Chloroplast Haplotype Diversity in the White Oak Populations of the Italian Peninsula, Sicily, and Sardinia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Sampling and Plant Material
2.2. DNA Extraction and cpSSR Amplification
2.3. Data Analysis
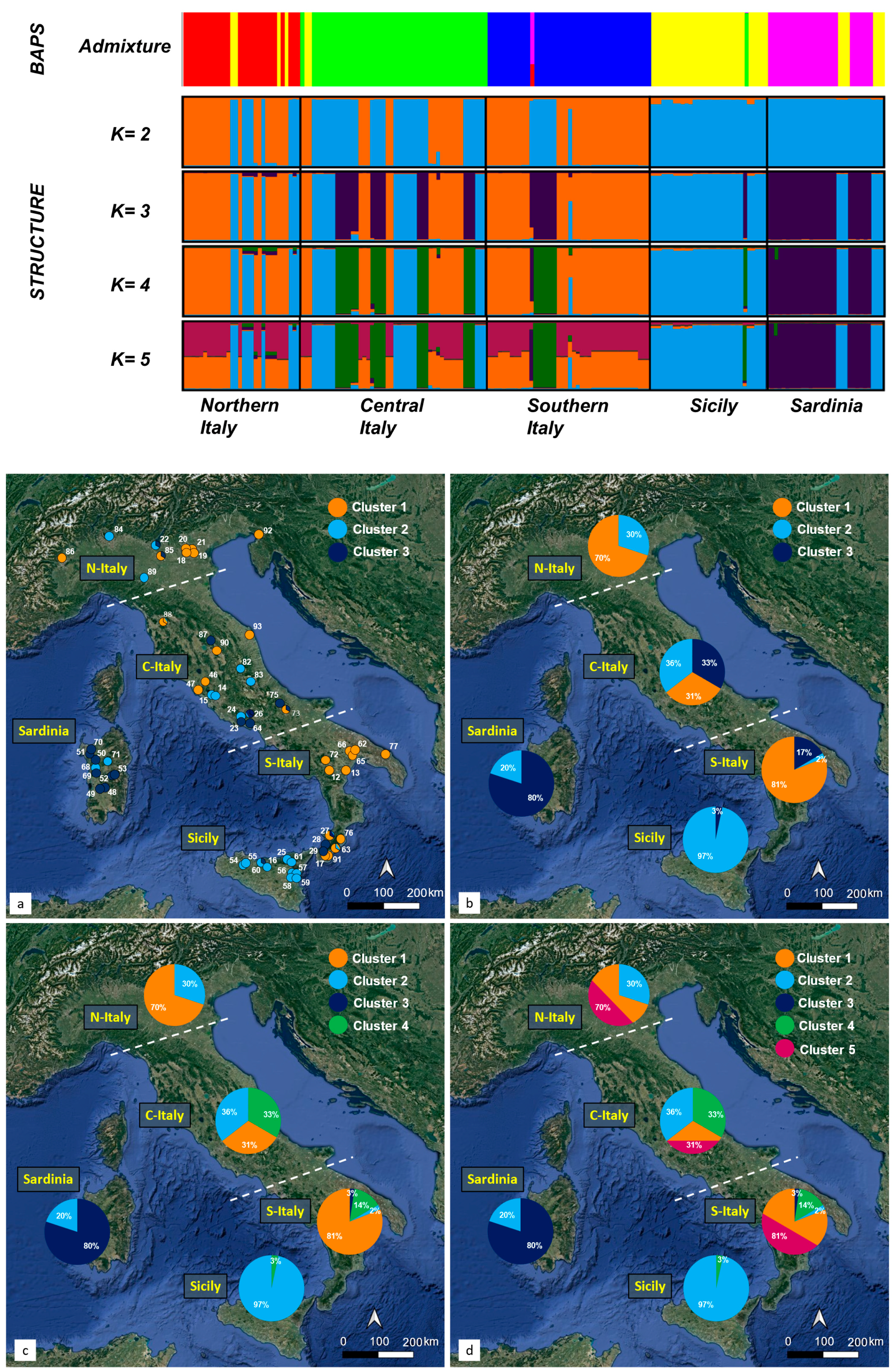
3. Results
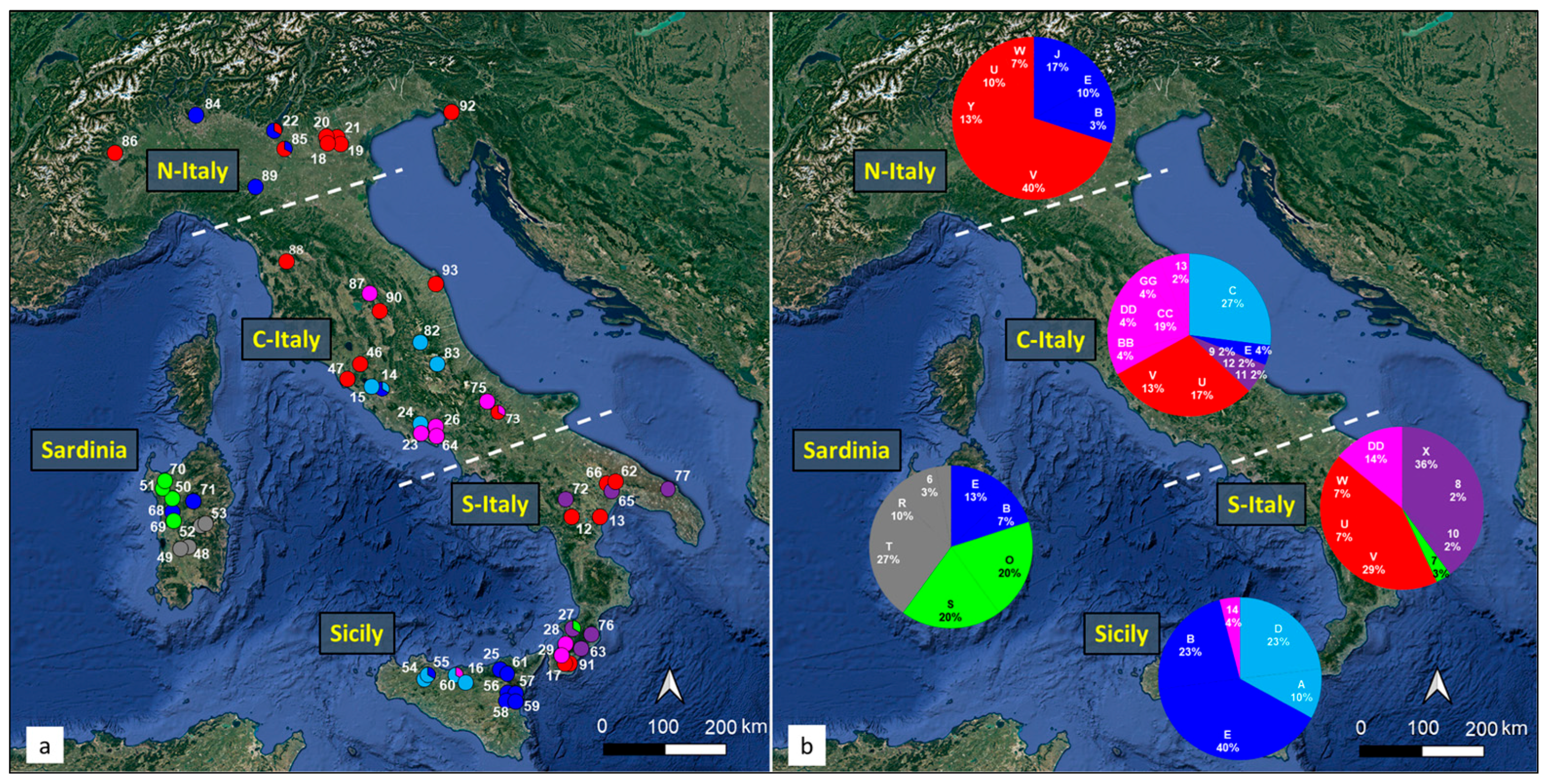
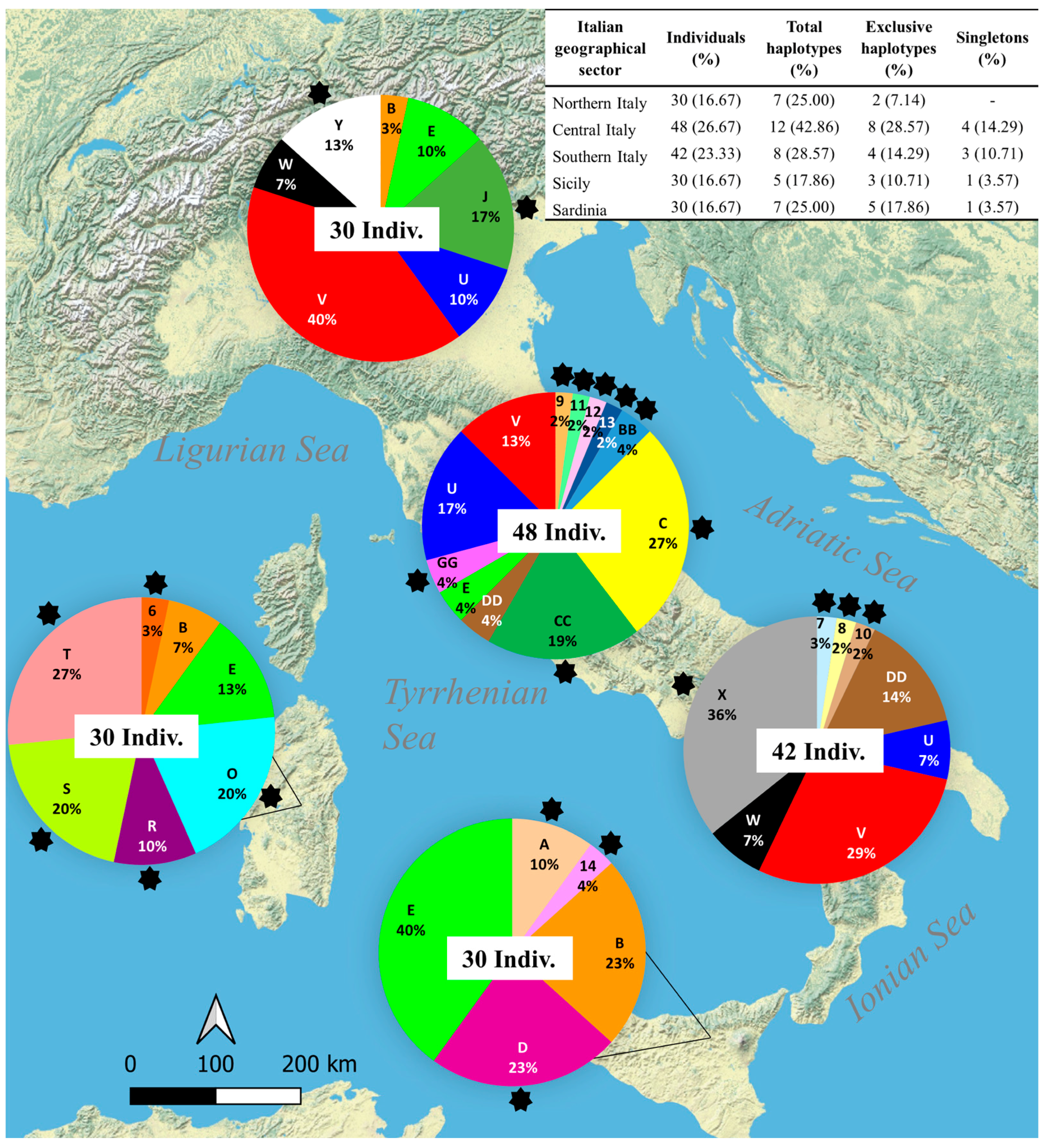
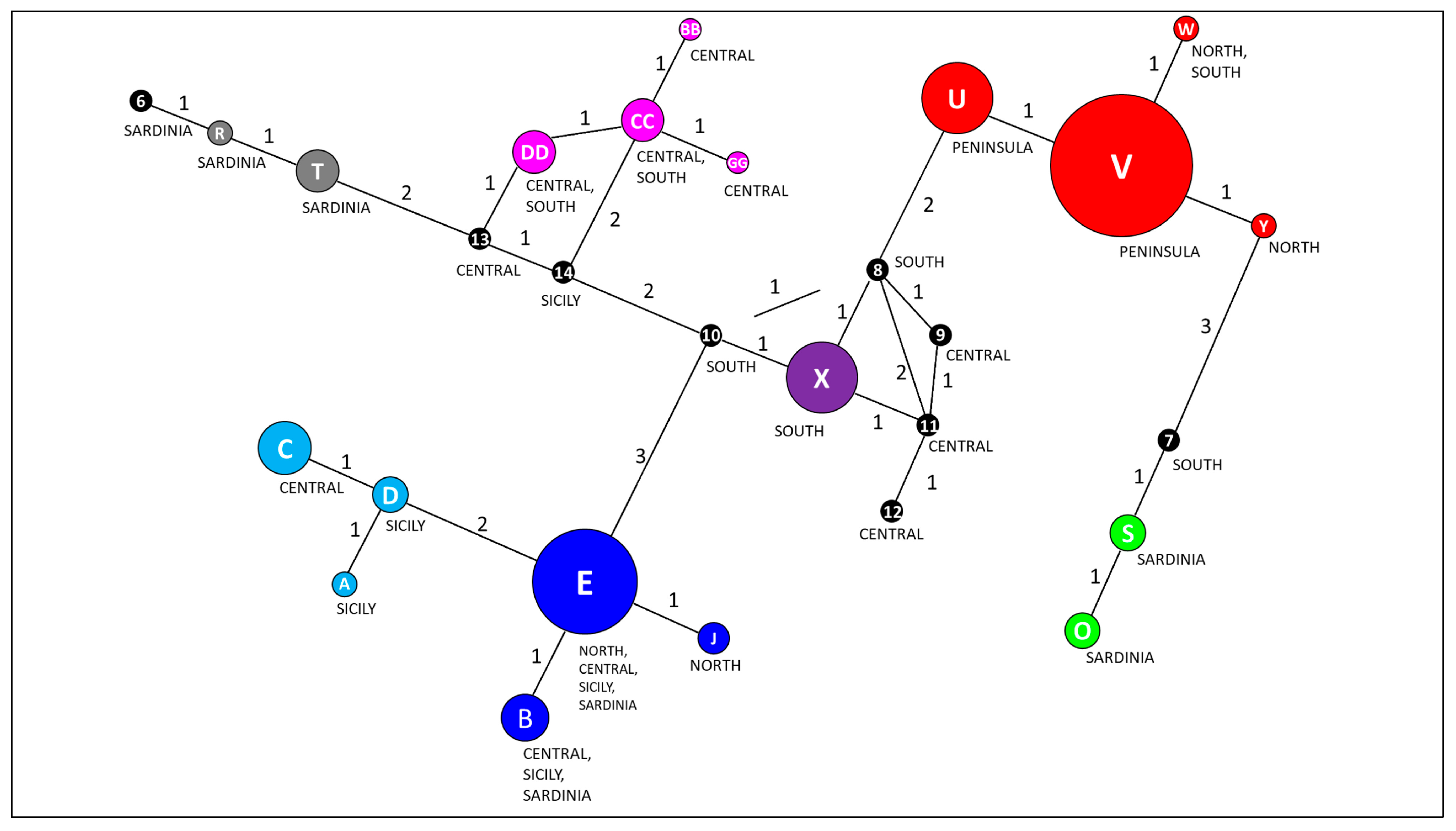
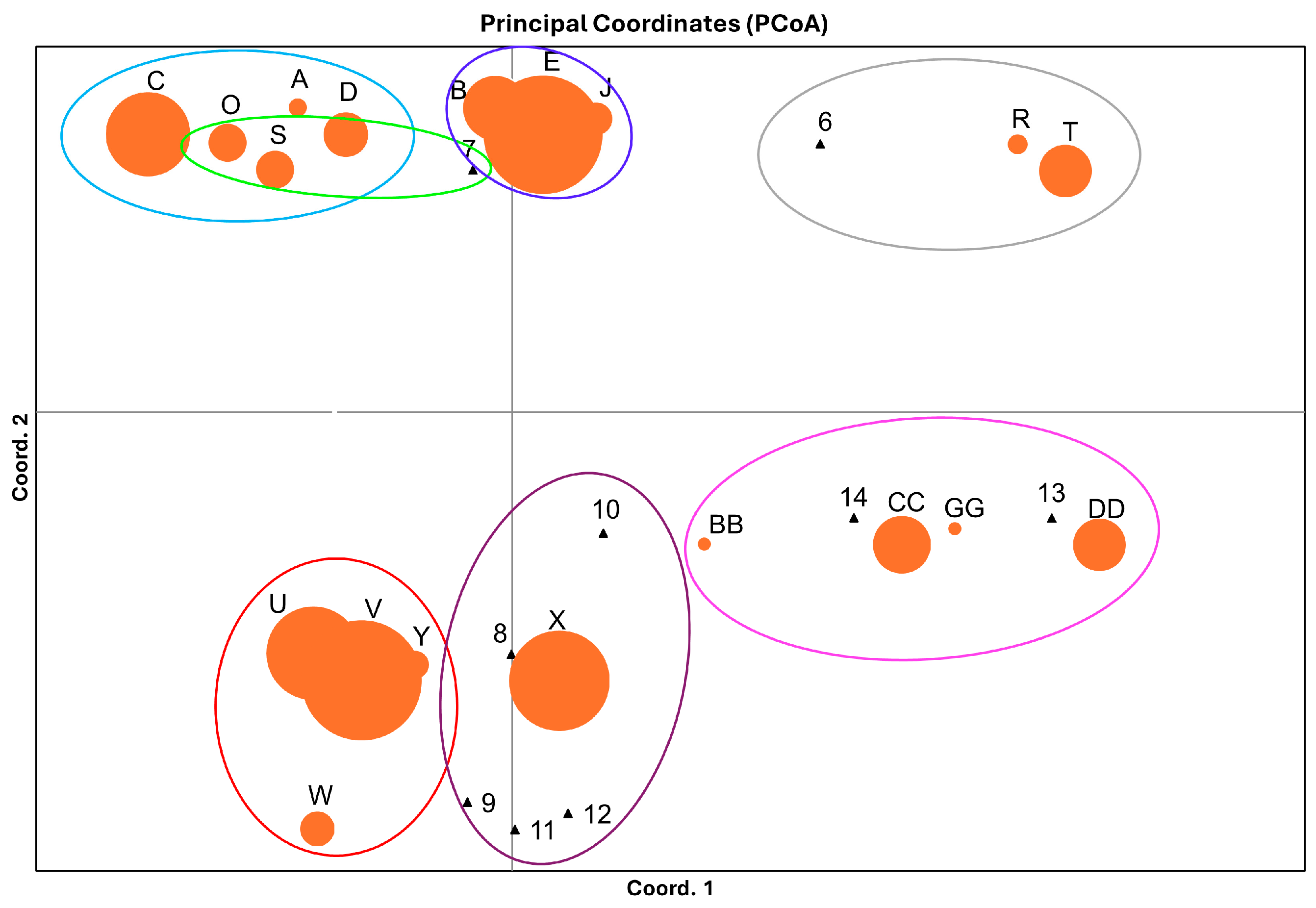
3.1. Overall Haplotype Distribution
3.2. Haplotype Diversity and Geographical Distribution
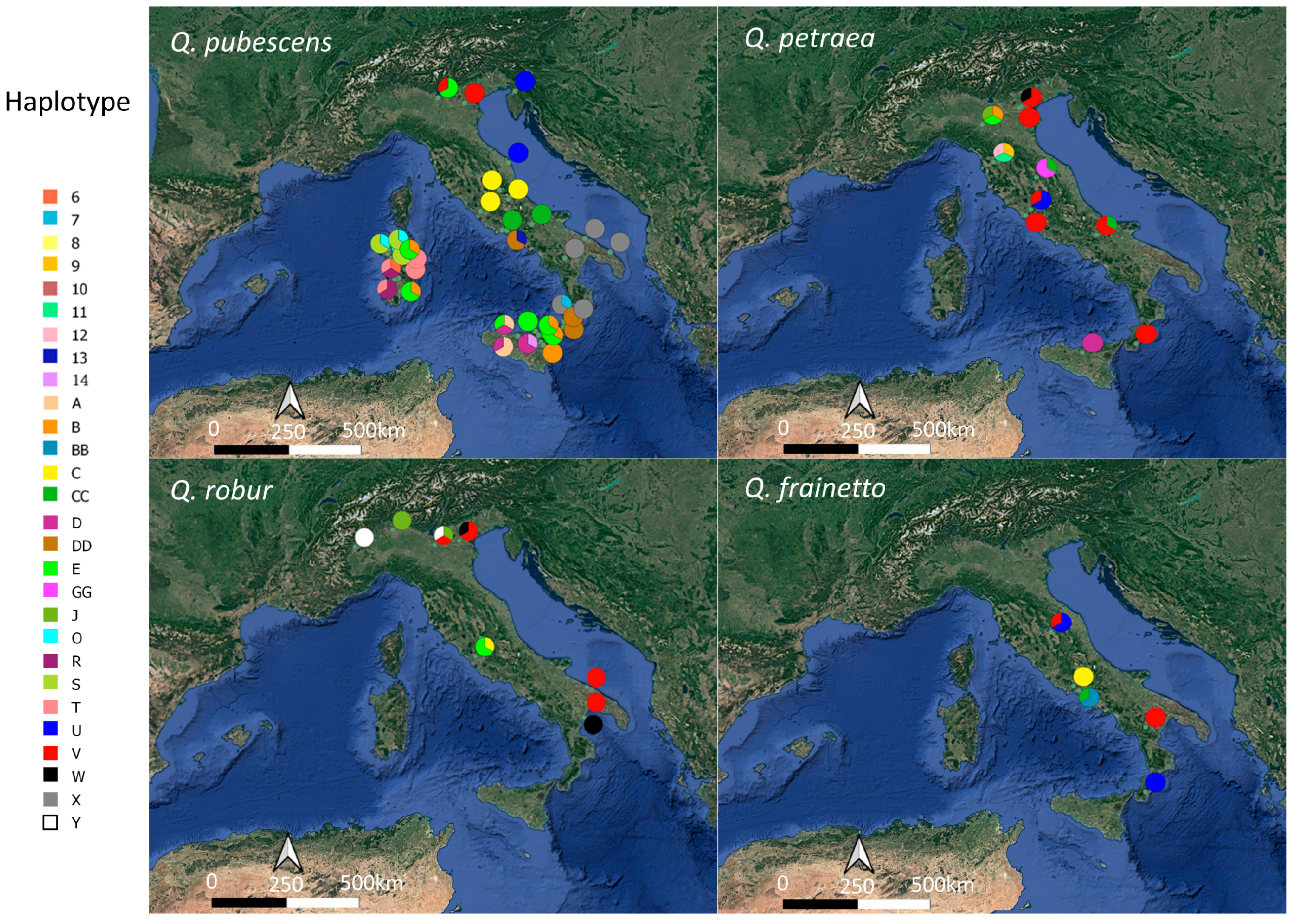
3.3. Genetic Diversity and Haplotype Distribution According to Taxonomy
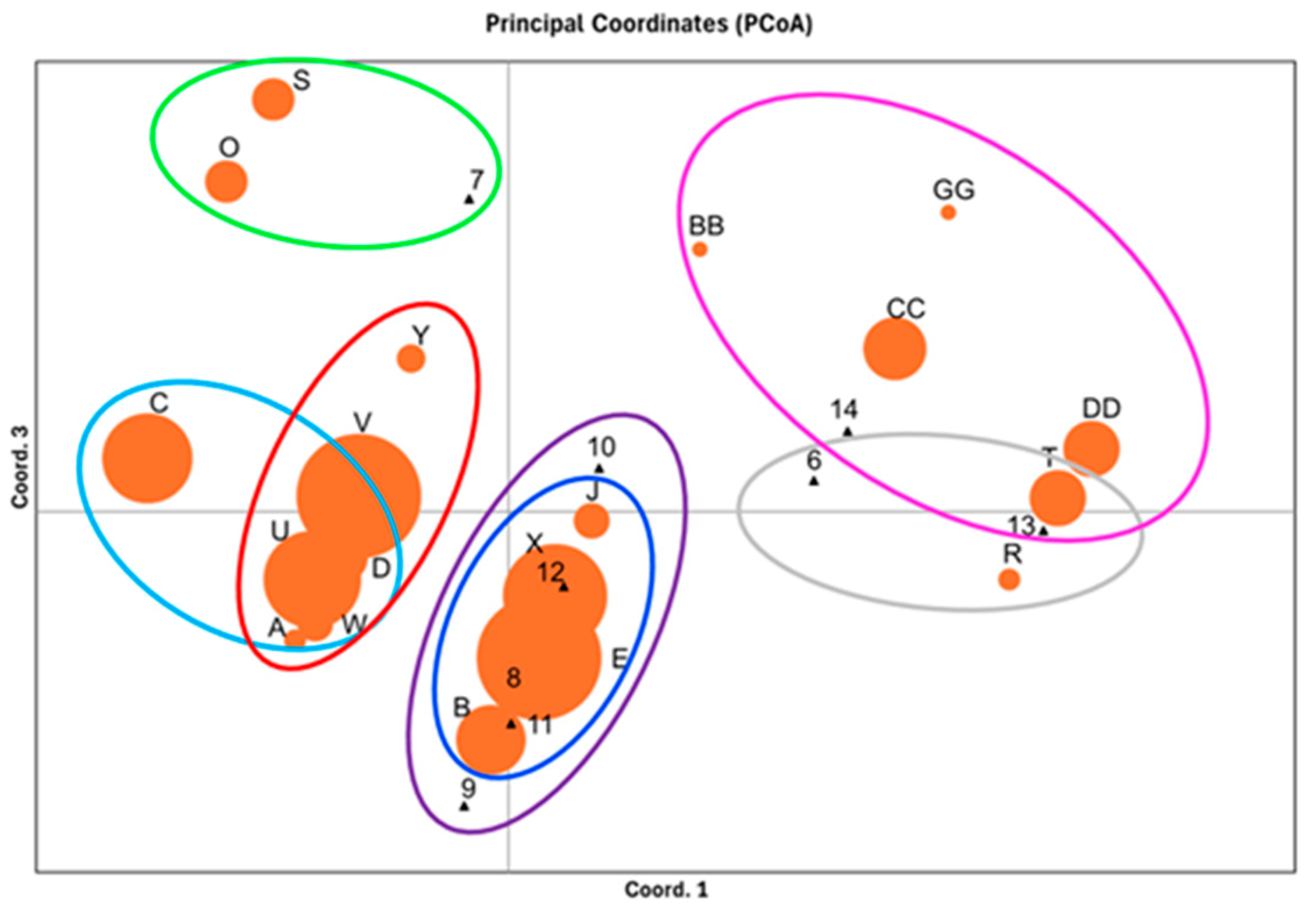
3.4. Genetic Structure
4. Discussion
4.1. Haplotype Distribution and Paleoecological and Paleogeographic Events
4.2. Haplotype Distribution vs. Taxonomical Identity
4.3. Haplotype Distribution vs. Geographical Structure
4.4. What Has Changed Compared to Previous Investigations?
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Pop | Species Complex | Taxon | BR 1 | Lith. 2 | Alt. | Latitude (N); Longitude (E) (WGS84) Adm. Region—Sector 3 |
|---|---|---|---|---|---|---|
| IT12 | - | Q. frainetto | To | Tes | 725 | 40.2422; 16.0551 Basilicata—S-IT |
| IT13 | - | Q. robur | Mo | Cls | 5 | 40.1741; 16.6997 Basilicata—S-IT |
| IT14 | - | Q. robur | tMo | Tcs | 112 | 41.5515; 12.2112 Lazio—C-IT |
| IT15 | Q. pub. | Q. pubescens | tMo | Ei | 105 | 41.9546; 12.4195 Lazio—C-IT |
| IT16 | Q. petr. | Q. petraea subsp. austrotyrrhenica | To | Cas | 1408 | 37.8583; 14.0580 Sicily—SIC |
| IT17 | Q. petr. | Q. petraea subsp. austrotyrrhenica | Mo | M | 195 | 38.14604; 16.0599 Calabria—S-IT |
| IT18 | Q. pub. | Q. pubescens | Ts | Cas | 42 | 45.2745; 11.7416 Veneto—N-IT |
| IT19 | Q. petr. | Q. dalechampii | Ts | Ei | 10 | 45.3430; 11.7617 Veneto—N-IT |
| IT20 | Q. petr. | Q. petraea | Ts | Ei | 242 | 45.3173; 11.6877 Veneto—N-IT |
| IT21 | - | Q. robur | Ts | Ei | 56 | 45.3227; 11.7408 Veneto—N-IT |
| IT22 | Q. pub. | Q. pubescens | Ts | Cls | 76 | 45.4319; 10.6256 Lombardia- N-IT |
| IT23 | Q. pub. | Q. pubescens | Mo | Tcs | 16 | 41.3640; 13.3391 Lazio—C-IT |
| IT24 | - | Q. frainetto | tMo | Ei | 89 | 41.4862; 13.3105 Lazio—C-IT |
| IT25 | Q. pub. | Q. virgiliana | tMo | Cas | 669 | 38.0689; 14.7373 Sicily—SIC |
| IT26 | - | Q. frainetto | tTo | Cas | 576 | 41.3588; 13.5265 Lazio—C-IT |
| IT27 | Q. pub. | Q. dalechampii | Mo | Tcs | 260 | 38.6265; 16.1628 Calabria—S-IT |
| IT28 | Q. pub. | Q. dalechampii | Mo | Tcs | 70 | 38.3778; 15.9396 Calabria—S-IT |
| IT29 | Q. pub. | Q. congesta | Mo | Ii | 980 | 38.2107; 15.9163 Calabria—S-IT |
| IT46 | Q. petr. | Q. petraea | To | Ei | 609 | 42.3236; 12.1302 Lazio—C-IT |
| IT47 | Q. petr. | Q. petraea | tMo | Ei | 358 | 42.1469; 11.9407 Lazio—C-IT |
| IT48 | Q. pub. | Q. ichnusae | tMo | Cas/Ei | 283 | 39.8158; 8.9486 Sardinia—SAR |
| IT49 | Q. pub. | Q. virgiliana | tMo | Cas | 567 | 39.7871; 8.7892 Sardinia—SAR |
| IT50 | Q. pub. | Q. ichnusae | Mo | M | 529 | 40.4685; 8.6721 Sardinia—SAR |
| IT51 | Q. pub. | Q. virgiliana | Mo | Ei | 246 | 40.6136; 8.5034 Sardinia—SAR |
| IT52 | Q. pub. | Q. congesta | tMo | Ii | 831 | 40.1096; 9.2489 Sardinia—SAR |
| IT53 | Q. pub. | Q. congesta | Mo | Ii | 921 | 40.1289; 9.3027 Sardinia—SAR |
| IT54 | Q. pub. | Q. leptobalana | Mo | Cas | 919 | 37.8841; 13.3831 Sicily—SIC |
| IT55 | Q. pub. | Q. virgiliana | tMo | Cas | 538 | 37.9563; 13.4239 Sicily—SIC |
| IT56 | Q. pub. | Q. congesta | tMo | Ei | 1298 | 37.7095; 14.9621 Sicily—SIC |
| IT57 | Q. pub. | Q. dalechampii | To | Ei | 1400 | 37.7062; 14.9672 Sicily—SIC |
| IT58 | Q. pub. | Q. virgiliana | tMo | M | 544 | 37.5917; 15.0447 Sicily—SIC |
| IT59 | Q. pub. | Q. virgiliana | tMo | M | 540 | 37.6086; 15.0723 Sicily—SIC |
| IT60 | Q. pub. | Q. congesta | tMo | Cas | 1179 | 37.9054; 13.9936 Sicily—SIC |
| IT61 | Q. pub. | Q. congesta | tMo | Cas | 1327 | 37.9415- 14.8762 Sicily—SIC |
| IT62 | - | Q. robur | Mo | Tcs | 342 | 40.6259; 16.8121 Puglia—S-IT |
| IT63 | Q. pub. | Q. congesta | tMo | Ii | 1190 | 38.4813; 16.3411 Calabria—S-IT |
| IT64 | Q. pub. | Q. pubescens | tTo | Cas | 576 | 41.3588; 13.5266 Lazio—C-IT |
| IT65 | - | Q. robur | Mo | Cas | 39 | 41.0925; 16.8223 Puglia—S-IT |
| IT66 | Q. pub. | Q. virgiliana | Mo | Cas | 39 | 41.0925; 16.8223 Puglia—S-IT |
| IT68 | Q. pub. | Q. pubescens | Mo | Cls | 91 | 38.0689; 9.0594 Sardinia—SAR |
| IT69 | Q. pub. | Q. ichnusae | Mo | M | 780 | 40.2382; 8.6978 Sardinia—SAR |
| IT70 | Q. pub. | Q. congesta | Mo | Ei | 147 | 40.7461; 8.5361 Sardinia—SAR |
| IT71 | Q. pub. | Q. congesta | Mo | Ii | 916 | 40.4277; 9.0069 Sardinia—SAR |
| IT72 | Q. pub. | Q. pubescens | To | Tes | 796 | 40.4600; 15.9745 Basilicata—S-IT |
| IT73 | Q. petr. | Q. petraea | To | C | 610 | 41.3316; 14.3650 Molise—C-IT |
| IT75 | Q. pub. | Q. pubescens | To | Cas | 610 | 41.3302; 14.3701 Molise—C-IT |
