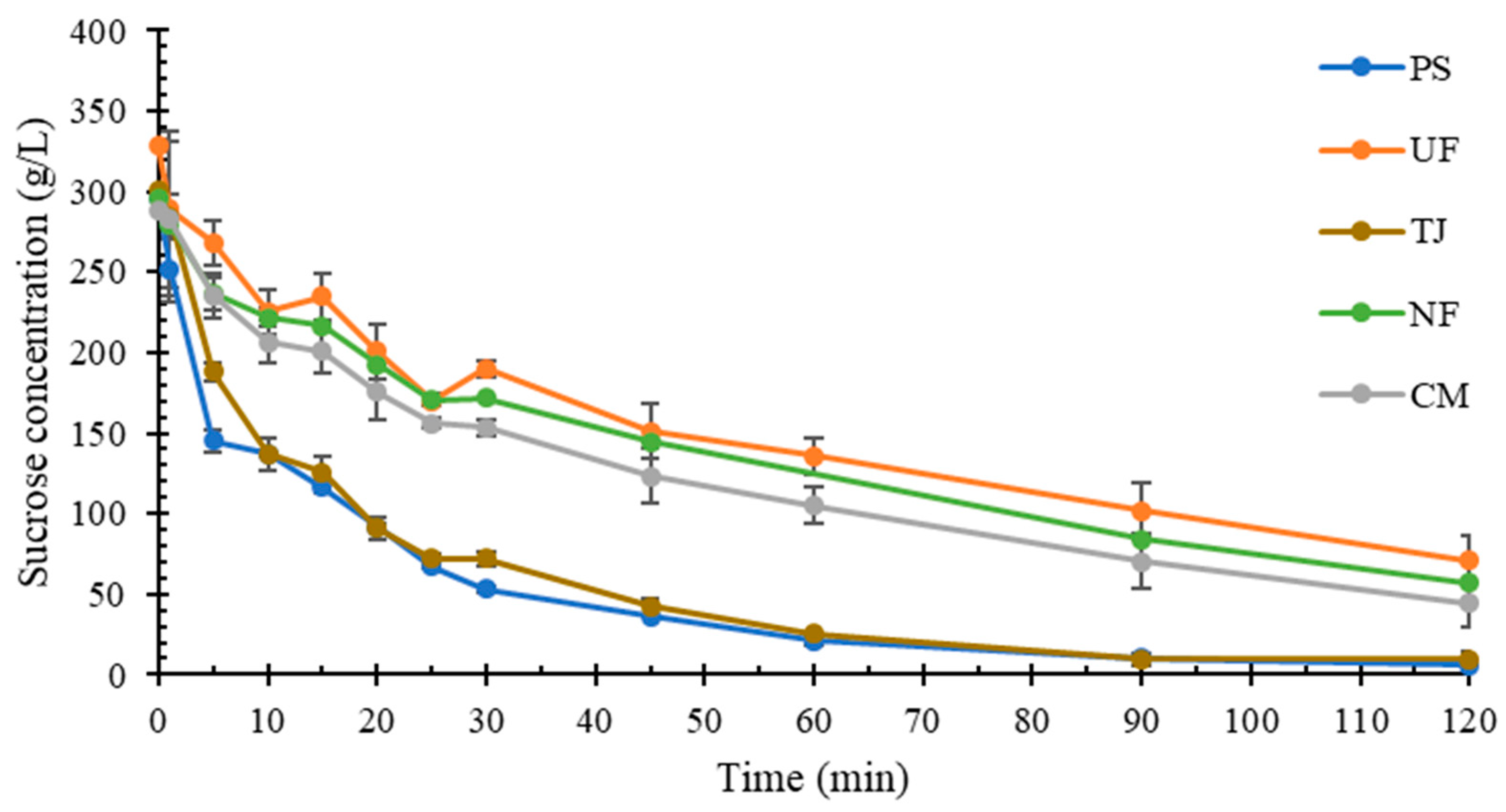
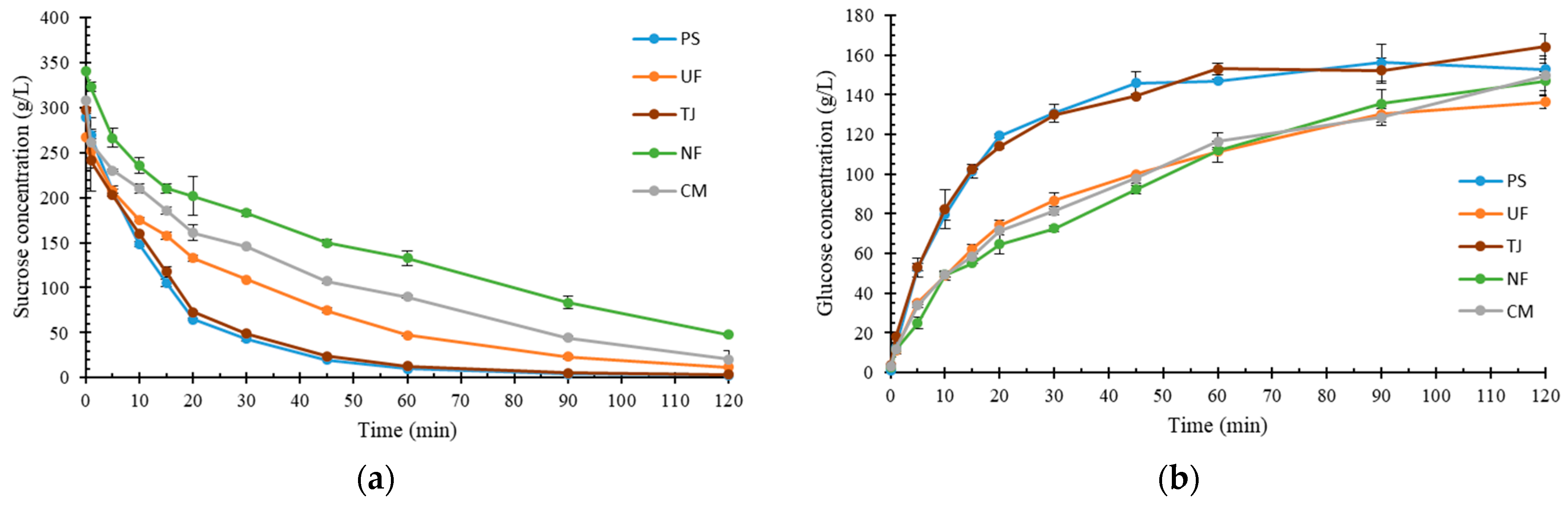
2.1. Molasses Composition
2.2. Impact of pH and Viscosity on Sucrose Hydrolysis
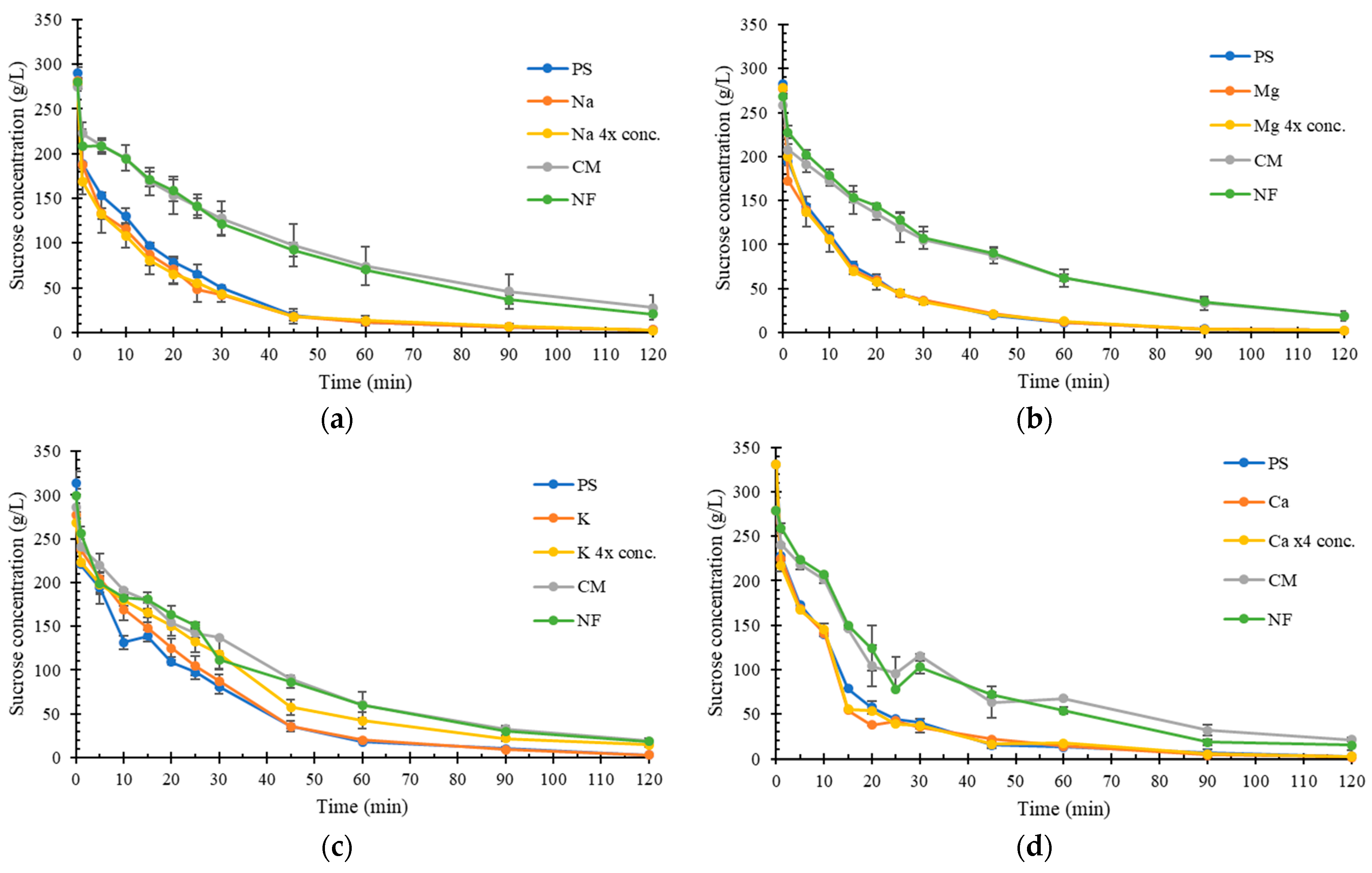
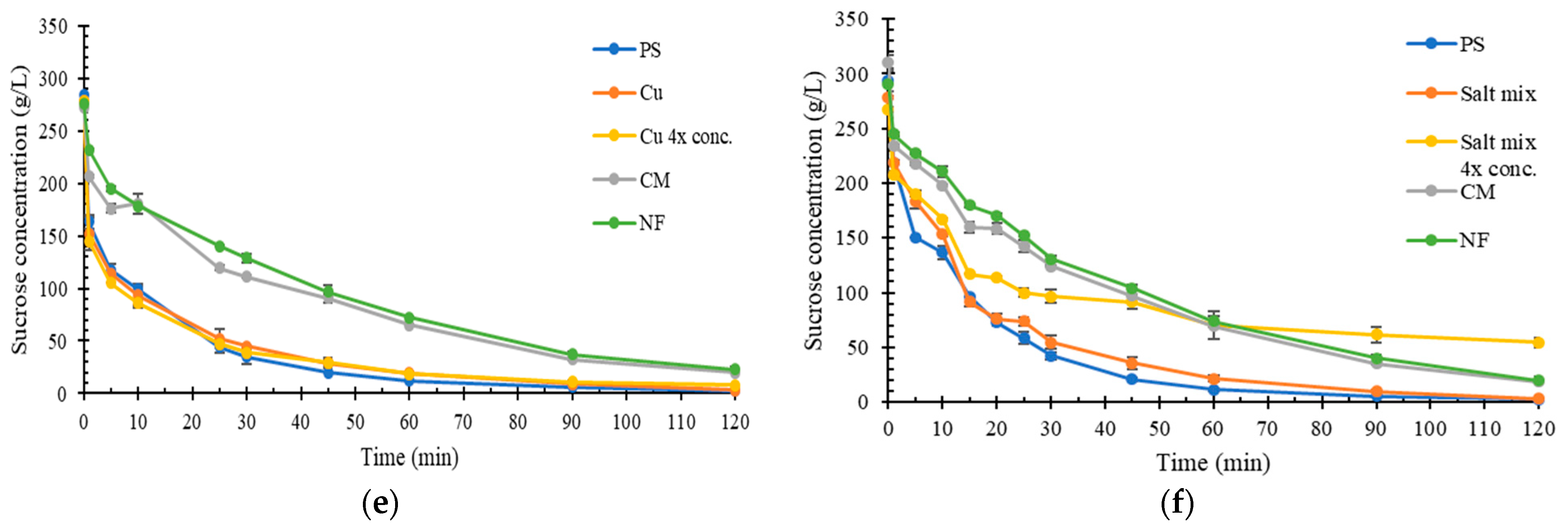
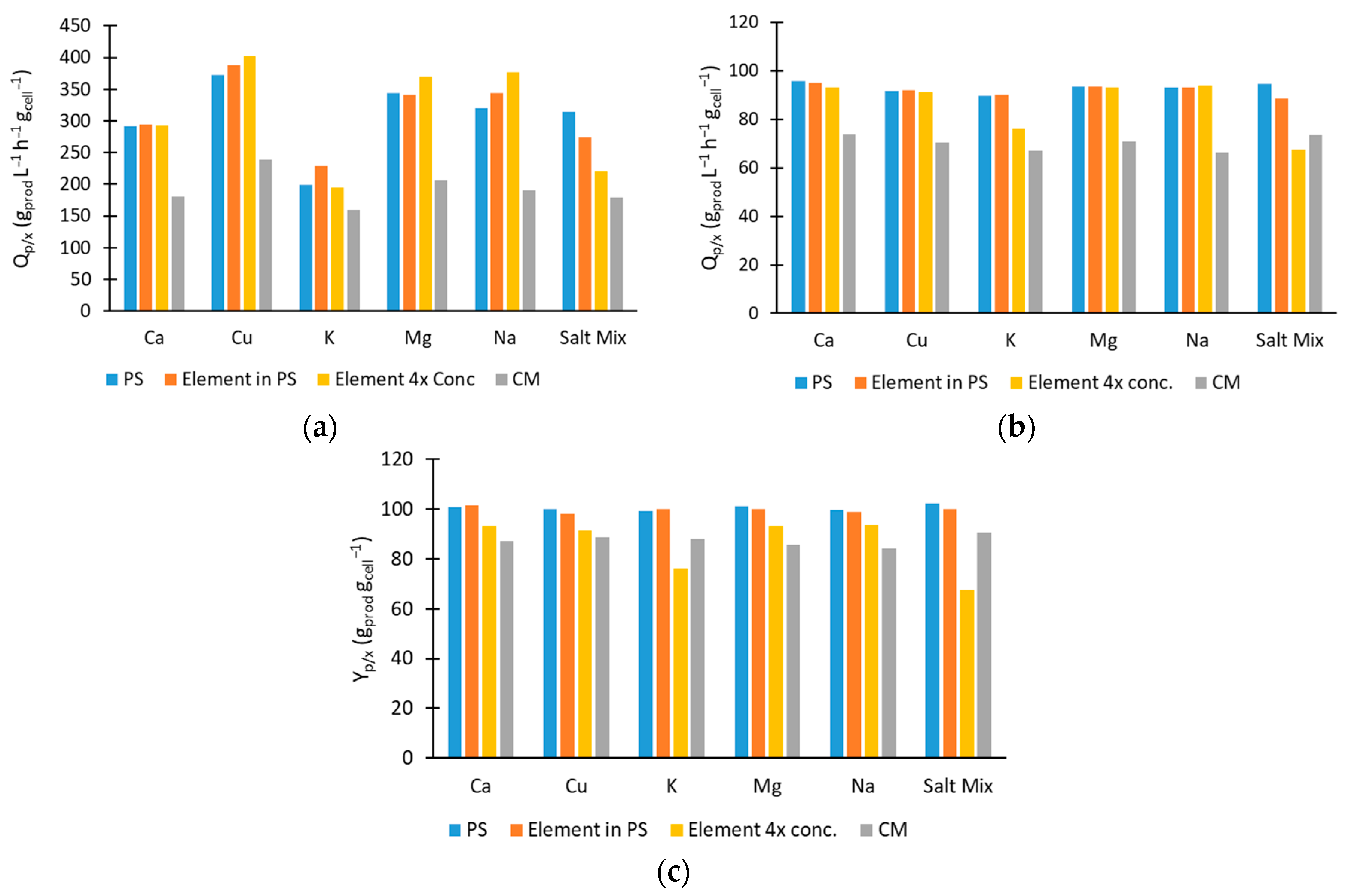
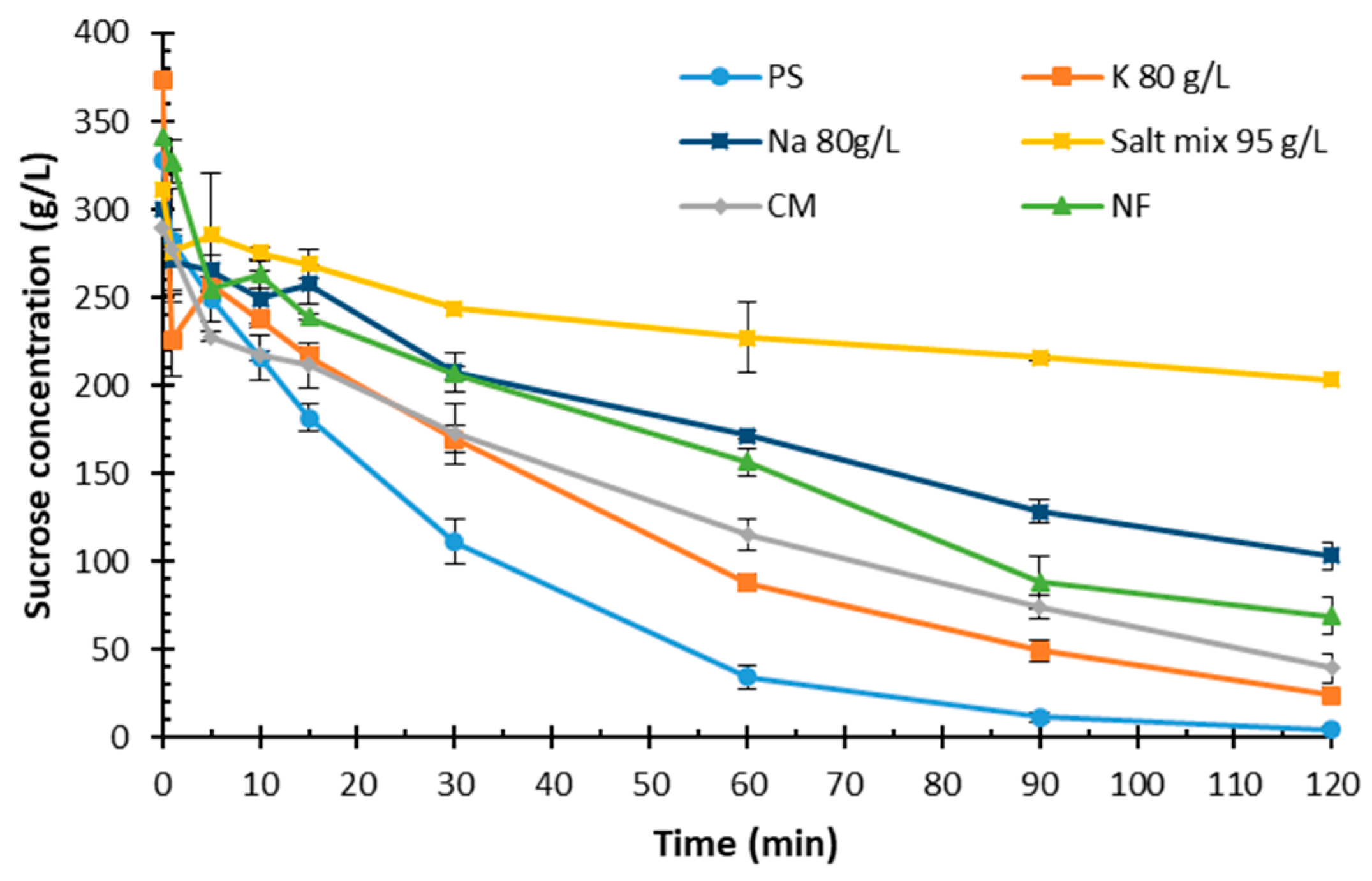
2.3. Impact of Inorganic Compounds on Sucrose Hydrolysis
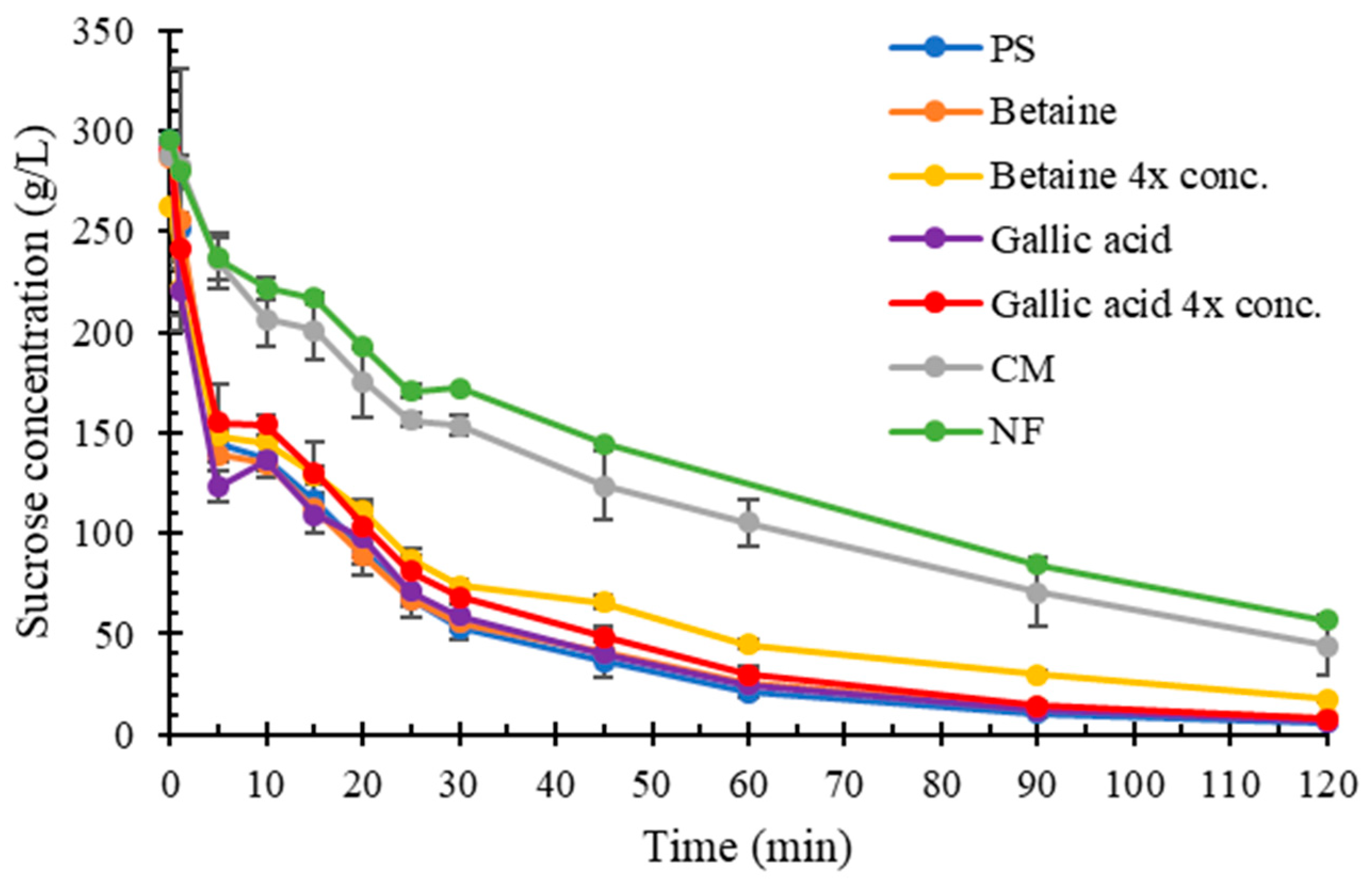
2.4. Impact of Organic Compounds on Sucrose Hydrolysis
2.5. Invertase as a Free Enzyme
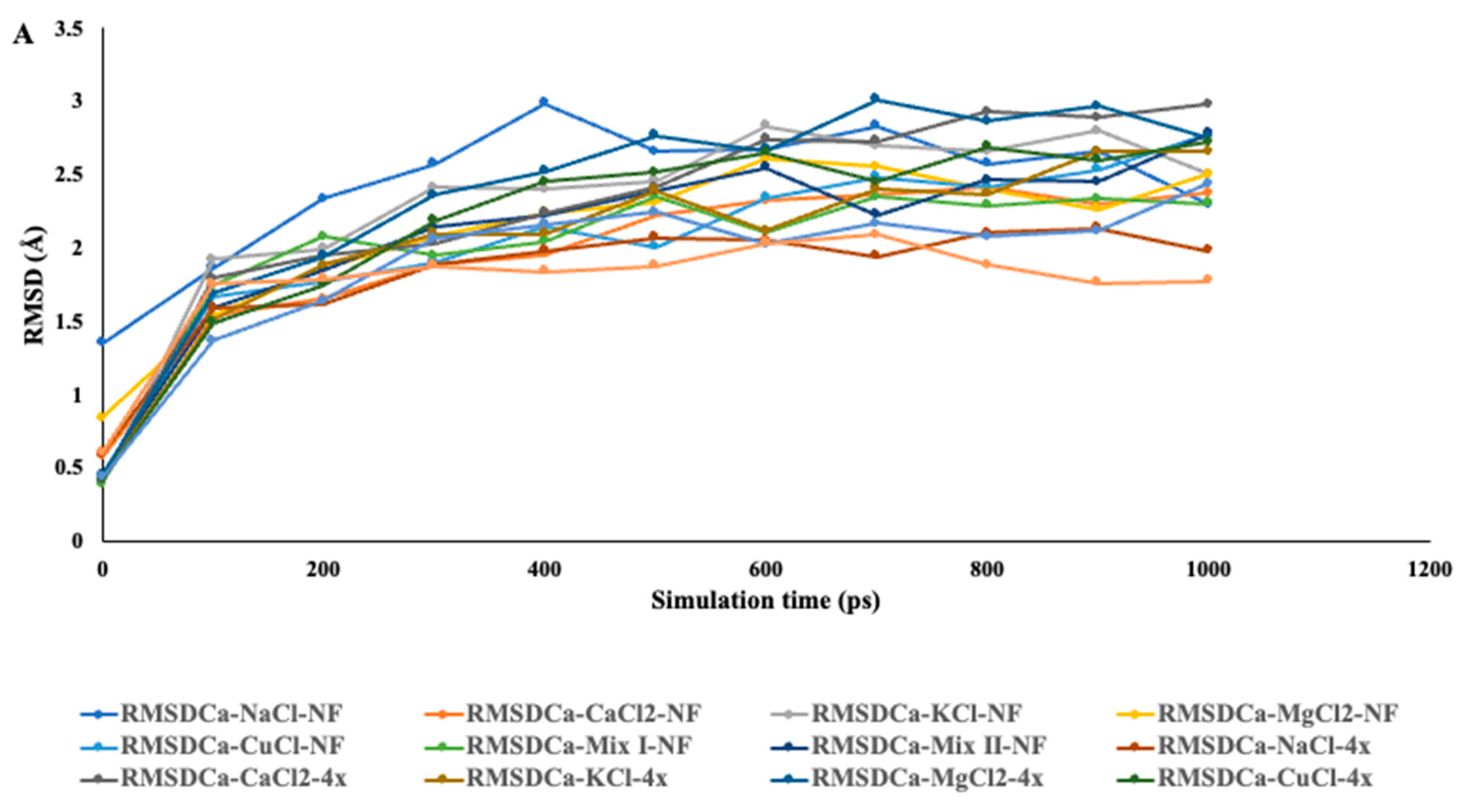
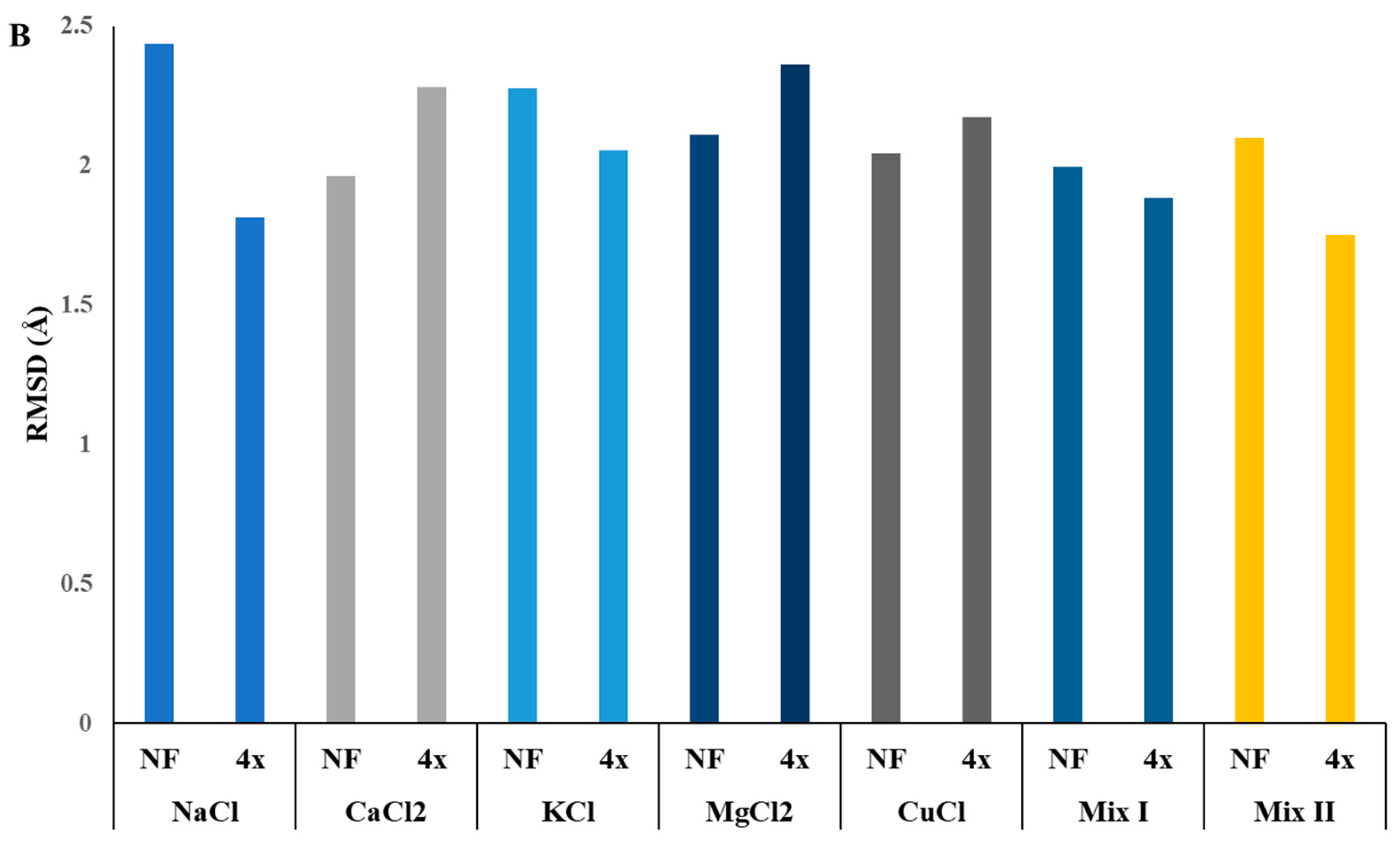
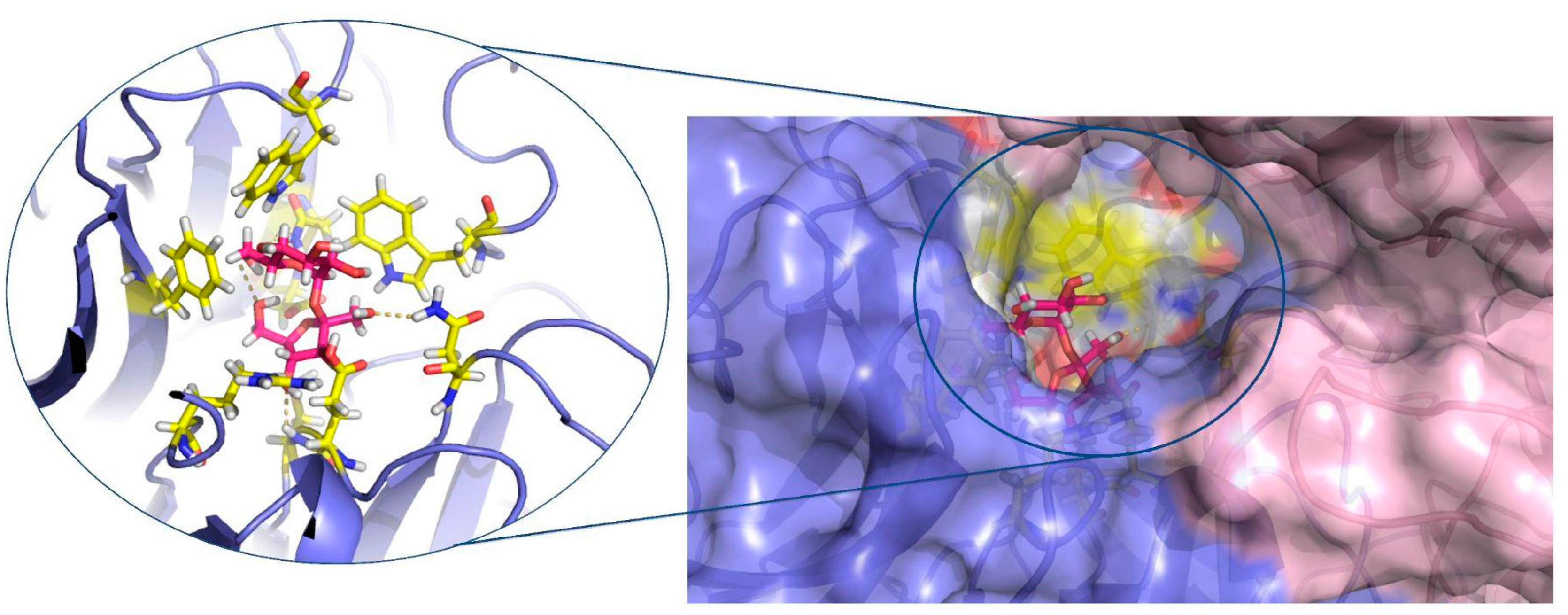
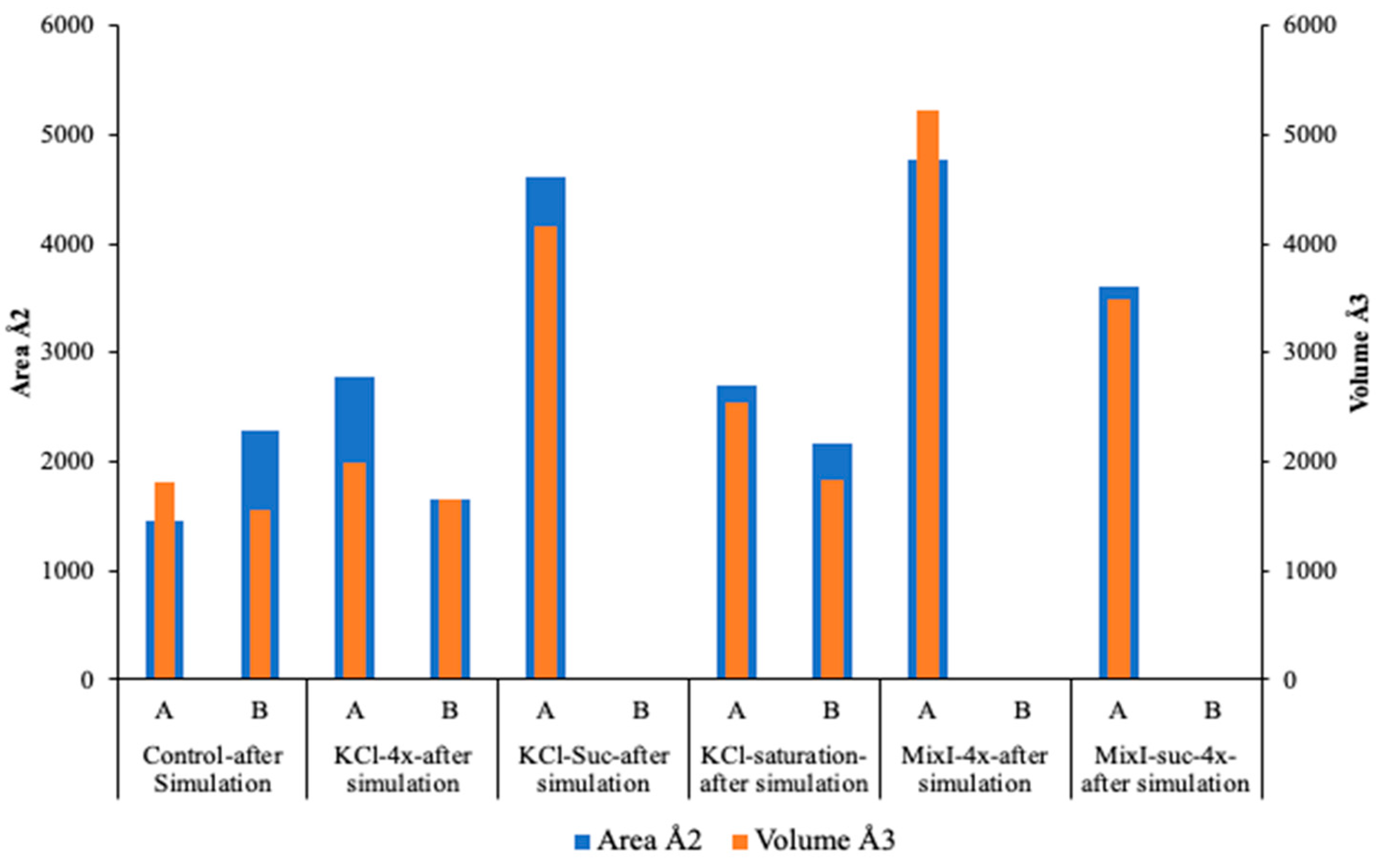
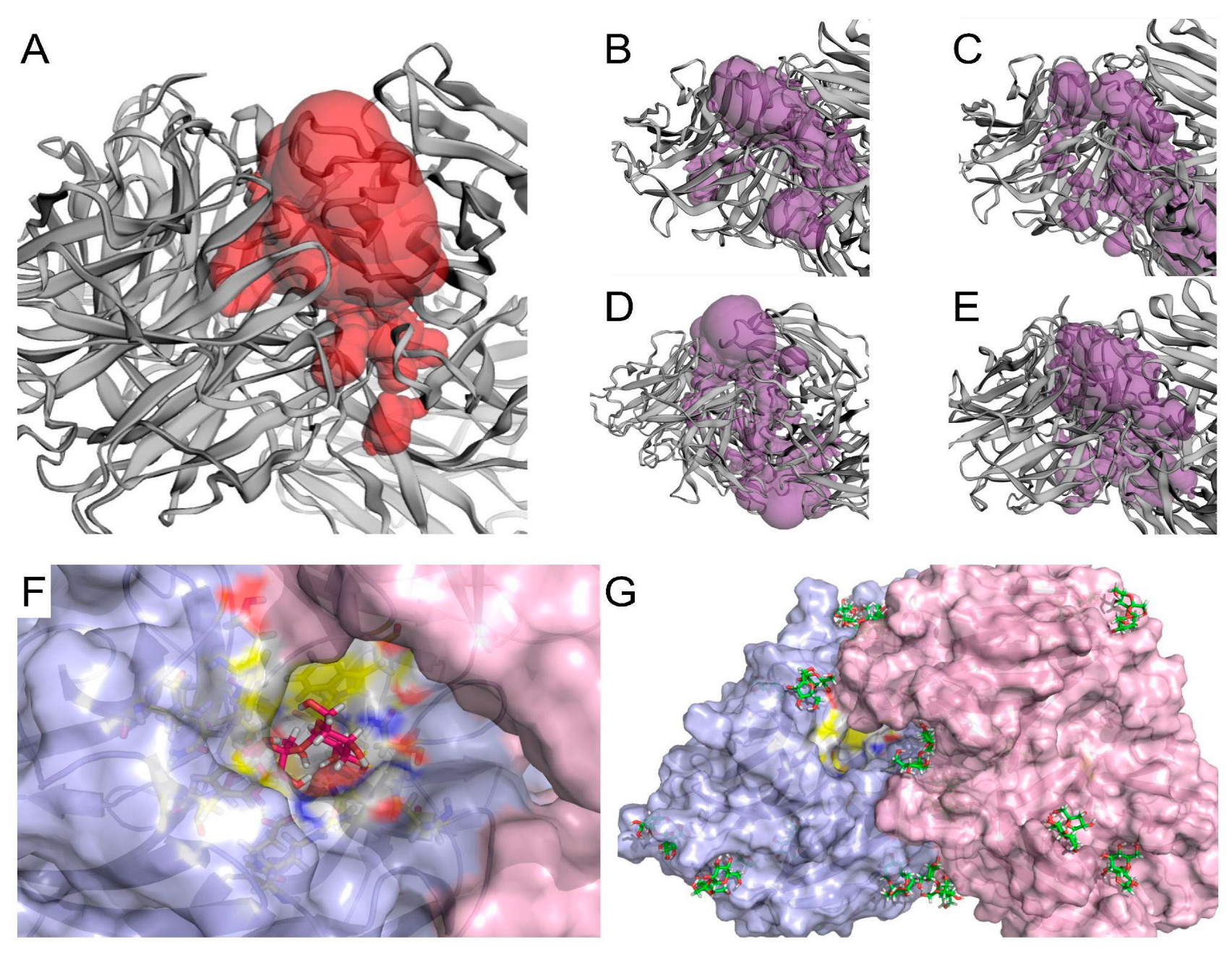
2.6. In Silico Investigation of Saccharomyces Cerevisiae Invertase
3. Materials and Methods
3.1. Molasses Pretreatment
3.2. Impact of pH and Viscosity on Sucrose Hydrolysis
3.3. Impact of Inorganic and Organic Compounds on Sucrose Hydrolysis
3.4. Invertase as a Free Enzyme
3.5. In Silico Investigation of Saccharomyces Cerevisiae Invertase
3.6. Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, S.; Wang, J.; Jiang, H. Microbial production of value-added bioproducts and enzymes from molasses, a by-product of sugar industry. Food Chem. 2021, 346, 128860. [Google Scholar] [CrossRef]
- Lipnizki, F. Membrane processes for sugar and starch processing. In Engineering Aspects of Membrane Separation and Application in Food Processing, 1st ed.; Field, R., Bekassy-Molnar, E., Lipnizki, F., Vatai, G., Eds.; Taylor & Francis Group, CRC Press: Boca Raton, FL, USA, 2016; pp. 241–267. [Google Scholar] [CrossRef]
- Beigbeder, J.B.; de Medeiros Dantas, J.M.; Lavoie, J.M. Optimization of yeast, sugar and nutrient concentrations for high ethanol production rate using industrial sugar beet molasses and response surface methodology. Fermentation 2021, 7, 86. [Google Scholar] [CrossRef]
