Analyzing the Functional Roles and Immunological Features of Chemokines in COAD
Abstract
:1. Introduction
2. Results
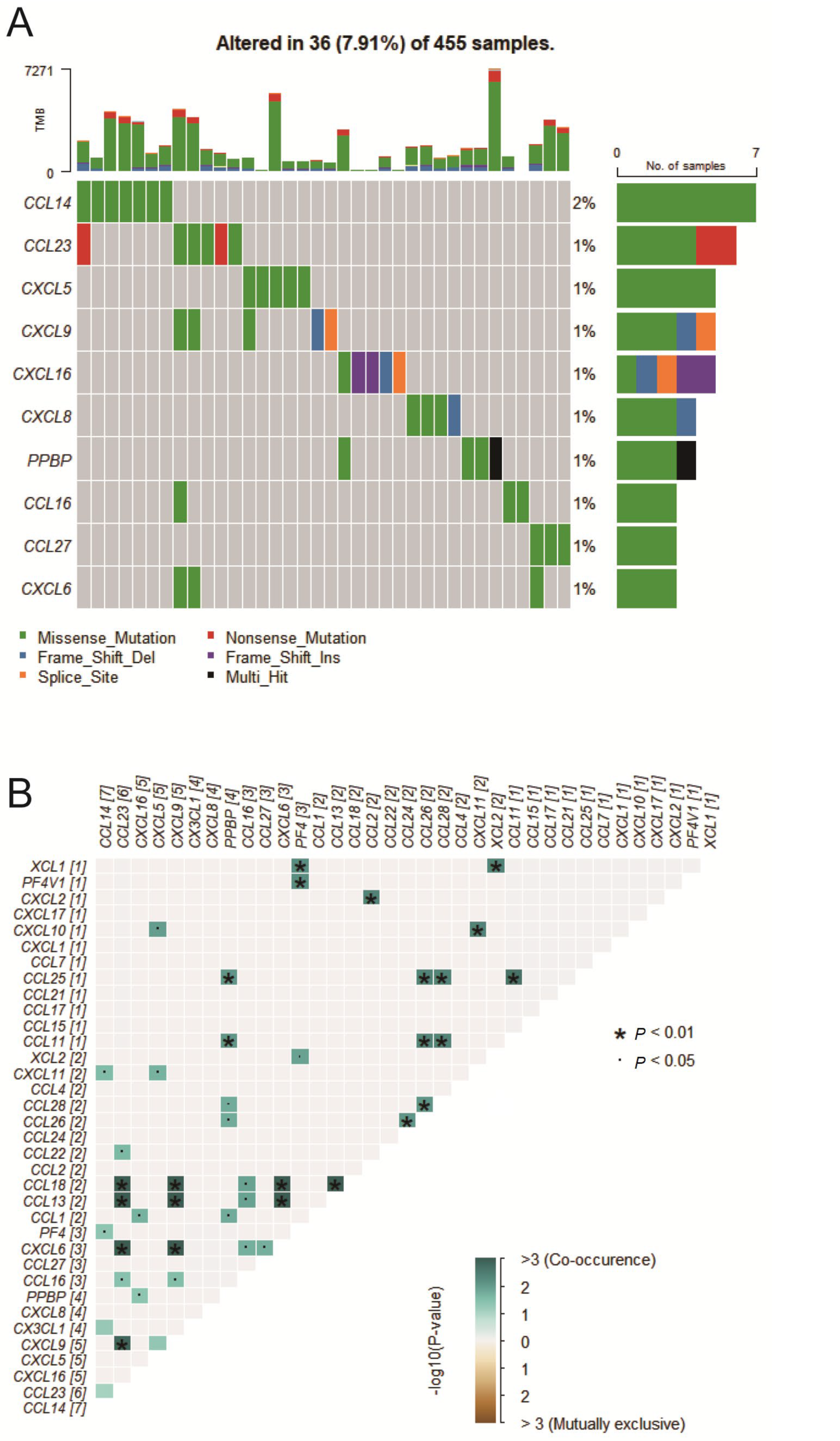
2.1. Chemokine Mutation Analysis
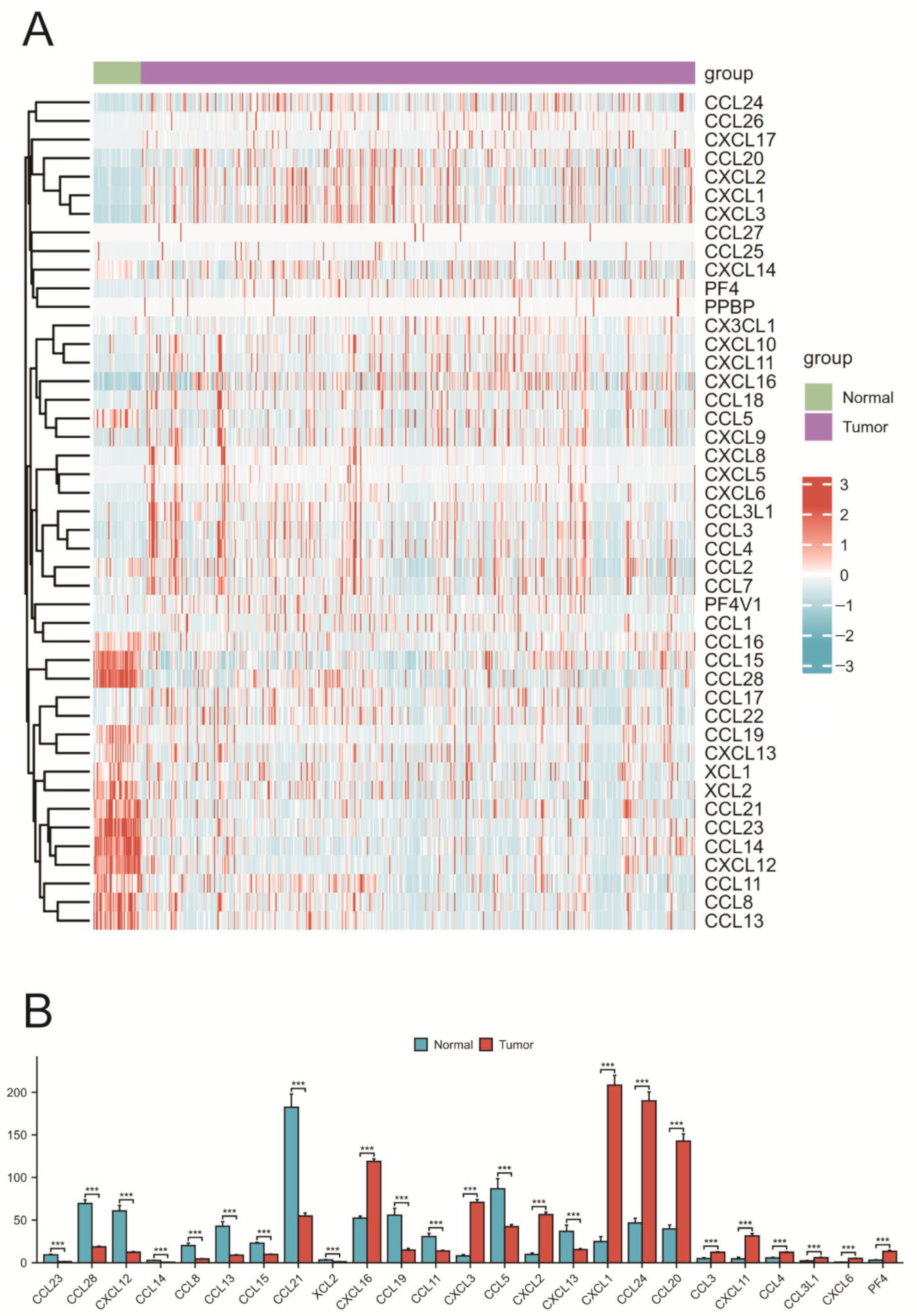
2.2. Expression of Chemokines in COAD
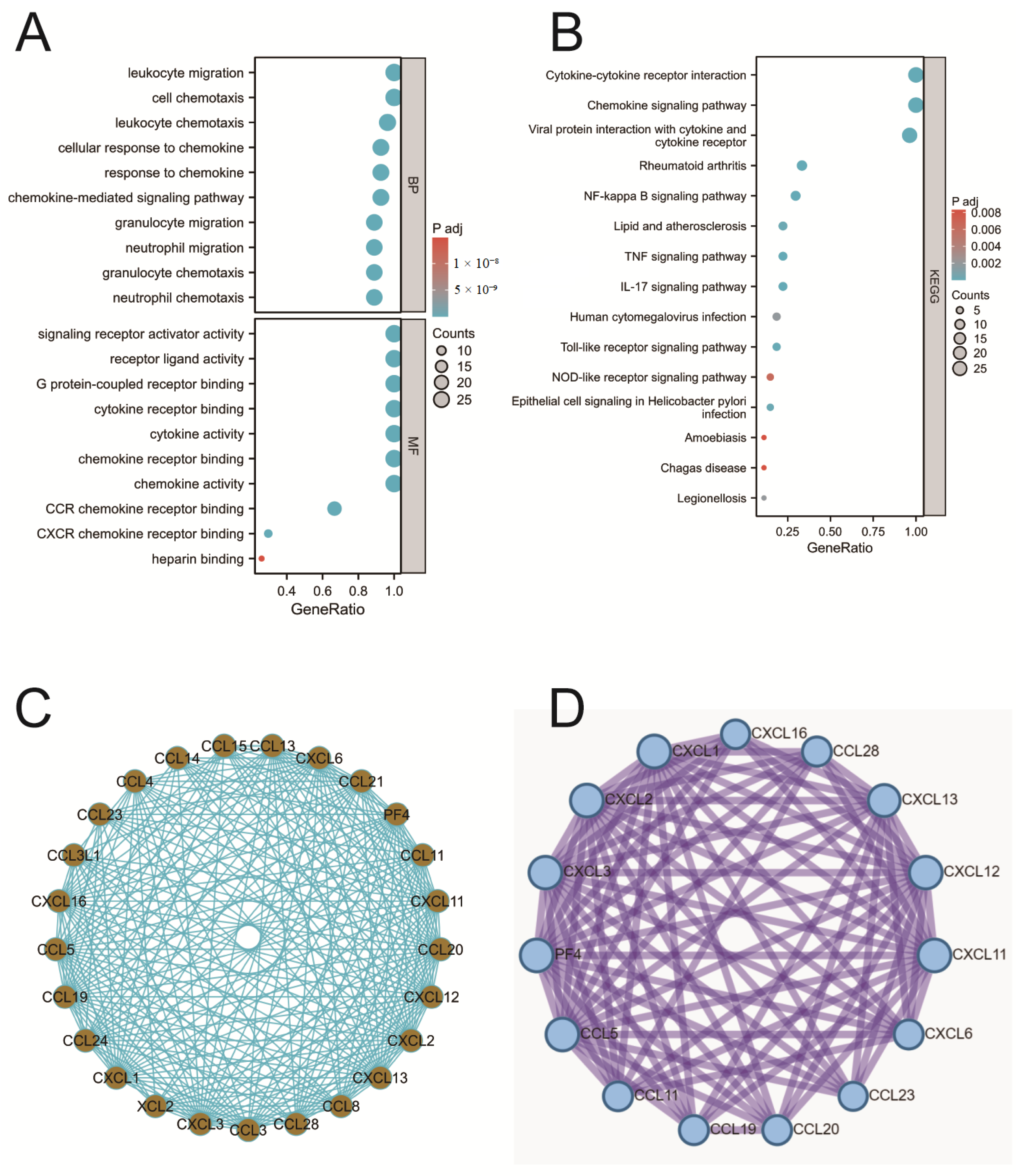
2.3. Enrichment Analysis of Chemokines
2.4. PPI Analysis
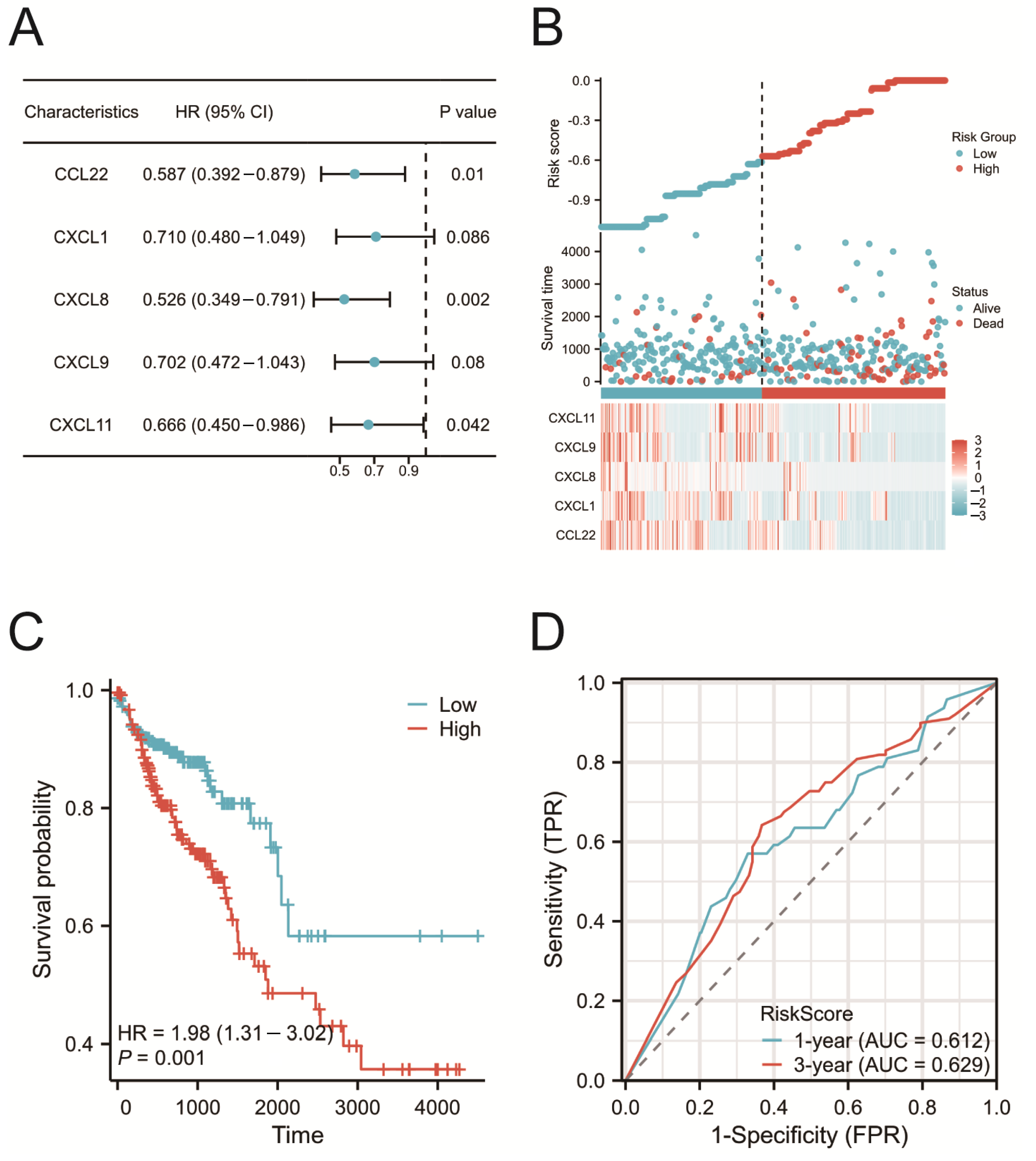
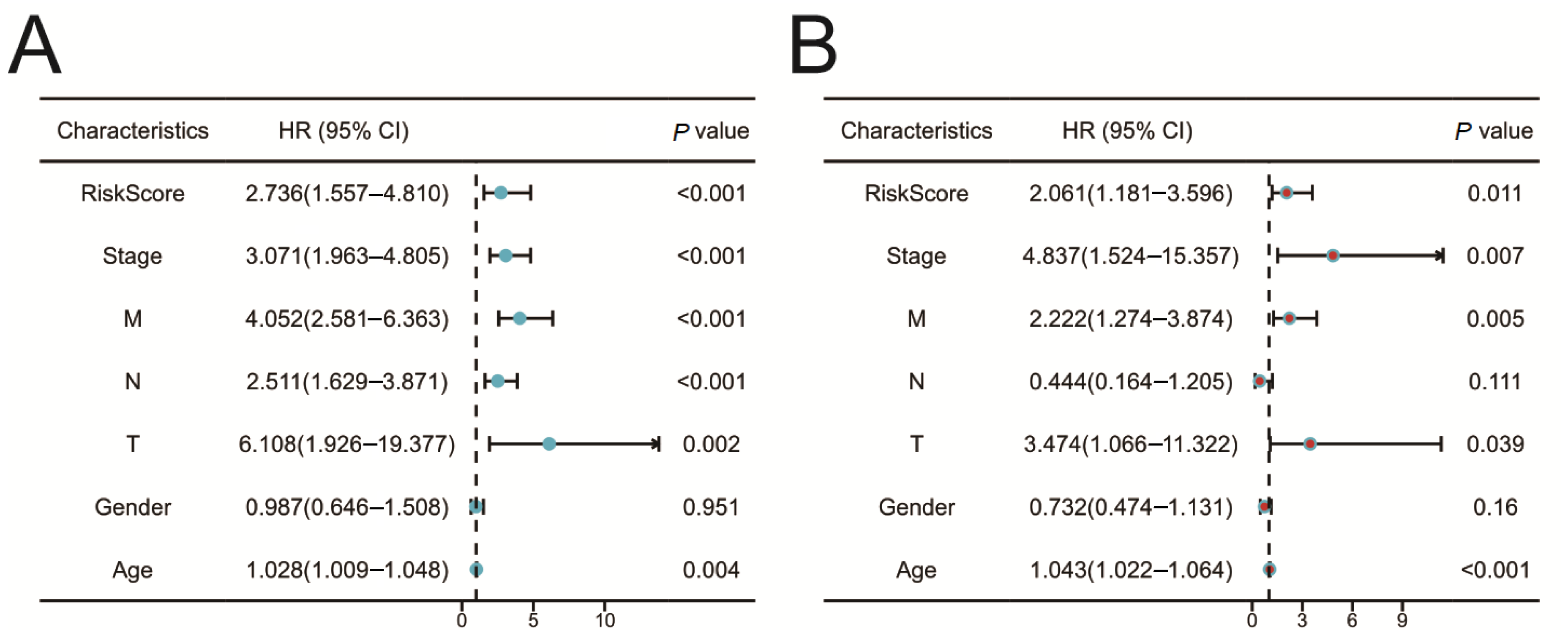
2.5. Construction of the Prognostic Risk Model
2.6. Validation of the Prognostic Risk Model
2.7. Protein and mRNA Expression of CCL22, CXCL1, CXCL8, CXCL9, and CXCL11
2.8. Correlation between Risk Score and Immune Cell Infiltration
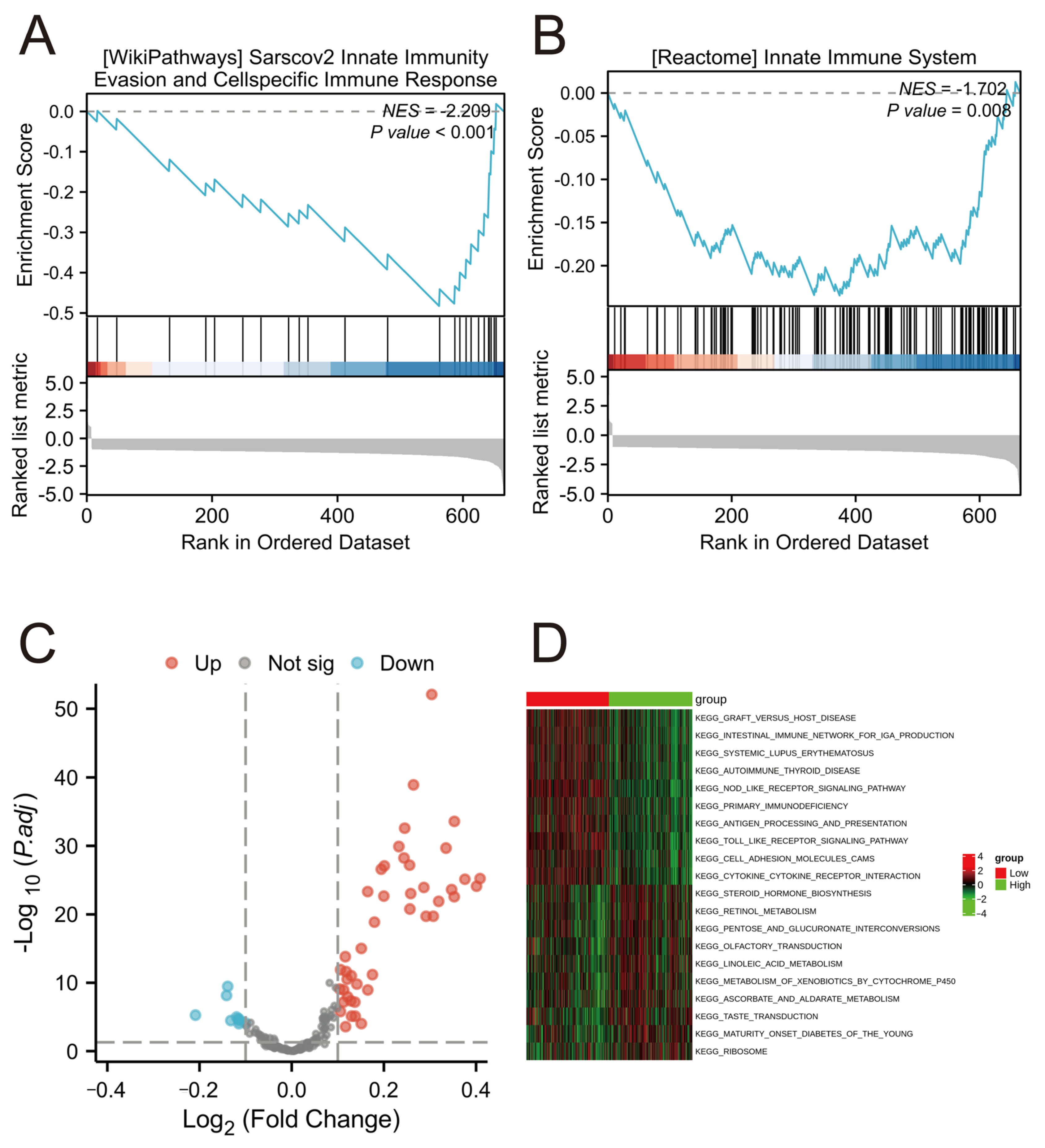
2.9. Functional Enrichment Analysis in Different Risk Groups
3. Discussion
4. Materials and Methods
4.1. Data Download and Processing
4.2. Expression of Chemokines in COAD
4.3. Enrichment Analysis of Chemokines
4.4. Protein–Protein Interaction Analysis of Chemokines
4.5. Construction of the Prognostic Risk Model
4.6. Immunohistochemical Analysis
4.7. Cell Culture
4.8. Quantitative Real-Time PCR (qRT-PCR)
4.9. Correlation between Risk Score and Immune Cell Infiltration
4.10. GSEA and GSVA Functional Annotation
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Karuniawati, H.; Jairoun, A.A.; Urbi, Z.; Ooi, J.; John, A.; Lim, Y.C.; Kibria, K.M.K.; Mohiuddin, A.K.M.; Ming, L.C.; et al. Colorectal Cancer: A Review of Carcinogenesis, Global Epidemiology, Current Challenges, Risk Factors, Preventive and Treatment Strategies. Cancers 2022, 14, 1732. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Collins, G.; Wang, H.; Toh, J.W.T. Pathological Features and Prognostication in Colorectal Cancer. Curr. Oncol. 2021, 28, 5356–5383. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef]