| IT76 | Q. pub. | Q. pubescens | To | Ii | 1091 | 38.4813;16.3411 Calabria—S-IT |
| IT77 | Q. pub. | Q. pubescens | Mo | Tcs | 23 | 40.6584;17.887 Puglia—S-IT |
| IT82 | Q. pub. | Q. pubescens | To | Cas | 1014 | 42.6205; 13.2173 Lazio—C-IT |
| IT83 | Q. pub. | Q. pubescens | To | Cas | 1303 | 42.3362; 13.5901 Abruzzo—C-IT |
| IT84 | - | Q. robur | Ts | Cls | 202 | 45.6213; 9.0943 Lombardia—N-IT |
| IT85 | - | Q. robur | Ts | Tcs | 21 | 45.2045; 10.7534 Lombardia—N-IT |
| IT86 | - | Q. robur | Ts | Cls | 276 | 45.1485; 7.5962 Piemonte—N-IT |
| IT87 | Q. petr. | Q. petraea | To | Tes | 346 | 43.2837; 12.3420 Umbria—C-IT |
| IT88 | Q. petr. | Q. petraea | To-s | Cls | 67 | 43.7353; 10.7334 Toscana—C-IT |
| IT90 | - | Q. frainetto | tTo-s | Tcs | 204 | 43.0946;12.4672 Umbria—C-IT |
| IT91 | - | Q. frainetto | Mo | Tcs | 347 | 38.22470- 16.0453 Calabria—S-IT |
| IT93 | Q. pub. | Q. pubescens | Ts | Cas | 458 | 45.6282;13.8784 Friuli—N-IT |
| IT93 | Q. pub. | Q. pubescens | tTo-s | Cls | 10 | 43.4676; 13.5917 Marche—C-IT |
| udt1 | udt1_81 | 0.678 |
| udt1_82 | 0.311 | |
| udt1_83 | 0.011 | |
| ucd5 | ucd5_74 | 0.217 |
| ucd5_75 | 0.667 | |
| ucd5_77 | 0.117 | |
| ucd4 | ucd4_94 | 0.139 |
| ucd4_95 | 0.356 | |
| ucd4_97 | 0.322 | |
| ucd4_98 | 0.072 | |
| ucd4_99 | 0.111 | |
| udt3 | udt3_125 | 0.139 |
| udt3_126 | 0.461 | |
| udt3_127 | 0.356 | |
| udt3_128 | 0.044 | |
| Cmcs6 | CmCs6_200 | 0.328 |
| CmCs6_202 | 0.139 | |
| CmCs6_203 | 0.533 | |
| Mean | 0.278 |
| Pop. | Species Complex | Taxon | Geograph. Sector | Site of Collection (Adm. Province) | Syntaxonomy |
|---|---|---|---|---|---|
| IT12 | - | Q. frainetto | S Italy | Rustico, San Mart. d’Agri (PZ) | Melittio albidae-Quercion frainetto Quezel et al. in Bonin & Gamisans 1976 |
| IT13 | - | Q. robur | S Italy | Oasi Pantano di Policoro (MT) | Rubio peregrinae-Fraxinetum oxycarpae Biondi & Allegrezza 2004 var with Q. robur |
| IT14 | - | Q. robur | C Italy | Insugherata (RM) | Rubio peregrinae-Quercetum cerris Di Pietro et al. 2010 var. with Q. robur |
| IT15 | Q. pub. | Q. pubescens | C Italy | Insugherata (RM) | Roso sempervirentis-Quercetum pubescentis Biondi 1986 |
| IT16 | Q. petr. | Q. petraea subsp. austrothyrr. | S Italy | Bosco Pomieri, Geraci Siculo (PA) | Ilici-Quercetum austrotyrrhenicae Brullo & Marcenò in Brullo 1984 corr. Brullo 2002 |
| IT17 | Q. petr. | Q. petraea subsp. austrothyrr. | S Italy | Pollia (Asprom.). San Luca | Aristolochio luteae-Quercetum austrotyrrhenicae Brullo et al. 1999 |
| IT18 | Q. pub. | Q. pubescens | N Italy | Colli Euganei (PD) | Fraxino orni-Ostryon carpinifoliae Tomazic 1940 (Quercetalia pubescenti-petraeae) |
| IT19 | Q. petr. | Q. petraea | N Italy | Colle S.Daniele (PD) | Physospermo-Quercion petraeae A.O. Horvat 1976 |
| IT20 | Q. petr. | Q. petraea | N Italy | M. Venda (PD) | Melampyro vulgati-Quercetum petraeae Puncer et Zupančić 1979 |
| IT21 | Q. robur | Q. robur | N Italy | Torreglia; M.Rua; Roccapendice (PD) | Asparago tenuifolii-Quercetum roboris (Lausi 1967). Marincek 1994 |
| IT22 | Q. pub. | Q. pubescens | N Italy | Ome, San Martino (BS) | Quercetalia pubescenti-petraeae Klika 1933 |
| IT23 | Q. pub. | Q. pubescens | C Italy | Monte San Biagio (LT) | Pistacio terebinthi-Quercetum pubescentis (Blasi et Di Pietro 1998) Allegrezza et al. 2003 |
| IT24 | - | Q. frainetto | C Italy | Selvapiana Amaseno (FR) | Quercus frainetto community (Crataego-Quercion cerridis Arrigoni 1998) |
| IT25 | Q. pub. | Q. virgiliana | S Italy | Valle del Fitalia (Frazzanò, ME) | Erico arborae-Quercetum virgilianae Brullo et Marcenò 1985 |
| IT26 | - | Q. frainetto | C Italy | Parco Monti Aurunci (LT) | Quercus frainetto community (Crataego-Quercion cerridis Arrigoni 1998) |
| IT27 | Q. pub. | Q. dalechampii | S Italy | Serre, Sant’Angelo-Pizzoni (VV) | indivisuals of Q. dalechampii in the Erico arborae-Quercetum virgilianae Brullo e Marcenò 1985 |
| IT28 | Q. pub. | Q. dalechampii | S Italy | Aspromonte, Croce Mammone (RC) | indivisuals of Q. dalechampii in the Oleo oleaster-Quercetum virgilianae Brullo 1984 |
| IT29 | Q. pub. | Q. congesta | S Italy | Aspromonte, Piani di Carmelia (RC) | Erico-Quercetum congestae Brullo et al. 2001 |
| IT46 | Q. petr. | Q. petraea | C Italy | Monti Cimini (VT) | individuals of Q. petraea in Coronillo emeri-Quercetum cerridis Blasi 1984 var. with Q. petraea |
| IT47 | Q. petr. | Q. petraea | C Italy | Tolfa mountains (Roma) | Carici olbiensis-Quercetum petraeae Di Pietro et al. 2010 |
| IT48 | Q. pub. | Q. ichnusae | Sardinia | Senis (OR) | Ornithogalo pyrenaici-Quercetum ichnusae Bacchetta et al. 2004 |
| IT49 | Q. pub. | Q. virgiliana | Sardinia | Pau (OR) | Lonicero implexae-Quercetum virgilianae Bacchetta et al. 2004 |
| IT50 | Q. pub. | Q. ichnusae | Sardinia | Monte Traessu (SS) | Ornithogalo pyrenaici-Quercetum ichnusae Bacchetta et al. 2004 |