- Milkessa, T.; Abate, D. Evaluation of Yeast Biomass Production Using Molasses and Supplements; LAP LAMBERT Academic Publishing: Saarbrücken, Germany, 2014. [Google Scholar]
- van Putten, R.-J.; Van Der Waal, J.C.; De Jong, E.; Rasrendra, C.B.; Heeres, H.J.; de Vries, J.G. Hydroxymethylfurfural, a versatile platform chemical made from renewable resources. Chem. Rev. 2013, 113, 1499–1597. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Brown, C.A.; Chen, R. Industrial production, application, microbial biosynthesis and degradation of furanic compound, hydroxymethylfurfural (HMF). AIMS Microbiol. 2018, 4, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Iris, K.M.; Tsang, D.C. Conversion of biomass to hydroxymethylfurfural: A review of catalytic systems and underlying mechanisms. Biores. Technol. 2017, 238, 716–732. [Google Scholar] [CrossRef]
- Chibbar, R.N.; Jaiswal, S.; Gangola, M.; Båga, M. Carbohydrate Metabolism. In Encyclopedia of Food Grains, 2nd ed.; Wrigley, C.C., Corke, H., Seetharaman, K., Faubion, J., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 161–173. [Google Scholar] [CrossRef]
- Bower, S.; Wickramasinghe, R.; Nagle, N.J.; Schell, D.J. Modeling sucrose hydrolysis in dilute sulfuric acid solutions at pretreatment conditions for lignocellulosic biomass. Biores. Technol. 2008, 99, 7354–7362. [Google Scholar] [CrossRef]
- Chen, H. Lignocellulose biorefinery feedstock engineering. In Lignocellulose Biorefinery Engineering, 1st ed.; Chen, H., Ed.; Woodhead Publishing: Sawston, UK, 2015; pp. 37–86. [Google Scholar] [CrossRef]
- Nakajima, M.; Nishizawa, K.; Nabetani, H. A forced flow membrane enzyme reactor for sucrose inversion using molasses. Bioprocess. Eng. 1993, 9, 31–35. [Google Scholar] [CrossRef]