- Sethi, S.; Ali, S.; Philip, P.A.; Sarkar, F.H. Clinical advances in molecular biomarkers for cancer diagnosis and therapy. Int. J. Mol. Sci. 2013, 14, 14771–14784. [Google Scholar] [CrossRef]
- Mousa, L.; Salem, M.E.; Mikhail, S. Biomarkers of Angiogenesis in Colorectal Cancer. Biomark. Cancer 2015, 7 (Suppl. S1), 13–19. [Google Scholar] [CrossRef] [PubMed]
- Boussios, S.; Ozturk, M.A.; Moschetta, M.; Karathanasi, A.; Zakynthinakis-Kyriakou, N.; Katsanos, K.H.; Christodoulou, D.K.; Pavlidis, N. The Developing Story of Predictive Biomarkers in Colorectal Cancer. J. Pers. Med. 2019, 9, 12. [Google Scholar] [CrossRef]
- Moser, B.; Willimann, K. Chemokines: Role in inflammation and immune surveillance. Ann. Rheum. Dis. 2004, 63 (Suppl. S2), ii84–ii89. [Google Scholar] [CrossRef]
- Balkwill, F.R. The chemokine system and cancer. J. Pathol. 2012, 226, 148–157. [Google Scholar] [CrossRef]
- Hughes, C.E.; Nibbs, R.J.B. A guide to chemokines and their receptors. FEBS J. 2018, 285, 2944–2971. [Google Scholar] [CrossRef]
- Korbecki, J.; Grochans, S.; Gutowska, I.; Barczak, K.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of Receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int. J. Mol. Sci. 2020, 21, 7619. [Google Scholar] [CrossRef] [PubMed]
- Scapini, P.; Lapinet-Vera, J.A.; Gasperini, S.; Calzetti, F.; Bazzoni, F.; Cassatella, M.A. The neutrophil as a cellular source of chemokines. Immunol. Rev. 2000, 177, 195–203. [Google Scholar] [CrossRef]
- Ozga, A.J.; Chow, M.T.; Luster, A.D. Chemokines and the immune response to cancer. Immunity 2021, 54, 859–874. [Google Scholar] [CrossRef] [PubMed]