| IT51 | Q. pub. | Q. virgiliana | Sardinia | Ittiri SS) | Lonicero implexae-Quercetum virgilianae Bacchetta et al. 2004 |
| IT52 | Q. pub. | Q. congesta | Sardinia | Fonni (Muggiana) (NU) | Glechomo sardoae-Quercetum congestae Bacchetta et al. 2004 |
| IT53 | Q. pub. | Q. congesta | Sardinia | Fonni (Govossai) (NU) | Glechomo sardoae-Quercetum congestae Bacchetta et al. 2004 |
| IT54 | Q. pub. | Q. leptobalana | Sicily | Bosco Ficuzza (PA) | Quercetum leptobalani Brullo 1984 |
| IT55 | Q. pub. | Q. virgiliana | Sicily | Marineo (PA) | Oleo-Quercetum virgilianae Brullo 1984 |
| IT56 | Q. pub. | Q. congesta | Sicily | Etna Volcano (CT) | Arabido turritae-Quercetum congestae Brullo e Marcenò 1985 |
| IT57 | Q. pub. | Q. dalechampii | Sicily | Etna Volcano (CT) | Quercenion dalechampii Brullo 1984 |
| IT58 | Q. pub. | Q. virgiliana | Sicily | Etna. M. Ceraulo, Mascalucia (CT) | Oleo-Quercetum virgilianae Brullo 1984 |
| IT59 | Q. pub. | Q. virgiliana | Sicily | Etna. Tre castagni, (CT) | Celtido aetnensis-Quercetum virgilianae Brullo e Marcenò 1985 |
| IT60 | Q. pub. | Q. congesta | Sicily | Madonie, Piani Torre-Zucchi (PA) | Conopodio capillifolii-Quercetum congestae Maniscalco & Raimondo 2009 |
| IT61 | Q. pub. | Q. congesta | Sicily | Valle del Flascio. Nebrodi (ME) | Festuco heterophyllae-Quercetum congestae Brullo & Marcenò 1986 |
| IT62 | Q. robur | Q. robur | S Italy | Tafuri (BA) | Quercus robur isolated individuals in Stipo-Quercetum dalechampii Biondi et al. 2004 |
| IT63 | Q. pub. | Q. congesta | S Italy | Serre, Sant’Angelo Nardodipace (VV) | Festuco heterophyllae-Quercetum congestae Brullo & Marcenò 1986 |
| IT64 | Q. pub. | Q. pubescens | C Italy | Parco Monti Aurunci (LT) | Pistacio terebinthi-Quercetum pubescentis (Blasi & Di Pietro 1998) Allegrezza et al. 2002 |
| IT65 | Q. robur | Q. robur | S Italy | Tafuri, Murgean Plateau (BA) | Quercus robur isolated individuals in Stipo-Quercetum dalechampii Biondi et al. 2004 |
| IT66 | Q. pub. | Q. virgiliana | S Italy | Selva S. Vito Gravina Laterza (TA) | individuals of Q. virgiliana in the Teucrio siculi-Quercetum trojanae Biondi et al. 2004 |
| IT68 | Q. pub. | Q. pubescens | Sardinia | M.te Zara. Monastir (CA) | Lonicero implexae-Quercetum virgilianae Bacchetta et al. 2004 |
| IT69 | Q. pub. | Q. ichnusae | Sardinia | Bosco S. Antonio. Macomer (NU) | Ornithogalo pyrenaici-Quercetum ichnusae Bacchetta et al. 2004 |
| IT70 | Q. pub. | Q. congesta | Sardinia | Sant’Orsola (SS) | Glechomo sardoae-Quercetum congestae Bacchetta et al. 2004 |
| IT71 | Q. pub. | Q. congesta | Sardinia | M. Rasu, Catena del Marghine (SS) | Glechomo sardoae-Quercetum congestae Bacchetta et al. 2004 |
| IT72 | Q. pub. | Q. pubescens | S Italy | Laurenzana (PZ) | Centaureo centaurii-Quercetum pubescentis Ubaldi & Zanotti 1995 |
| IT73 | Q. petr. | Q. petraea | C Italy | Monte Vairano (CB) | individuals of Q. petraea in Aremonio-Quercetum cerridis Blasi et al. ex Terzi et al. 2022 |
| IT75 | Q. pub. | Q. pubescens | C Italy | Monte Vairano (CB) | Cytiso sessilifolii-Quercetum pubescentis Blasi et al. 1982 |
| IT76 | Q. pub. | Q. pubescens | S Italy | Mar di Pace—Nardodipace (VV) | Erico-Quercetum congestae Brullo et al. 2001 |
| IT77 | Q. pub. | Q. pubescens | S Italy | Bosco del Compare (BR) | Individuals of Q. pubescens in Fraxino orni-Quercetum ilicis Horvatic (1956) 1958 |
| IT82 | Q. pub. | Q. pubescens | C Italy | Torrita, Monti della Laga (RI) | Cytiso sessilifolii-Quercetum pubescentis Blasi et al. 1982 |
| IT83 | Q. pub. | Q. pubescens | C Italy | Barisciano, San Colombo (AQ) | Cytiso sessilifolii-Quercetum pubescentis Blasi et al. 1982 |
| IT84 | - | Q. robur | N Italy | Parco delle Groane—Solaro (MI) | Polygonato multiflori—Quercetum roboris Sartori 1984 |
| IT85 | - | Q. robur | N Italy | Bosco Fontana—Marmirolo (MN) | Polygonato multiflori—Quercetum roboris Sartori 1984 |
| IT86 | - | Q. robur | N Italy | Parco La Mandria Ven. Reale (TO) | Quercus robur community belonging to Carpinion betuli Issler 1931 |
| IT87 | Q. petr. | Q. petraea | C Italy | Montecorona Umbertide (PG) | Hieracio racemosi-Quercetum petraeae Pedrotti et al. 1982 |
| IT88 | Q. petr. | Q. petraea | C Italy | Parco Castelfranco di Sotto (PI) | individuals of Q. petraea in Melico-Quercetum cerridis Arrigoni, in Arrigoni et al. 1990 |
| IT89 | Q. petr. | Q. petraea | N Italy | Parco dei boschi di Carrega (PR) | Physospermo cornubiensis-Quercetum petraeae Oberdorfer & Hofmann 1967 |
| IT90 | - | Q. frainetto | C Italy | Collestrada (PG) | Malo florentinae-Quercetum frainetto Biondi et al. 2001 ex Terzi et al. 2022 |
| IT91 | - | Q. frainetto | S Italy | Parco Naz. Aspromonte—Platì (RC) | Cytiso-Quercetum frainetto Scelsi & Spampinato 1996 |
| IT92 | Q. pub. | Q. pubescens | N Italy | Basovizza (TS) | Ostryo-Quercetum pubescentis (Horvat 1959). Trinajstić 1974. |
| IT93 | Q. pub. | Q. pubescens | C Italy | Selva di Castelfidardo (AN) | Roso sempervirentis-Quercetum pubescentis Biondi 1986 |