- Combes, D.; Monsan, P. Sucrose hydrolysis by invertase. Characterization of products and substrate inhibition. Carbohydr. Res. 1983, 117, 215–228. [Google Scholar] [CrossRef]
- Zech, M.; Görisch, H. Invertase from Saccharomyces cerevisiae: Reversible inactivation by components of industrial molasses media. Enzyme Microb. Technol. 1995, 17, 41–46. [Google Scholar] [CrossRef]
- Sjölin, M.; Sayed, M.; Thuvander, J.; Lipnizki, F.; Hatti-Kaul, R.; Wallberg, O. Effect of membrane purification and concentration of sucrose in sugar beet molasses for the production of 5-hydroxymethylfurfural. Chem. Eng. Res. Des. 2022, 179, 365–373. [Google Scholar] [CrossRef]
- Plascencia-Espinosa, M.; Santiago-Hernández, A.; Pavón-Orozco, P.; Vallejo-Becerra, V.; Trejo-Estrada, S.; Sosa-Peinado, A.; Benitez-Cardoza, C.G.; Hidalgo-Lara, M.E. Effect of deglycosylation on the properties of thermophilic invertase purified from the yeast Candida guilliermondii MpIIIa. Process Biochem. 2014, 49, 1480–1487. [Google Scholar] [CrossRef]
- Essel, K.K.; Osei, Y.D. Investigation of Some Kinetic Properties of Commercial Invertase from Yeast. Nat. Prod. Chem. Res. 2014, 2, 6. [Google Scholar] [CrossRef]
- Šarić, L.C.; Filipčev, B.V.; Šimurina, O.D.; Plavšić, D.V.; Šarić, B.M.; Lazarević, J.M.; Milovanović, I.L. Sugar beet molasses: Properties and applications in osmotic dehydration of fruits and vegetables. Food Feed. Res. 2016, 43, 135–144. [Google Scholar] [CrossRef]
- Kumar, R.; Kesavapillai, B. Stimulation of extracellular invertase production from spent yeast when sugarcane pressmud used as substrate through solid state fermentation. SpringerPlus 2012, 1, 81. [Google Scholar] [CrossRef] [PubMed]