- Mukaida, N.; Baba, T. Chemokines in tumor development and progression. Exp. Cell Res. 2012, 318, 95–102. [Google Scholar] [CrossRef]
- Bule, P.; Aguiar, S.I.; Aires-Da-Silva, F.; Dias, J.N.R. Chemokine-Directed Tumor Microenvironment Modulation in Cancer Immunotherapy. Int. J. Mol. Sci. 2021, 22, 9804. [Google Scholar] [CrossRef] [PubMed]
- Kalbasi, A.; Komar, C.; Tooker, G.M.; Liu, M.; Lee, J.W.; Gladney, W.L.; Ben-Josef, E.; Beatty, G.L. Tumor-Derived CCL2 Mediates Resistance to Radiotherapy in Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2017, 23, 137–148. [Google Scholar] [CrossRef]
- Tan, M.C.; Goedegebuure, P.S.; Belt, B.A.; Flaherty, B.; Sankpal, N.; Gillanders, W.E.; Eberlein, T.J.; Hsieh, C.S.; Linehan, D.C. Disruption of CCR5-dependent homing of regulatory T cells inhibits tumor growth in a murine model of pancreatic cancer. J. Immunol. 2009, 182, 1746–1755. [Google Scholar] [CrossRef]
- Sun, H.; Zhao, L.; Pan, K.; Zhang, Z.; Zhou, M.; Cao, G. Integrated analysis of mRNA and miRNA expression profiles in pancreatic ductal adenocarcinoma. Oncol. Rep. 2017, 37, 2779–2786. [Google Scholar] [CrossRef]
- Rivas-Fuentes, S.; Salgado-Aguayo, A.; Pertuz Belloso, S.; Gorocica Rosete, P.; Alvarado-Vasquez, N.; Aquino-Jarquin, G. Role of Chemokines in Non-Small Cell Lung Cancer: Angiogenesis and Inflammation. J. Cancer 2015, 6, 938–952. [Google Scholar] [CrossRef]
- Singh, S.; Wu, S.; Varney, M.; Singh, A.P.; Singh, R.K. CXCR1 and CXCR2 silencing modulates CXCL8-dependent endothelial cell proliferation, migration and capillary-like structure formation. Microvasc. Res. 2011, 82, 318–325. [Google Scholar] [CrossRef]