References
- Hampe, A.; Petit, R.J. Conserving biodiversity under climate change: The rear edge matters. Ecol. Lett. 2005, 8, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Denk, T.; Grimm, G.W.; Manos, P.S.; Deng, M.; Hipp, A.L. An updated infrageneric classification of the oaks: Review of previous taxonomic schemes and synthesis of evolutionary patterns. In Oaks Physiological Ecology. Exploring the Functional Diversity of Genus Quercus L.; Gil-Pelegrín, E., Peguero-Pina, J.J., Sancho-Knapik, D., Eds.; Tree Physiology; Springer International Publishing: Cham, Switzerland, 2017; pp. 13–38. ISBN 978-3-319-69099-5. [Google Scholar]
- Govaerts, R.; Nic Lughadha, E.; Black, N.; Turner, R.; Paton, A. The world checklist of vascular plants, a continuously updated resource for exploring global plant diversity. Sci. Data 2021, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Mucina, L.; Bültmann, H.; Dierßen, K.; Theurillat, J.-P.; Raus, T.; Čarni, A.; Šumberová, K.; Willner, W.; Dengler, J.; García, R.G.; et al. vegetation of Europe: Hierarchical floristic classification system of vascular plant, bryophyte, lichen, and algal communities. Appl. Veg. Sci. 2016, 19, 3–264. [Google Scholar] [CrossRef]
- Plomion, C.; Martin, F. Oak genomics is proving its worth. New Phytol. 2020, 226, 943–946. [Google Scholar] [CrossRef] [PubMed]
- Brullo, S.; Guarino, R.; Siracusa, G. Taxonomical revision about the deciduous oaks of Sicily. Webbia 1999, 54, 1–72. [Google Scholar] [CrossRef]
- Di Pietro, R.; Viscosi, V.; Peruzzi, L.; Fortini, P. A Review of the application of the name Quercus dalechampii. Taxon 2012, 61, 1311–1316. [Google Scholar] [CrossRef]
- Fortini, P.; Di Marzio, P.; Di Pietro, R. Differentiation and hybridization of Quercus frainetto, Q. petraea, and Q. pubescens (Fagaceae): Insights from macro-morphological leaf traits and molecular data. Plant Syst. Evol. 2015, 301, 375–385. [Google Scholar] [CrossRef]
- Wellstein, C.; Spada, F. The status of Quercus pubescens Willd. in Europe. In Warm-Temperate Deciduous Forests around the Northern Hemisphere; Box, E.O., Fujiwara, K., Eds.; Geobotany Studies; Springer International Publishing: Cham, Switzerland, 2015; pp. 153–163. ISBN 978-3-319-01261-2. [Google Scholar]
- Kučera, P. New name for central europaean oak formerly labelled as Quercus dalechampii. Biologia 2018, 73, 313–317. [Google Scholar] [CrossRef]
- Grossoni, P.; Bruschi, P.; Bussotti, F.; Pollastrini, M.; Selvi, F. The taxonomic interpretation of Mediterranean oaks of Quercus Sect. Quercus (Fagaceae): Uncertainties and diverging concepts. Flora Mediterr. 2021, 31, 271–278. [Google Scholar] [CrossRef]
- Gasparini, P.; Papitto, G. The italian forest inventory in brief. In Italian National Forest Inventory—Methods and Results of the Third Survey: Inventario Nazionale delle Foreste e dei Serbatoi Forestali di Carbonio—Metodi e Risultati della Terza Indagine; Gasparini, P., Di Cosmo, L., Floris, A., De Laurentis, D., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 1–15. ISBN 978-3-030-98678-0. [Google Scholar]
- Presl, C.B. Quercus. In Deliciae Pragensis Historiam Naturalem Spectantes; Presl, J.S., Presl, C.B., Eds.; Sumtibus Calve: Pragae, Czech Republic, 1822; Volume 1, p. 32. [Google Scholar]
- Gussone, G. Florae Siculae Synopsis; Ex Typis Tramater: Naples, Italy, 1842; Volume 2. [Google Scholar]
- Dimopoulos, P.; Raus, T.; Bergmeier, E.; Constantinidis, T.; Iatrou, G.; Kokkini, S.; Strid, A.; Tzanoudakis, D. Vascular Plants of Greece: An Annotated Checklist; Botanic Garden and Botanical Museum Berlin-Dahlem: Berlin, Germany; Hellenic Botanical Society: Athens, Greece, 2013. [Google Scholar]
- Pignatti, S.; Guarino, R.; La Rosa, M. Flora d’Italia, 2nd ed.; Edagricole: Bologna, Italy, 2017; Volume 4, ISBN 88-506-5242-9. [Google Scholar]
- Bartolucci, F.; Peruzzi, L.; Galasso, G.; Albano, A.; Alessandrini, A.; Ardenghi, N.M.G.; Astuti, G.; Bacchetta, G.; Ballelli, S.; Banfi, E.; et al. An updated checklist of the vascular flora native to Italy. Plant Biosyst. 2018, 152, 179–303. [Google Scholar] [CrossRef]
- Kaplan, Z.; Danihelka, J.; Chrtek, J.; Prančl, J.; Grulich, V.; Jelinek, B.; Úradníček, L.; Řepka, R.; Smarda, P.; Vašut, R.; et al. Distributions of vascular plants in the Czech Republic. Part 11. Preslia 2022, 94, 335–427. [Google Scholar] [CrossRef]
- Tenore, M. Semina Anno 1830 Collecta, Quae in Horto Botanico Neapolitano pro Mutua Commutatione Offeruntur; Fibreni: Naples, Italy, 1830. [Google Scholar]
- Tenore, M. Florae Neapolitanae Sylloge; Fibreni: Naples, Italy, 1831. [Google Scholar]
- Franjić, J.; Liber, Z.; Škvorc, Ž.; Idžojtić, M.; Šoštarić, R.; Stančić, Z. Morphological and molecular differentiation of the croatian populations of Quercus pubescens Willd. (Fagaceae). Acta Soc. Bot. Pol. 2006, 75, 123–130. [Google Scholar] [CrossRef]