- Takeshige, K.; Ouchi, K. Factors affecting the ethanol productivity of yeast in molasses. J. Ferment. Bioeng. 1995, 79, 449–452. [Google Scholar] [CrossRef]
- Kaur, N.; Sharma, A.D. Production, optimization and characterization of extracellular invertase by an actinomycete strain. J. Sci. Ind. Res. 2005, 64, 515–519. [Google Scholar]
- Cuezzo de Ginés, S.; Maldonado, M.; Font de Valdez, G. Purification and characterization of invertase from lactobacillus reuteri CRL 1100. Curr. Microbiol. 2000, 40, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Hargono, H.; Jos, B.; Abdullah, A.; Riyanto, T. Inhibition effect of Ca2+ ions on sucrose hydrolysis using invertase. Bull. Chem. React. Eng. Catal. 2019, 14, 646–653. [Google Scholar] [CrossRef]
- Maruyama, Y.; Onodera, K. Production and some properties of invertase isozymes of Fusarium oxysporum. J. Gen. Appl. Microbiol. 1979, 25, 361–366. [Google Scholar] [CrossRef]
- Rashad, M.M.; Nooman, M.U. Production, purification and characterization of extracellular invertase from Saccharomyses Cerevisiae NRRL Y-12632 by solid-state fermentation of red carrot residue. Aust. J. Basic. Appl. Sci. 2009, 3, 1910–1919. [Google Scholar]
- Bellido, C.; Lucas, S.; González-Benito, G.; García-Cubero, M.T.; Coca, M. Synergistic positive effect of organic acids on the inhibitory effect of phenolic compounds on Acetone-Butanol-Ethanol (ABE) production. Food Bioprod. Process. 2018, 108, 117–125. [Google Scholar] [CrossRef]
- Guimaraes, C.M.; GIao, M.S.; Martinez, S.S.; Pintado, A.I.; Pintado, M.E.; Bento, L.S.; Malcata, F.X. Antioxidant activity of sugar molasses, including protective effect against DNA oxidative damage. J. Food. Sci. 2007, 72, C039–C043. [Google Scholar] [CrossRef]