- Azenshtein, E.; Meshel, T.; Shina, S.; Barak, N.; Keydar, I.; Ben-Baruch, A. The angiogenic factors CXCL8 and VEGF in breast cancer: Regulation by an array of pro-malignancy factors. Cancer Lett. 2005, 217, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.-N.; Han, Y.-B.; Yang, R.; Yang, Z.-C. Chemokines in colon cancer progression. Semin. Cancer Biol. 2022, 86, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Erreni, M.; Bianchi, P.; Laghi, L.; Mirolo, M.; Fabbri, M.; Locati, M.; Mantovani, A.; Allavena, P. Expression of chemokines and chemokine receptors in human colon cancer. Methods Enzymol. 2009, 460, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, L.; Chu, F.; Zhang, L.; Zhang, L.; Wu, H.; Li, K. The tumor microenvironment in gastrointestinal adenocarcinomas revealed a prognostic and immunotherapeutic biomarker. Aging 2022, 14, 10171–10216. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Huang, X.; Han, X.; Li, Z.; Zhu, Q.; Yan, J.; Yu, S.; Jin, Z.; Wang, Z.; Zheng, Q.J.B.; et al. The chemokine CXCL9 expression is associated with better prognosis for colorectal carcinoma patients. Biomed. Pharmacother. 2016, 78, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Yang, C.; Wang, S.; Shi, D.; Zhang, C.; Lin, X.; Xiong, B. M2 macrophages confer resistance to 5-fluorouracil in colorectal cancer through the activation of CCL22/PI3K/AKT signaling. OncoTargets Ther. 2019, 12, 3051–3063. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Yang, Z.; Cheng, X.; Xiao, Y.; Yu, K.; Cai, X.; Xia, C.; Li, Y. CXCL8 induces epithelial-mesenchymal transition in colon cancer cells via the PI3K/Akt/NF-kappaB signaling pathway. Oncol. Rep. 2017, 37, 2095–2100. [Google Scholar] [CrossRef]
- Ahmad, R.; Kochumon, S.; Chandy, B.; Shenouda, S.; Koshy, M.; Hasan, A.; Arefanian, H.; Al-Mulla, F.; Sindhu, S. TNF-alpha Drives the CCL4 Expression in Human Monocytic Cells: Involvement of the SAPK/JNK and NF-kappaB Signaling Pathways. Cell Physiol. Biochem. 2019, 52, 908–921. [Google Scholar] [CrossRef]