- Enescu, C.; Curtu, A.; Șofletea, N. Is Quercus virgiliana a distinct morphological and genetic entity among European white oaks? Turk. J. Agric. For. 2013, 37, 632–641. [Google Scholar] [CrossRef]
- Di Pietro, R.; Di Marzio, P.; Medagli, P.; Misano, G.; Silletti, G.N.; Wagensommer, R.P.; Fortini, P. Evidence from multivariate morphometric study of the Quercus pubescens complex in southeast Italy. Bot. Serbica 2016, 40, 83–100. [Google Scholar]
- Di Pietro, R.; Di Marzio, P.; Antonecchia, G.; Conte, A.; Fortini, P. Preliminary characterization of the Quercus pubescens complex in southern italy using molecular markers. Acta Bot. Croat. 2020, 79, 15–25. [Google Scholar] [CrossRef]
- Di Pietro, R.; Conte, A.L.; Di Marzio, P.; Gianguzzi, L.; Spampinato, G.; Caldarella, O.; Fortini, P. A multivariate morphometric analysis of diagnostic traits in southern Italy and Sicily pubescent oaks. Folia Geobot. 2020, 55, 163–183. [Google Scholar] [CrossRef]
- Di Pietro, R.; Conte, A.L.; Di Marzio, P.; Fortini, P.; Farris, E.; Gianguzzi, L.; Müller, M.; Rosati, L.; Spampinato, G.; Gailing, O. Does the genetic diversity among pubescent white oaks in southern Italy, Sicily and Sardinia islands support the current taxonomic classification? Eur. J. For. Res. 2021, 140, 355–371. [Google Scholar] [CrossRef]
- Fortini, P.; Di Marzio, P.; Conte, A.L.; Antonecchia, G.; Proietti, E.; Di Pietro, R. Morphological and molecular results from a geographical transect focusing on Quercus pubescens/Q. virgiliana ecological-altitudinal vicariance in peninsular Italy. Plant Biosyst. 2022, 156, 1498–1511. [Google Scholar] [CrossRef]
- Brewer, S.; Cheddadi, R.; de Beaulieu, J.L.; Reille, M. The spread of deciduous Quercus throughout Europe since the last glacial period. For. Ecol. Manag. 2002, 156, 27–48. [Google Scholar] [CrossRef]
- Dumolin, S.; Demesure, B.; Petit, R.J. Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theoret. Appl. Genet. 1995, 91, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Dumolin-Lapègue, S.; Demesure, B.; Fineschi, S.; Le Come, V.; Petit, R.J. Phylogeographic structure of white oaks throughout the european continent. Genetics 1997, 146, 1475–1487. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Csaikl, U.M.; Bordács, S.; Burg, K.; Coart, E.; Cottrell, J.; van Dam, B.; Deans, J.D.; Dumolin-Lapègue, S.; Fineschi, S.; et al. Chloroplast DNA variation in european white oaks: Phylogeography and patterns of diversity based on data from over 2600 populations. For. Ecol. Manag. 2002, 156, 5–26. [Google Scholar] [CrossRef]
- Petit, R.J.; Brewer, S.; Bordács, S.; Burg, K.; Cheddadi, R.; Coart, E.; Cottrell, J.; Csaikl, U.M.; van Dam, B.; Deans, J.D.; et al. Identification of refugia and post-glacial colonisation routes of european white oaks based on chloroplast DNA and fossil pollen evidence. For. Ecol. Manag. 2002, 156, 49–74. [Google Scholar] [CrossRef]
- Bennett, K.D.; Tzedakis, P.C.; Willis, K.J. Quaternary refugia of north european trees. J. Biogeogr. 1991, 18, 103–115. [Google Scholar] [CrossRef]
- Fineschi, S.; Taurchini, D.; Grossoni, P.; Petit, R.J.; Vendramin, G.G. Chloroplast DNA variation of white oaks in Italy. For. Ecol. Manag. 2002, 156, 103–114. [Google Scholar] [CrossRef]
- Lupini, A.; Aci, M.M.; Mauceri, A.; Luzzi, G.; Bagnato, S.; Menguzzato, G.; Mercati, F.; Sunseri, F. Genetic diversity in old populations of sessile oak from Calabria assessed by nuclear and chloroplast SSR. J. Mt. Sci. 2019, 16, 1111–1120. [Google Scholar] [CrossRef]
- Provan, J.; Powell, W.; Hollingsworth, P.M. Chloroplast microsatellites: New tools for studies in plant ecology and evolution. Trends Ecol. Evol. 2001, 16, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Deguilloux, M.-F.; Dumolin-Lapègue, S.; Gielly, L.; Grivet, D.; Petit, R.J. A set of primers for the amplification of chloroplast microsatellites in Quercus. Mol. Ecol. Notes 2003, 3, 24–27. [Google Scholar] [CrossRef]
- Neophytou, C.; Michiels, H.-G. Upper rhine valley: A migration crossroads of middle european oaks. For. Ecol. Manag. 2013, 304, 89–98. [Google Scholar] [CrossRef]
- Chmielewski, M.; Meyza, K.; Chybicki, I.J.; Dzialuk, A.; Litkowiec, M.; Burczyk, J. Chloroplast microsatellites as a tool for phylogeographic studies: The case of white oaks in Poland. iForest 2015, 8, 765. [Google Scholar] [CrossRef]
- Magri, D.; Fineschi, S.; Bellarosa, R.; Buonamici, A.; Sebastiani, F.; Schirone, B.; Simeone, M.C.; Vendramin, G.G. The distribution of Quercus suber chloroplast haplotypes matches the palaeogeographical history of the western Mediterranean. Mol. Ecol. 2007, 16, 5259–5266. [Google Scholar] [CrossRef] [PubMed]
- Bagnoli, F.; Tsuda, Y.; Fineschi, S.; Bruschi, P.; Magri, D.; Zhelev, P.; Paule, L.; Simeone, M.C.; González-Martínez, S.C.; Vendramin, G.G. Combining molecular and fossil data to infer demographic history of Quercus cerris: Insights on european eastern glacial refugia. J. Biogeogr. 2016, 43, 679–690. [Google Scholar] [CrossRef]