- Payet, B.; Shum Cheong Sing, A.; Smadja, J. Comparison of the concentrations of phenolic constituents in cane sugar manufacturing products with their antioxidant activities. J. Agric. Food Chem. 2006, 54, 7270–7276. [Google Scholar] [CrossRef]
- Polakovič, M.; Handriková, G.; Košík, M. Inhibitory effects of some phenolic compounds on enzymatic hydrolysis of sucrose. Biomass Bioenerg. 1992, 3, 369–371. [Google Scholar] [CrossRef]
- Stamogiannou, I.; Van Camp, J.; Smagghe, G.; Van de Walle, D.; Dewettinck, K.; Raes, K. Impact of phenolic compound as activators or inhibitors on the enzymatic hydrolysis of cellulose. Int. J. Biol. Macromol. 2021, 186, 174–180. [Google Scholar] [CrossRef]
- Uribe, S.; Sampedro, J.G. Measuring solution viscosity and its effect on enzyme activity. Biol. Proced. Online 2003, 5, 108–115. [Google Scholar] [CrossRef]
- Van der Bruggen, B.; Koninckx, A.; Vandecasteele, C. Separation of monovalent and divalent ions from aqueous solution by electrodialysis and nanofiltration. Water Res. 2004, 38, 1347–1353. [Google Scholar] [CrossRef]
- Sjölin, M.; Thuvander, J.; Wallberg, O.; Lipnizki, F. Purification of sucrose in sugar beet molasses by utilizing ceramic nanofiltration and ultrafiltration membranes. Membranes 2019, 10, 5. [Google Scholar] [CrossRef]
- Nilsson, U.; Öste, R.; Jägerstad, M. Cereal fructans: Hydrolysis by yeast invertase, in vitro and during fermentation. J. Cereal Sci. 1987, 6, 53–60. [Google Scholar] [CrossRef]
- Kulshrestha, S.; Tyagi, P.; Sindhi, V.; Yadavilli, K.S. Invertase and its applications–a brief review. J. Pharm. Res. 2013, 7, 792–797. [Google Scholar] [CrossRef]
- Kramers, H.A. Brownian motion in a field of force and the diffusion model of chemical reactions. Physica 1940, 7, 284–304. [Google Scholar] [CrossRef]