- Lu, C.; Zhang, C. Oxaliplatin inhibits colorectal cancer progression by inhibiting CXCL11 secreted by cancer-associated fibroblasts and the CXCR3/PI3K/AKT pathway. Clin. Transl. Oncol. 2023, 25, 160–172. [Google Scholar] [CrossRef]
- Huang, W.-S.; Hsieh, M.-C.; Huang, C.-Y.; Kuo, Y.-H.; Tung, S.-Y.; Shen, C.-H.; Hsieh, Y.-Y.; Teng, C.-C.; Lee, K.-F.; Chen, T.-C.; et al. The Association of CXC Receptor 4 Mediated Signaling Pathway with Oxaliplatin-Resistant Human Colorectal Cancer Cells. PLoS ONE 2016, 11, e0159927. [Google Scholar] [CrossRef]
- Xun, Y.; Yang, H.; Li, J.; Wu, F.; Liu, F. CXC Chemokine Receptors in the Tumor Microenvironment and an Update of Antagonist Development. Rev. Physiol. Biochem. Pharmacol. 2020, 178, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Meng, J.; Wu, X.; Zhang, Y. Overexpression of CXCL12 chemokine up-regulates connexin and integrin expression in mesenchymal stem cells through PI3K/Akt pathway. Cell Commun. Adhes. 2013, 20, 67–72. [Google Scholar]
- O’Hayre, M.; Salanga, C.L.; Handel, T.M.; Allen, S.J. Chemokines and cancer: Migration, intracellular signalling and intercellular communication in the microenvironment. Biochem. J. 2008, 409, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Hanna, D.N.; Smith, P.M.; Novitskiy, S.V.; Washington, M.K.; Zi, J.; Weaver, C.J.; Hamaamen, J.A.; Lewis, K.B.; Zhu, J.; Yang, J.J.G. SMAD4 suppresses colitis-associated carcinoma through inhibition of CCL20/CCR6-mediated inflammation. Gastroenterology 2022, 163, 1334–1350.e14. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.-Y.; Gao, Z.-H.; Chu, J.-H.; Han, X.-Z.; Qu, X.