- Grivet, D.; Deguilloux, M.-F.; Petit, R.J.; Sork, V.L. Contrasting patterns of historical colonization in white oaks (Quercus spp.) in California and Europe. Mol. Ecol. 2006, 15, 4085–4093. [Google Scholar] [CrossRef]
- Neophytou, C.; Semizer-Cuming, D.; Michiels, H.-G.; Kremer, A.; Jansen, S.; Fussi, B. Relict stands of central european oaks: Unravelling autochthony and genetic structure based on a multi-population study. For. Ecol. Manag. 2024, 551, 121554. [Google Scholar] [CrossRef]
- Curtu, A.L.; Gailing, O.; Leinemann, L.; Finkeldey, R. Genetic variation and differentiation within a natural community of five oak species (Quercus spp.). Plant Biol. 2007, 9, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Danusevičius, D.; Baliuckas, V.; Buchovska, J.; Kembrytė, R. Geographical Structuring of Quercus robur L. Chloroplast DNA haplotypes in Lithuania: Recolonization, adaptation, or overexploitation effects? Forests 2021, 12, 831. [Google Scholar] [CrossRef]
- Popović, M.; Katičić Bogdan, I.; Varga, F.; Šatović, Z.; Bogdan, S.; Ivanković, M. Genetic diversity in peripheral pedunculate oak (Quercus robur L.) provenances—Potential climate change mitigators in the center of distribution despite challenges in natural populations. Forests 2023, 14, 2290. [Google Scholar] [CrossRef]
- Gailing, O.; Wachter, H.; Schmitt, H.-P.; Curtu, A.-L.; Finkeldey, R. Characterization of different provenances of slavonian pedunculate oaks (Quercus robur L.) in Münsterland (Germany) with chloroplast DNA markers: PCR-RFLPs and chloroplast microsatellites. Allg. Forst. Jagdztg. 2007, 178, 85–90. [Google Scholar]
- Burger, K.; Gailing, O. Genetic Variability of indigenous (Quercus robur L.) and late flushing oak (Quercus robur L. subsp. slavonica (Gáyer) Mátyás) in adult stands compared with their natural regeneration. Eur. J. For. Res. 2022, 141, 1073–1088. [Google Scholar] [CrossRef]
- Semerikova, S.A.; Isakov, I.Y.; Semerikov, V.L. Chloroplast DNA variation and phylogeography of pedunculate oak Quercus robur L. in the eastern part of the range. Russ. J. Genet. 2021, 57, 47–60. [Google Scholar] [CrossRef]
- Semerikova, S.A.; Podergina, S.M.; Tashev, A.N.; Semerikov, V.L. Phylogeography of oaks in the crimea reveals pleistocene refugia and migration routes. Russ. J. Ecol. 2023, 54, 197–212. [Google Scholar] [CrossRef]
- Médail, F.; Diadema, K. Glacial refugia influence plant diversity patterns in the Mediterranean basin. J. Biogeogr. 2009, 36, 1333–1345. [Google Scholar] [CrossRef]
- Fortini, P.; Di Pietro, R.; Proietti, E.; Cardoni, S.; Quaranta, L.; Simeone, M.C. Dissecting the continuum and unravelling the phylogeographic knot of plastid DNA in european white oaks (Quercus sect. Quercus): Ancient signatures and multiple diversity reservoirs. Eur. J. For. Res. 2024, 143, 107–127. [Google Scholar] [CrossRef]
- Proietti, E.; Filesi, L.; Di Marzio, P.; Di Pietro, R.; Masin, R.; Conte, A.L.; Fortini, P. Morphology, geometric morphometrics, and taxonomy in relict deciduous oaks woods in northern Italy. Rend. Fis. Acc. Lincei 2021, 32, 549–564. [Google Scholar] [CrossRef]
- Avanzi, C.; Bagnoli, F.; Romiti, E.; Spanu, I.; Tsuda, Y.; Vajana, E.; Vendramin, G.G.; Piotti, A. The latitudinal trend in genetic diversity and distinctiveness of Quercus robur rear edge forest remnants calls for new conservation priorities. bioRxiv, 2023; advance online publication. [Google Scholar] [CrossRef]
- Pignatti, S. La flora dell’Appennino meridionale: Distribuzione attuale ed ipotesi sull’origine. Biogeographia 1986, 10, 89–100. [Google Scholar] [CrossRef]
- Blasi, C.; Filibek, G.; Burrascano, S.; Copiz, R.; Di Pietro, R.; Ercole, S.; Lattanzi, E.; Rosati, L.; Tilia, A. Primi risultati per una nuova regionalizzazione fitogeografia del territorio italiano. Biogeographia 2007, 28, 9–23. [Google Scholar] [CrossRef]
- Brullo, S. Contributo alla conoscenza della vegetazione delle Madonie (Sicilia settentrionale). Boll. Acc. Gioenia Sci. Nat. 1984, 16, 351–420. [Google Scholar]
- Brullo, S.; Marcenò, C. Contributo alla conoscenza della classe Quercetea ilicis in Sicilia. Not. Fitosoc. 1985, 19, 183–229. [Google Scholar]
- Brullo, S.; Scelsi, F.; Spampinato, G. La Vegetazione dell’Aspromonte; Laruffa: Reggio Calabria, Italy, 2001; ISBN 88-7221-160-3. [Google Scholar]
- Brullo, S.; Gianguzzi, L.; La Mantia, A.; Siracusa, G. La classe Quercetea ilicis in Sicilia. Boll. Acc. Gioenia Sci. Nat 2008, 41, 1–24. [Google Scholar]
- Bacchetta, G.; Biondi, E.; Farris, E.; Filigheddu, R.; Mossa, L. A phytosociological study of the deciduous oak woods of Sardinia (Italy). Fitosociologia 2004, 41, 53–64. [Google Scholar]
- Thiers, B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. New York Botanical Garden’s Virtual Herbarium. 2015. Available online: http://sweetgum.nybg.org/ih (accessed on 20 March 2024).