- Jacob, M.; Schmid, F.X. Protein folding as a diffusional process. Biochemistry 1999, 38, 13773–13779. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.S.; Ewing, E.E.; Senesac, A.H. Inhibition of potato tuber invertase by an endogenous inhibitor: Effects of salts, pH, temperature, and sugars on binding. Plant Physiol. 1980, 66, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Escudero, I.; Ruiz, M.O. Extraction of betaine from beet molasses using membrane contactors. J. Membr. Sci. 2011, 372, 258–268. [Google Scholar] [CrossRef]
- Ghazi, I.; Fernandez-Arrojo, L.; Gomez De Segura, A.; Alcalde, M.; Plou, F.J.; Ballesteros, A. Beet sugar syrup and molasses as low-cost feedstock for the enzymatic production of fructo-oligosaccharides. J. Agric. Food Chem. 2006, 54, 2964–2968. [Google Scholar] [CrossRef] [PubMed]
- Sainz-Polo, M.A.; Ramírez-Escudero, M.; Lafraya, A.; González, B.; Marín-Navarro, J.; Polaina, J.; Sanz-Aparicio, J. Three-dimensional structure of Saccharomyces invertase: Role of a non-catalytic domain in oligomerization and substrate specificity. J. Biol. Chem. 2013, 288, 9755–9766. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Bagia, C.; Mpourmpakis, G. Determination of proton affinities and acidity constants of sugars. J. Phys. Chem. A. 2013, 117, 5211–5219. [Google Scholar] [CrossRef]
- Krieger, E.; Joo, K.; Lee, J.; Lee, J.; Raman, S.; Thompson, J.; Tyka, M.; Baker, D.; Karplus, K. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins Struct. Funct. Bioinf. 2009, 77, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Tian, W.; Chen, C.; Lei, X.; Zhao, J.; Liang, J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res. 2018, 46, W363–W367. [Google Scholar] [CrossRef]