-J. Downregulation of the CXCR4/CXCL12 axis blocks the activation of the Wnt/beta-catenin pathway in human colon cancer cells. Biomed. Pharmacother. 2015, 71, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Rohrle, N.; Knott, M.M.L.; Anz, D. CCL22 Signaling in the Tumor Environment. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2020; Volume 1231, pp. 79–96. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, X.; Feng, Y.; Zhang, Y.; Liu, Z. The CC ligand chemokine family members CCL17/CCL22 predict the survival and response to immune checkpoint blockade therapy of patients with head and neck squamous cell carcinoma. Curr. Probl. Cancer 2022, 46, 100896. [Google Scholar] [CrossRef]
- Zhou, C.; Gao, Y.; Ding, P.; Wu, T.; Ji, G. The role of CXCL family members in different diseases. Cell Death Discov. 2023, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Barczak, K.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. CXCL1: Gene, Promoter, Regulation of Expression, mRNA Stability, Regulation of Activity in the Intercellular Space. Int. J. Mol. Sci. 2022, 23, 792. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Liu, W.; Zheng, Y.; Wang, S.; Yang, B.; Li, M.; Song, J.; Zhang, F.; Zhang, X.; Wang, Q.; et al. CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-kappaB/SOX4 signaling. Cell Death Dis. 2018, 9, 880. [Google Scholar] [CrossRef]
- Zhuo, C.; Ruan, Q.; Zhao, X.; Shen, Y.; Lin, R. CXCL1 promotes colon cancer progression through activation of NF-κB/P300 signaling pathway. Biol. Direct 2022, 17, 34. [Google Scholar] [CrossRef]
