Speeding up Glioblastoma Cancer Research: Highlighting the Zebrafish Xenograft Model
Abstract
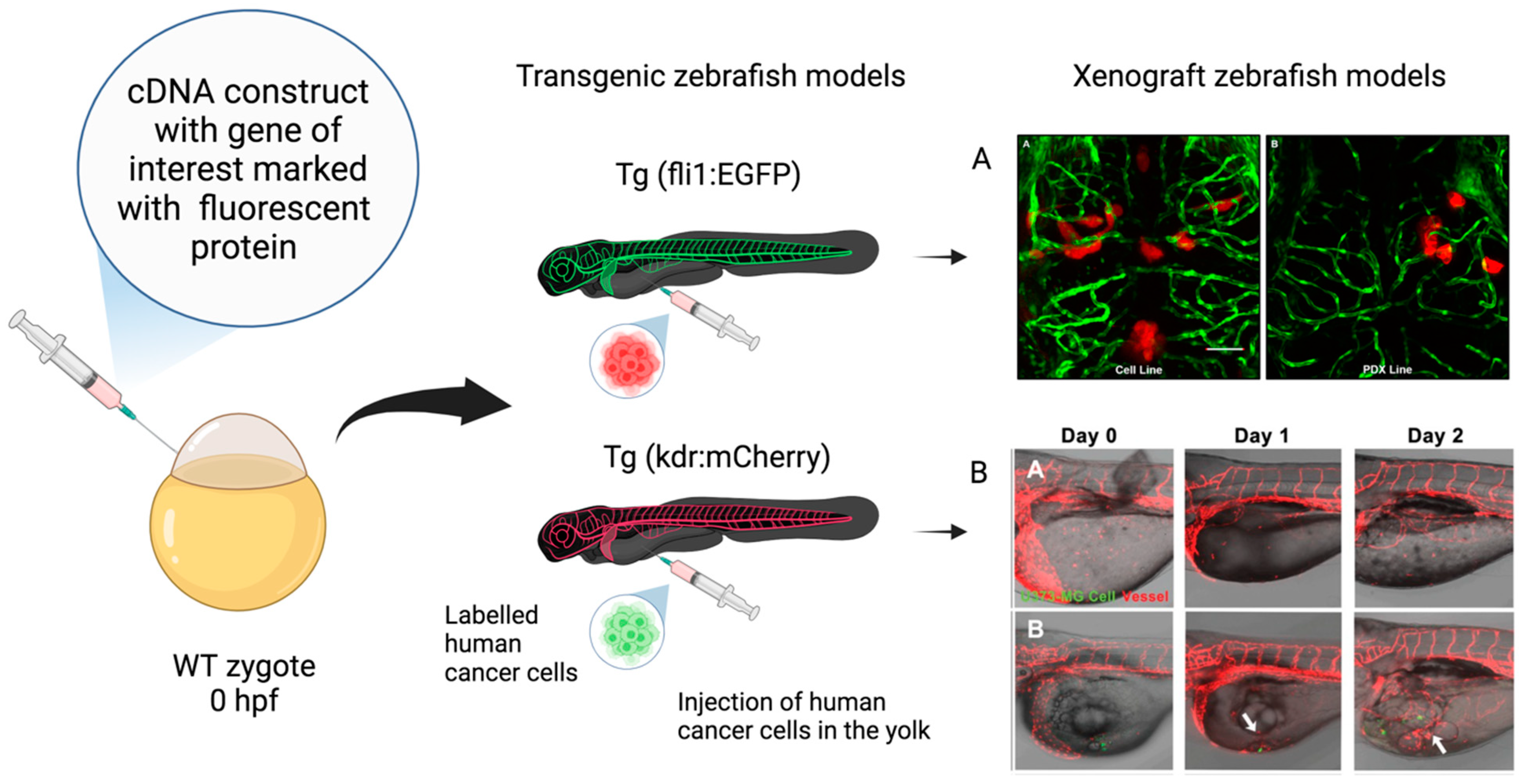
:1. Introduction
2. Multifaceted Aspects of GBM
3. Traditional Animal Models for In Vivo GBM Research
3.1. Mouse Models
3.2. Canine Models
3.3. Porcine Models
3.4. Non-Human Primate Models
3.5. Drosophila Melanogaster Model
4. Zebrafish (Danio rerio) Models in Cancer Research
Transgenic and Transplantation (Xenograft) Zebrafish Models
5. Comparative Analysis: Zebrafish vs. Traditional In Vivo Models for GBM Research
6. Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro Oncol. 2019, 21, v1–v100. [Google Scholar] [CrossRef] [PubMed]
- Karschnia, P.; Young, J.S.; Dono, A.; Häni, L.; Sciortino, T.; Bruno, F.; Juenger, S.T.; Teske, N.; Morshed, R.A.; Haddad, A.F.; et al. Prognostic validation of a new classification system for extent of resection in glioblastoma: A report of the RANO resect group. Neuro Oncol. 2023, 25, 940–954. [Google Scholar] [CrossRef] [PubMed]
- Korja, M.; Raj, R.; Seppä, K.; Luostarinen, T.; Malila, N.; Seppälä, M.; Mäenpää, H.; Pitkäniemi, J. Glioblastoma survival is improving despite increasing incidence rates: A nationwide study between 2000 and 2013 in Finland. Neuro Oncol. 2019, 21, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Omuro, A.; Brandes, A.A.; Carpentier, A.F.; Idbaih, A.; Reardon, D.A.; Cloughesy, T.; Sumrall, A.; Baehring, J.; van den Bent, M.; Bähr, O.; et al. Radiotherapy combined with nivolumab or temozolomide for newly diagnosed glioblastoma with unmethylated MGMT promoter: An international randomized phase III trial. Neuro Oncol. 2023, 25, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Blakstad, H.; Brekke, J.; Rahman, M.A.; Arnesen, V.S.; Miletic, H.; Brandal, P.; Lie, S.A.; Chekenya, M.; Goplen, D. Survival in a consecutive series of 467 glioblastoma patients: Association with prognostic factors and treatment at recurrence at two independent institutions. PLoS ONE 2023, 18, e0281166. [Google Scholar] [CrossRef] [PubMed]
- Virtuso, A.; D’Amico, G.; Scalia, F.; De Luca, C.; Papa, M.; Maugeri, G.; D’Agata, V.; Caruso Bavisotto, C.; D’Amico, G.A. The Interplay between Glioblastoma Cells and Tumor Microenvironment: New Perspectives for Early Diagnosis and Targeted Cancer Therapy. Brain Sci. 2024, 14, 331. [Google Scholar] [CrossRef] [PubMed]
- Burko, P.; D’Amico, G.; Miltykh, I.; Scalia, F.; Conway de Macario, E.; Macario, A.J.L.; Giglia, G.; Cappello, F.; Caruso Bavisotto, C. Molecular Pathways Implicated in Radioresistance of Glioblastoma Multiforme: What Is the Role of Extracellular Vesicles? Int. J. Mol. Sci. 2023, 24, 4883. [Google Scholar] [CrossRef] [PubMed]
- Sahu, U.; Barth, R.F.; Otani, Y.; McCormack, R.; Kaur, B. Rat and Mouse Brain Tumor Models for Experimental Neuro-Oncology Research. J. Neuropathol. Exp. Neurol. 2022, 81, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Antonica, F.; Aiello, G.; Soldano, A.; Abballe, L.; Miele, E.; Tiberi, L. Modeling Brain Tumors: A Perspective Overview of in vivo and Organoid Models. Front. Mol. Neurosci. 2022, 15, 818696. [Google Scholar] [CrossRef]