- Everette, J.D.; Bryant, Q.M.; Green, A.M.; Abbey, Y.A.; Wangila, G.W.; Walker, R.B. Thorough study of reactivity of various compound classes toward the Folin− Ciocalteu reagent. J. Agric. Food Chem. 2010, 58, 8139–8144. [Google Scholar] [CrossRef]
| Reference | Invertase Origin | Reaction Conditions | Type of Element Effects on Invertase | |
|---|---|---|---|---|
| Inhibition | Promotion | |||
| Kumar and Kesavapillai (2012) [18] | S. cerevisiae | 5 mM salt, pH 4–10, 10–60 °C | Zn2+, Mg2+, Cu2+ | Mn2+, Al3+, Fe3+ |
| Plascencia-Espinosa et al. (2014) [15] | Candida guilliermondii | pH 5, 65 °C, 1–10 mM salt, 10% sucrose | Cu2+, Zn2+, Hg2+ | Mn2+, Co2+ |
| Takeshige and Ouchi (1995) [19] | S. cerevisiae | 10% sucrose, pH 5, 30 °C | K+, Na+, Ca2+, Mg2+, Fe3+, Mn2+, Zn2+, Cu2+ | |
| Essel and Osei (2014) [16] | S. cerevisiae | 0.1 mM salt, 40 °C, pH 5.5 | Ag+ | |
| Kaur and Sharma (2005) [20] | Streptomyces | 1 mM salt, pH 5, 50–60 °C | Cu2+, Na+, Hg2+, Ca2+ | Co2+ |
| Ginés, et al. (2000) [21] | Lactobacillus reuteri | 1 mM salt, 0.1 M sucrose, pH 5, 30 °C | Ca2+, Cu2+, Cd2+, Hg2+ | |
| Hargono, et al. (2019) [22] | S. cerevisiae | 10 g/L sucrose, 50 °C, pH 5 | Ca2+ | |
| Maruyama and Onodera (1979) [23] | Fusarium oxysporum | 20 mM salt, 3% sucrose, 30 °C | Ca2+, Zn2+, Cu2+ | (Ni2+ and Co2+ neutral effect) |
| Rashad and Nooman (2009) [24] | S. cerevisiae | 1 mM salt, 0.1 M sucrose, 30 °C | Hg2+, Ba2+, Zn2+, Fe2+ | Co2+, (Ni2+, Cu2+, Ca2+, and Mg2+ neutral effect) |
| Element | Analysis Wavelength (nm) | Crude Molasses (mg L−1 gsucrose−1) | Ultrafiltered Molasses (mg L−1 gsucrose−1) | Nanofiltered Molasses (mg L−1 gsucrose−1) | Thick Juice (mg L−1 gsucrose−1) |
|---|---|---|---|---|---|
| Ca | 317.933 | 2.17 | 1.71 | 3.33 | 0.15 |
| Cd | 228.802 | 0.00 | 0.00 | 0.00 | 0.00 |
| Cu | 327.393 | 0.01 | 0.01 | 0.07 | 0.00 |
| Fe | 238.204 | 0.01 | 0.00 | 0.05 | 0.00 |