- Huang, R.; Wang, Z.; Hong, J.; Wu, J.; Huang, O.; He, J.; Chen, W.; Li, Y.; Chen, X.; Shen, K. Targeting cancer-associated adipocyte-derived CXCL8 inhibits triple-negative breast cancer progression and enhances the efficacy of anti-PD-1 immunotherapy. Cell Death Dis. 2023, 14, 703. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Liao, X.; Qiu, S.; Xu, H.; Zhang, S.; Wang, S.; Ai, J.; Yang, L. CXCL8 in Tumor Biology and Its Implications for Clinical Translation. Front. Mol. Biosci. 2022, 9, 723846. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Choi, I.; Ning, Y.; Kim, N.Y.; Khatchadourian, V.; Yang, D.; Chung, H.K.; Choi, D.; LaBonte, M.J.; Ladner, R.D.; et al. Interleukin-8 and its receptor CXCR2 in the tumour microenvironment promote colon cancer growth, progression and metastasis. Br. J. Cancer 2012, 106, 1833–1841. [Google Scholar] [CrossRef]
- Tan, S.; Wang, K.; Sun, F.; Li, Y.; Gao, Y. CXCL9 promotes prostate cancer progression through inhibition of cytokines from T cells. Mol. Med. Rep. 2018, 18, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Neo, S.Y.; Lundqvist, A. The multifaceted roles of CXCL9 within the tumor microenvironment. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2020; pp. 45–51. [Google Scholar]
- Gao, Q.; Zhang, Y. CXCL11 Signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2021, 1302, 41–50. [Google Scholar] [CrossRef]
- Li, Y.; Han, S.; Wu, B.; Zhong, C.; Shi, Y.; Lv, C.; Fu, L.; Zhang, Y.; Lang, Q.; Liang, Z.; et al. CXCL11 Correlates with Immune Infiltration and Impacts Patient Immunotherapy Efficacy: A Pan-Cancer Analysis. Front. Immunol. 2022, 13, 951247. [Google Scholar] [CrossRef]
- Cao, Y.; Jiao, N.; Sun, T.; Ma, Y.; Zhang, X.; Chen, H.; Hong, J.; Zhang, Y. CXCL11 Correlates With Antitumor Immunity and an Improved Prognosis in Colon Cancer. Front. Cell Dev. Biol. 2021, 9, 646252. [Google Scholar] [CrossRef]