- Sebastiani, F.; Carnevale, S.; Vendramin, G.G. A new set of mono- and dinucleotide chloroplast microsatellites in Fagaceae. Mol. Ecol. Notes 2004, 4, 259–261. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. FSTAT (Version 2.9.4), a Program (for Windows 95 and Above) to Estimate and Test Population Genetics Parameters; Department of Ecology & Evolution, Lausanne University: Vaud, Switzerland, 2003; Volume 53. [Google Scholar]
- Pons, O.; Petit, R. Measuring and testing genetic differentiation with ordered vs. unordered alleles. Genetics 1996, 144, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Corander, J.; Cheng, L.; Marttinen, P.; Sirén, J.; Tang, J. BAPS: Bayesian Analysis of Population Structure, Manual v. 6.0, 14 February 2013. Available online: https://github.com/ocbe-uio/BAPS/blob/develop/BAPS6manual.pdf (accessed on 20 October 2023).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Petit, R.J.; Duminil, J.; Fineschi, S.; Hampe, A.; Salvini, D.; Vendramin, G.G. Invited review: Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 2005, 14, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.J.C. Climatic changes in Europe during the last glacial/interglacial transition. Quatern. Int. 1995, 28, 63–76. [Google Scholar] [CrossRef]
- Schenk, F.; Väliranta, M.; Muschitiello, F.; Tarasov, L.; Heikkilä, M.; Björck, S.; Brandefelt, J.; Johansson, A.V.; Näslund, J.-O.; Wohlfarth, B. Warm summers during the younger dryas cold reversal. Nat. Commun. 2018, 9, 1634. [Google Scholar] [CrossRef] [PubMed]
- Ravazzi, C.; Peresani, M.; Pini, R.; Vescovi, E. Il tardoglaciale nelle Alpi Italiane e in Pianura Padana. Evoluzione stratigrafica, storia della vegetazione e del popolamento antropico. Alp. Mediterr. Quat. 2007, 20, 163–184. [Google Scholar]
- Vescovi, E. Long-Term Population Dynamics of Major Forest Trees under Strongly Changing Climatic Conditions. Ph.D. Thesis, University of Bern, Bern, Switzerland, 2007. [Google Scholar]
- Kaltenrieder, P.; Belis, C.A.; Hofstetter, S.; Ammann, B.; Ravazzi, C.; Tinner, W. Environmental and climatic conditions at a potential glacial refugial site of tree species near the southern alpine glaciers. new insights from multiproxy sedimentary studies at Lago Della Costa (Euganean hills, northeastern Italy). Quat. Sci. Rev. 2009, 28, 2647–2662. [Google Scholar] [CrossRef]
- Acquafredda, P.; Palmentola, G. Il glacialismo quaternario nell’Italia meridionale dal massiccio del Matese all’Aspromonte. Biogeographia 1986, 10, 13–18. [Google Scholar] [CrossRef]
- Dramis, F.; Kotarba, A. Southern Limit of Relict Rock Glaciers, Central Apennines, Italy. Permafr. Periglac. 1992, 3, 257–260. [Google Scholar] [CrossRef]
- Di Pietro, R.; Terzi, M.; Fortini, P. A revision of the high-altitude acidophilous and chionophilous grasslands of the Apennines (peninsular Italy), a long-lasting intricate syntaxonomic issue. Phytocoenologia 2017, 47, 261–304. [Google Scholar] [CrossRef]
- Follieri, M.; Magri, D.; Sadori, L. 250,000-year pollen record from Valle Di Castiglione (Roma). Pollen Spores 1988, 30, 329–356. [Google Scholar]
- Di Pasquale, G.; Saracino, A.; Bosso, L.; Russo, D.; Moroni, A.; Bonanomi, G.; Allevato, E. Coastal pine-oak glacial refugia in the Mediterranean basin: A biogeographic approach based on charcoal analysis and spatial modelling. Forests 2020, 11, 673. [Google Scholar] [CrossRef]
- Lowe, J.J. Lateglacial and early holocene lake sediments from the northern Apennines, Italy—Pollen stratigraphy and radiocarbon dating. Boreas 1992, 21, 193–208. [Google Scholar] [CrossRef]
- Bertoldi, R.; Chelli, A.; Roma, R.; Tellini, C. New data from northern Apennines (Italy) pollen sequences spanning the last 30,000 yrs. Il Quat. 2007, 20, 3–20. [Google Scholar]
- Vescovi, E.; Ammann, B.; Ravazzi, C.; Tinner, W. A new late-glacial and holocene record of vegetation and fire history from Lago Del Greppo, northern Apennines, Italy. Veget. Hist. Archaeobot. 2010, 19, 219–233. [Google Scholar] [CrossRef]
- Follieri, M.; Giardini, M.; Magri, D.; Sadori, L. Palynostratigraphy of the last glacial period in the volcanic region of central Italy. Quatern. Int. 1998, 47, 3–20. [Google Scholar] [CrossRef]
- Magri, D. Late quaternary vegetation history at Lagaccione near Lago Di Bolsena (Central Italy). Rev. Palaeobot. Palyno. 1999, 106, 171–208. [Google Scholar] [CrossRef]
- Magri, D.; Sadori, L. Late Pleistocene and Holocene pollen stratigraphy at Lago Di Vico, Central Italy. Veg. Hist. Archaeobot. 1999, 8, 247–260. [Google Scholar] [CrossRef]
- Sadori, L. The lateglacial and holocene vegetation and climate history of Lago Di Mezzano (central Italy). Quat. Sci. Rev. 2018, 202, 30–44. [Google Scholar] [CrossRef]
- Di Rita, F.; Anzidei, A.P.; Magri, D. A lateglacial and early holocene pollen record from Valle Di Castiglione (Rome): Vegetation dynamics and climate implications. Quatern. Int. 2013, 288, 73–80. [Google Scholar] [CrossRef]
- Magri, D. Lineamenti della vegetazione tardoglaciale in Italia peninsulare e in Sicilia. In Il Tardiglaciale in Italia–Lavori in Corso; British Archaeological Reports, International Series 1859; Mussi, M., Ed.; Archaeopress: Oxford, UK, 2008; pp. 1–8. ISBN 1-4073-0342-2. [Google Scholar]
- Mattioni, C.; Martin, M.A.; Chiocchini, F.; Cherubini, M.; Gaudet, M.; Pollegioni, P.; Velichkov, I.; Jarman, R.; Chambers, F.M.; Paule, L.; et al. Landscape genetics structure of european sweet chestnut (Castanea Sativa Mill): Indications for conservation priorities. Tree Genet. Genomes 2017, 13, 39. [Google Scholar] [CrossRef]
- Mattioni, C.; Ranzino, L.; Cherubini, M.; Leonardi, L.; La Mantia, T.; Castellana, S.; Villani, F.; Simeone, M.C. Monuments unveiled: Genetic characterization of large old chestnut (Castanea sativa Mill.) trees using comparative nuclear and chloroplast DNA analysis. Forests 2020, 11, 1118. [Google Scholar] [CrossRef]
- Mossa, L.; Bacchetta, G.; Brullo, S. Quercus ichnusae (Fagaceae), a new species from Sardinia. Isr. J. Plant Sci. 1999, 47, 199–207. [Google Scholar] [CrossRef]
- Giuli, C.D.; Masini, F.; Valleri, G. Paleogeographic evolution of the adriatic area since oligocene to pleistocene. Riv. Ital. Paleontol. Stratigr. 1987, 93, 109–126. [Google Scholar] [CrossRef] [PubMed]
- Di Pietro, R.; Fortini, P.; Ciaschetti, G.; Rosati, L.; Viciani, D.; Terzi, M. A revision of the syntaxonomy of the Apennine-Balkan Quercus cerris and Q. frainetto forests and correct application of the name Melittio-Quercion frainetto. Plant Biosyst. 2020, 154, 887–909. [Google Scholar] [CrossRef]
- Fineschi, S.; Vendramin, G. Choroplastic diversity in italian oaks: Evidence of a higher genetic richness in southern and insular populations. Forests 2004, 1, 82. [Google Scholar] [CrossRef]
- Kerkez Janković, I.; Jovanović, M.; Milovanović, J.; Aleksić, J.; Šijačić-Nikolić, M. Genetic diversity and genetic structure of three sympatric oak species in serbian landscape of outstanding features “Kosmaj” assessed by nuclear microsatellites. Seefor 2023, 14, 117–127. [Google Scholar] [CrossRef]
- Fortini, P.; Viscosi, V.; Maiuro, L.; Fineschi, S.; Vendramin, G.G. Comparative leaf surface morphology and molecular data of five oaks of the subgenus Quercus Oerst (Fagaceae). Plant Biosyst. 2009, 143, 543–554. [Google Scholar] [CrossRef]
- Blasi, C.; Di Pietro, R. Two New Phytosociological Types of Quercus pubescens s.l. woodland communities in southern Latium. Plant Biosyst. 1998, 132, 207–223. [Google Scholar] [CrossRef]
- Allegrezza, M.; Baldoni, M.; Biondi, E.; Taffetani, F.; Zuccarello, V. Studio fitosociologico dei boschi a Quercus pubescens s.l. delle Marche e delle zone contigue dell’Appennino centro-settentrionale (Italia centrale). Fitosociologia 2002, 39, 161–171. [Google Scholar]
- Blasi, C.; Di Pietro, R.; Filesi, L. Syntaxonomical revision of Quercetalia pubescenti-Petraeae in the italian peninsula. Fitosociologia 2004, 41, 87–164. [Google Scholar]
- Schaal, B.A.; Hayworth, D.A.; Olsen, K.M.; Rauscher, J.T.; Smith, W.A. Phylogeographic studies in plants: Problems and prospects. Mol. Ecol. 1998, 7, 465–474. [Google Scholar] [CrossRef]
- Andrello, M.; D’Aloia, C.; Dalongeville, A.; Escalante, M.A.; Guerrero, J.; Perrier, C.; Torres-Florez, J.P.; Xuereb, A.; Manel, S. Evolving spatial conservation prioritization with intraspecific genetic data. Trends Ecol. Evol. 2022, 37, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Fady, B.; Esposito, E.; Abulaila, K.; Aleksic, J.M.; Alia, R.; Alizoti, P.; Apostol, E.-N.; Aravanopoulos, P.; Ballian, D.; Kharrat, M.B.D.; et al. Forest Genetics Research in the Mediterranean Basin: Bibliometric Analysis, Knowledge Gaps, and Perspectives. Curr. For. Rep. 2022, 8, 277–298. [Google Scholar] [CrossRef]