| K | 766.490 | 67.62 | 64.86 | 60.05 | 5.74 |
| Mg | 285.213 | 0.06 | 0.07 | 0.07 | 0.00 |
| Na | 589.592 | 9.50 | 9.07 | 8.28 | 0.60 |
| P | 213.617 | 0.52 | 0.47 | 0.58 | 0.07 |
| Pb | 220.353 | 0.00 | 0.00 | 0.00 | 0 |
| Zn | 206.200 | 0.02 | 0.02 | 0.03 | 0.01 |
| PS | K ×4 Conc. | Na 80 g/L | Salt Mix 4× conc. | CM | NF | |
|---|---|---|---|---|---|---|
| Qp/x,10min | 14,567 | 8548 | 6039 | 3680 | 10,788 | 12,391 |
| Qp/x,60min | 5569 | 3727 | 2509 | 1577 | 3780 | 4389 |
| Yp/x,120min | 6415 | 5459 | 3863 | 1891 | 4997 | 5658 |
| NaCl (g L−1) | CaCl2 (g L−1) | KCl (g L−1) | MgCl2 (g L−1) | CuCl2 (g L−1) | Salt Mix I (g L−1) | Betaine (g L−1) | Gallic Acid (g L−1) | Salt Mix II (g L−1) | |
|---|---|---|---|---|---|---|---|---|---|
| NF molasses | 7.25 | 2.38 | 38.71 | 0.16 | 0.004 | 48.51 | 25.21 | 2.17 | 75.89 |
| NF 4× conc. | 25.44 | 14.65 | 152.67 | 0.83 | 0.26 | 193.87 | 100.84 | 8.68 | 303.56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sjölin, M.; Djärf, M.; Ismail, M.; Schagerlöf, H.; Wallberg, O.; Hatti-Kaul, R.; Sayed, M. Investigating the Inhibitory Factors of Sucrose Hydrolysis in Sugar Beet Molasses with Yeast and Invertase. Catalysts 2024, 14, 330. https://doi.org/10.3390/catal14050330
Sjölin M, Djärf M, Ismail M, Schagerlöf H, Wallberg O, Hatti-Kaul R, Sayed M. Investigating the Inhibitory Factors of Sucrose Hydrolysis in Sugar Beet Molasses with Yeast and Invertase. Catalysts. 2024; 14(5):330. https://doi.org/10.3390/catal14050330
Chicago/Turabian StyleSjölin, Mikael, Maria Djärf, Mohamed Ismail, Herje Schagerlöf, Ola Wallberg, Rajni Hatti-Kaul, and Mahmoud Sayed. 2024. "Investigating the Inhibitory Factors of Sucrose Hydrolysis in Sugar Beet Molasses with Yeast and Invertase" Catalysts 14, no. 5: 330. https://doi.org/10.3390/catal14050330