- Liu, M.; Fu, X.; Jiang, L.; Ma, J.; Zheng, X.; Wang, S.; Guo, H.; Tian, T.; Nan, K.; Wang, W. Colon cancer cells secreted CXCL11 via RBP-Jkappa to facilitated tumour-associated macrophage-induced cancer metastasis. J. Cell. Mol. Med. 2021, 25, 10575–10590. [Google Scholar] [CrossRef]
- Saxena, M.; Yeretssian, G. NOD-Like Receptors: Master Regulators of Inflammation and Cancer. Front. Immunol. 2014, 5, 327. [Google Scholar] [CrossRef]
- Kallingal, A.; Olszewski, M.; Maciejewska, N.; Brankiewicz, W.; Baginski, M. Cancer immune escape: The role of antigen presentation machinery. J. Cancer Res. Clin. Oncol. 2023, 149, 8131–8141. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. STRING 8—A global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37, D412–D416. [Google Scholar] [CrossRef] [PubMed]
- Ponten, F.; Jirstrom, K.; Uhlen, M. The Human Protein Atlas—A tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Subramanian, A.; Kuehn, H.; Gould, J.; Tamayo, P.; Mesirov, J.P. GSEA-P: A desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007, 23, 3251–3253. [Google Scholar] [CrossRef]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013, 14, 7. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Song, Y. Analyzing the Functional Roles and Immunological Features of Chemokines in COAD. Int. J. Mol. Sci. 2024, 25, 5410. https://doi.org/10.3390/ijms25105410
Xu H, Song Y. Analyzing the Functional Roles and Immunological Features of Chemokines in COAD. International Journal of Molecular Sciences. 2024; 25(10):5410. https://doi.org/10.3390/ijms25105410
Chicago/Turabian StyleXu, Houxi, and Yihua Song. 2024. "Analyzing the Functional Roles and Immunological Features of Chemokines in COAD" International Journal of Molecular Sciences 25, no. 10: 5410. https://doi.org/10.3390/ijms25105410