- Médail, F. Plant biogeography and vegetation patterns of the Mediterranean islands. Bot. Rev. 2022, 88, 63–129. [Google Scholar] [CrossRef]
- Allen, C.D.; Macalady, A.K.; Chenchouni, H.; Bachelet, D.; McDowell, N.; Vennetier, M.; Kitzberger, T.; Rigling, A.; Breshears, D.D.; Hogg, E.H. (Ted); et al. A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. For. Ecol. Manag. 2010, 259, 660–684. [Google Scholar] [CrossRef]
- Natalini, F.; Alejano, R.; Vázquez-Piqué, J.; Cañellas, I.; Gea-Izquierdo, G. The role of climate change in the widespread mortality of holm oak in open woodlands of southwestern Spain. Dendrochronologia 2016, 38, 51–60. [Google Scholar] [CrossRef]
- Conte, A.L.; Di Pietro, R.; Iamonico, D.; Di Marzio, P.; Cillis, G.; Lucia, D.; Fortini, P. Oak decline in the Mediterranean basin: A study case from the southern Apennines (Italy). Plant Sociol. 2019, 56, 69–80. [Google Scholar]
- Blasi, C.; Capotorti, G.; Copiz, R.; Guida, D.; Barbara, M.; Smiraglia, D.; Zavattero, L. Terrestrial Ecoregions of Italy Explanatory Notes. Global Map S.r.l.: Firenze, Italy, 2019; ISBN 978-88-97091-04-2. [Google Scholar]

| Locus | Location | Repeat Motif | Primers Sequence (5′–3′) Sense and Antisense | Tm (°C) | PCR Product Size (bp) |
|---|---|---|---|---|---|
| μdt1 | trnE-trnT intergenic | (A)11 | ATCTTACACTAAGCTCGGAA TTCAATAACTTGTTGATCCC | 48 | 81–83 |
| μdt3 | trnD-trnY intergenic | (A)11 | TGTTAGTAATCCTTTCGTTT AGGTATAAAGTCTAAGGTAA | 46 | 125–128 |
| μcd4 | ycf6-psbM intergenic | (T)12 | TTATTTGTTTTTGGTTTCACC TTTCCCATAGAGAGTCTGTAT | 45 | 94–99 |
| μcd5 | ycf6-psbM intergenic | (A)8 | CCCCCGGATCTCTGTCAACTG TAATAAACGAGAATCACATAA | 45 | 74–77 |
| Cmcs6 | ndhG-ndhI intergenic | (T)10 | GAAAAAGGACCCTTCCTAAT CTTATGATCGTCACGAATTG | 55 | 200–203 |
| Locus | Na | Ne | i | h | Uh |
|---|---|---|---|---|---|
| udt1 | 3 | 1.798 | 0.677 | 0.444 | 0.446 |
| ucd5 | 3 | 1.980 | 0.852 | 0.495 | 0.498 |
| ucd4 | 5 | 3.744 | 1.441 | 0.733 | 0.737 |
| udt3 | 4 | 2.775 | 1.137 | 0.640 | 0.643 |
| CmCs6 | 3 | 2.432 | 0.975 | 0.589 | 0.592 |
| Mean (se) | 3.600 (0.400) | 2.546 (0.345) | 1.000 (0.130) | 0.580 (0.051) | 0.583 (0.052) |
| H | N | Freq. | Pops | Gs | Species | Species Complex |
|---|---|---|---|---|---|---|
| 6 | 1 | 0.56 | 1 | Sardinia | Q. ichnusae | Q. pubescens |
| 7 | 1 | 0.56 | 1 | South | Q. dalechampii | Q. pubescens |
| 8 | 1 | 0.56 | 1 | South | Q. congesta | Q. pubescens |
| 9 | 1 | 0.56 | 1 | Central | Q. petraea | Q. petraea |
| 10 | 1 | 0.56 | 1 | South | Q. congesta | Q. pubescens |
| 11 | 1 | 0.56 | 1 | Central | Q. petraea | Q. petraea |
| 12 | 1 | 0.56 | 1 | Central | Q. petraea | Q. petraea |
| 13 | 1 | 0.56 | 1 | Central | Q. pubescens | Q. pubescens |
| 14 | 1 | 0.56 | 1 | Sicily | Q. congesta | Q. pubescens |
| A | 3 | 1.67 | 2 | Sicily | Q. leptobalana, Q. virgiliana | Q. pubescens |
| B | 10 | 5.56 | 8 | North, Sicily, and Sardinia | Q. congesta, Q. dalechampii, Q. petraea, Q. pubescens, Q. virgiliana | Q. petraea, Q. pubescens |
| BB | 2 | 1.11 | 1 | Central | Q. frainetto | Q. frainetto |
| C | 13 | 7.22 | 5 | Central | Q. frainetto, Q. pubescens, Q. robur | Q. frainetto, Q. pubescens, Q. robur |
| CC | 9 | 5 | 5 | Central | Q. frainetto, Q. petraea, Q. pubescens | Q. frainetto, Q. petraea, Q. pubescens |
| D | 7 | 3.89 | 4 | Sicily | Q. congesta, Q. leptobalana, Q. petraea subsp. austrotyrrhenica, Q. virgiliana | Q. petraea, Q. pubescens |
| DD | 8 | 4.44 | 3 | Central and South | Q. congesta, Q. dalechampii, Q. pubescens | Q. pubescens |
| E | 21 | 11.67 | 11 | North, Central, Sicily, and Sardinia | Q. congesta, Q. dalechampii, Q. petraea, Q. pubescens, Q. robur, Q. virgiliana | Q. petraea, Q. pubescens, Q. robur |
| GG | 2 | 1.11 | 1 | Central | Q. petraea | Q. petraea |
| J | 5 | 2.78 | 3 | North | Q. petraea, Q. robur | Q. petraea, Q. robur |
| O | 6 | 3.33 | 4 | Sardinia | Q. congesta, Q. ichnusae, Q. virgiliana | Q. pubescens |
| R | 3 | 1.67 | 2 | Sardinia | Q. ichnusae, Q. virgiliana | Q. pubescens |
| S | 6 | 3.33 | 3 | Sardinia | Q. congesta, Q. ichnusae, Q. virgiliana | Q. pubescens |
| T | 8 | 4.44 | 4 | Sardinia | Q. congesta, Q. ichnusae, Q. virgiliana | Q. pubescens |
| U | 14 | 7.78 | 5 | North, Central, and South | Q. frainetto, Q. petraea, Q. pubescens | Q. frainetto, Q. petraea, Q. pubescens |
| V | 30 | 16.67 | 13 | North, Central, and South | Q. frainetto, Q. petraea, Q. petraea subsp. austrotyrrhenica, Q. pubescens, Q. robur | Q. frainetto, Q. petraea, Q. pubescens, Q. robur |
| W | 5 | 2.78 | 3 | North and South | Q. petraea, Q. robur | Q. petraea, Q. robur |
| X | 15 | 8.33 | 6 | South | Q. congesta, Q. dalechampii, Q. pubescens, Q. virgiliana | Q. pubescens |
| Y | 4 | 2.22 | 2 | North | Q. robur | Q. robur |
| Geographical Sector | N | Na | Ne | Np | h | uh | HS | HT | GST | RST |
|---|---|---|---|---|---|---|---|---|---|---|
| Northern Italy | 30 | 2.200 (0.200) | 1.698 (0.115) | 0.000 (0.000) | 0.399 (0.045) | 0.413 (0.047) | 0.333 (0.136) | 0.799 (0.084) | 0.583 (0.190) | 0.628 * (0.217) |
| Central Italy | 48 | 3.200 (0.490) | 2.503 (0.457) | 0.200 (0.200) | 0.558 (0.060) | 0.570 (0.061) | 0.381 (0.110) | 0.908 (0.045) | 0.581 (0.110) | 0.861 ** (0.080) |
| Southern Italy | 42 | 2.800 (0.374) | 1.675 (0.253) | 0.000 (0.000) | 0.349 (0.093) | 0.358 (0.095) | 0.137 (0.070) | 0.851 (0.047) | 0.839 (0.090) | 0.876 ns (0.090) |
| Sicily | 30 | 2.000 (0.316) | 1.377 (0.205) | 0.000 (0.000) | 0.212 (0.104) | 0.219 (0.108) | 0.500 (0.114) | 0.763 (0.055) | 0.345 (0.166) | 0.398 ns (0.233) |
| Sardinia | 30 | 2.400 (0.400) | 1.863 (0.293) | 0.000 (0.000) | 0.421 (0.068) | 0.435 (0.071) | 0.500 (0.114) | 0.869 (0.036) | 0.425 (0.123) | 0.961 ** (0.011) |
| Mean (among geogr. sectors) | 2.520 (0.356) | 1.823 (0.265) | 0.040 (0.040) | 0.388 (0.074) | 0.399 (0.076) | 0.370 (0.110) | 0.838 (0.053) | 0.555 (0.136) | 0.745 (0.126) |
| Species Complex | Na | Ne | Np | h | uh | HS | HT | GST | RST |
|---|---|---|---|---|---|---|---|---|---|
| Q. frainetto | 2.600 (0.400) | 2.002 (0.256) | 0.000 (0.000) | 0.468 (0.065) | 0.501 (0.070) | 0.133 (0.133) | 0.900 (0.117) | 0.852 (0.138) | 0.988 ns (0.006) |
| Q. petraea | 2.800 (0.200) | 1.827 (0.175) | 0.200 (0.200) | 0.434 (0.050) | 0.449 (0.052) | 0.467 (0.133) | 0.800 (0.116) | 0.417 (0.126) | 0.841 ** (0.111) |
| Q. pubescens | 3.200 (0.490) | 2.661 (0.349) | 0.600 (0.245) | 0.600 (0.047) | 0.606 (0.048 | 0.351 (0.062) | 0.940 (0.011) | 0.626 (0.066) | 0.880 ** (0.045) |
| Q. robur | 2.200 (0.374) | 1.747 (0.232) | 0.000 (0.000) | 0.377 (0.100) | 0.393 (0.104) | 0.292 (0.146) | 0.849 (0.078) | 0.657 (0.181) | 0.721 ns (0.172) |
| Mean | 2.700 (0.366) | 2.059 (0.253) | 0.200 (0.111) | 0.470 (0.066) | 0.487 (0.069) | 0.311 (0.1189) | 0.872 (0.080) | 0.638 (0.128) | 0.858 (0.084) |
| Taxa | Individuals | Haplotypes | Haplotypes/ Individuals | Exclusive Haplotypes | Singletons |
|---|---|---|---|---|---|
| Q. congesta | 27 {15.0} | 11 (39.3) | 3 (10.7) | 3 (10.7) | |
| Q. dalechampii | 9 {5.0} | 5 (17.9) | 1 (3.6) | 1 (3.6) | |
| Q. ichnusae | 9 {5.0} | 5 (17.9) | 1 (3.6) | 1 (3.6) | |
| Q. leptobalana | 3 {1.7} | 2 (7.1) | - | - | |
| Q. pubescens | 42 {23.3} | 9 (32.1) | 1 (3.6) | 1 (3.6) | |
| Q. virgiliana | 21 {11.7} | 9 (32.1) | - | - | |
| Q. pubescens complex | 111 {61.7} | 20 (71.4) | 0.18 | 13 (46.4) | 6 (21.4) |
| Q. petraea | 24 {13.3} | 11 (39.3) | 4 (14.3) | 3 (10.7) | |
| Q. petraea subsp. austrotyrrhenica | 6 {3.3} | 2 (7.1) | - | - | |
| Q. petraea complex | 30 {16.7} | 12 (42.9) | 0.40 | 4 (14.3) | 3 (10.7) |
| Q. frainetto | 15 {8.3} | 5 (17.9) | 0.33 | 1 (3.6) | - |
| Q. robur | 24 {13.3} | 6 (21.4) | 0.25 | 1 (3.6) | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Pietro, R.; Quaranta, L.; Mattioni, C.; Simeone, M.C.; Di Marzio, P.; Proietti, E.; Fortini, P. Chloroplast Haplotype Diversity in the White Oak Populations of the Italian Peninsula, Sicily, and Sardinia. Forests 2024, 15, 864. https://doi.org/10.3390/f15050864
Di Pietro R, Quaranta L, Mattioni C, Simeone MC, Di Marzio P, Proietti E, Fortini P. Chloroplast Haplotype Diversity in the White Oak Populations of the Italian Peninsula, Sicily, and Sardinia. Forests. 2024; 15(5):864. https://doi.org/10.3390/f15050864
Chicago/Turabian StyleDi Pietro, Romeo, Luca Quaranta, Claudia Mattioni, Marco Cosimo Simeone, Piera Di Marzio, Elisa Proietti, and Paola Fortini. 2024. "Chloroplast Haplotype Diversity in the White Oak Populations of the Italian Peninsula, Sicily, and Sardinia" Forests 15, no. 5: 864. https://doi.org/10.3390/f15050864