- Reimunde, P.; Pensado-López, A.; Carreira Crende, M.; Lombao Iglesias, V.; Sánchez, L.; Torrecilla-Parra, M.; Ramírez, C.M.; Anfray, C.; Torres Andón, F. Cellular and Molecular Mechanisms Underlying Glioblastoma and Zebrafish Models for the Discovery of New Treatments. Cancers 2021, 13, 1087. [Google Scholar] [CrossRef]
- Pliakopanou, A.; Antonopoulos, I.; Darzenta, N.; Serifi, I.; Simos, Y.V.; Katsenos, A.P.; Bellos, S.; Alexiou, G.A.; Kyritsis, A.P.; Leonardos, I.; et al. Glioblastoma research on zebrafish xenograft models: A systematic review. Clin. Transl. Oncol. 2024, 26, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Luckett, P.H.; Olufawo, M.; Lamichhane, B.; Park, K.Y.; Dierker, D.; Verastegui, G.T.; Yang, P.; Kim, A.H.; Chheda, M.G.; Snyder, A.Z.; et al. Predicting survival in glioblastoma with multimodal neuroimaging and machine learning. J. Neurooncol. 2023, 164, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Aaroe, A.; Liang, J.; Puduvalli, V.K. Tumor microenvironment in glioblastoma: Current and emerging concepts. Neurooncol. Adv. 2023, 5, vdad009. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Gu, L.; Li, Y.; Zheng, Z.; Chen, W.; Wang, Y.; Wang, Y.; Xing, H.; Shi, Y.; Liu, D.; et al. Histological and molecular glioblastoma, IDH-wildtype: A real-world landscape using the 2021 WHO classification of central nervous system tumors. Front. Oncol. 2023, 13, 1200815. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity. Cell 2020, 180, 188–204.e22. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lage, M.; Lynch, T.M.; Bi, Y.; Cocito, C.; Way, G.P.; Pal, S.; Haller, J.; Yan, R.E.; Ziober, A.; Nguyen, A.; et al. Immune landscapes associated with different glioblastoma molecular subtypes. Acta Neuropathol. Commun. 2019, 7, 203. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.L.V.; Gomes, I.N.F.; Carloni, A.C.; Rosa, M.N.; da Silva, L.S.; Evangelista, A.F.; Reis, R.M.; Silva, V.A.O. Role of glioblastoma stem cells in cancer therapeutic resistance: A perspective on antineoplastic agents from natural sources and chemical derivatives. Stem Cell Res. Ther. 2021, 12, 206. [Google Scholar] [CrossRef]
- Kim, S.M.; Lim, E.J.; Yoo, K.C.; Zhao, Y.; Kang, J.H.; Lim, E.J.; Shin, I.; Kang, S.G.; Lim, H.W.; Lee, S.J. Glioblastoma-educated mesenchymal stem-like cells promote glioblastoma infiltration via extracellular matrix remodelling in the tumour microenvironment. Clin. Transl. Med. 2022, 12, e997. [Google Scholar] [CrossRef]
- Folkins, C.; Shaked, Y.; Man, S.; Tang, T.; Lee, C.R.; Zhu, Z.; Hoffman, R.M.; Kerbel, R.S. Glioma tumor stem-like cells promote tumor angiogenesis and vasculogenesis via vascular endothelial growth factor and stromal-derived factor 1. Cancer Res. 2009, 69, 7243–7251. [Google Scholar] [CrossRef]
- Alberti, G.; Sánchez-López, C.M.; Marcilla, A.; Barone, R.; Bavisotto, C.C.; Graziano, F.; de Macario, E.C.; Macario, A.J.; Bucchieri, F.; Cappello, F.; et al. Hsp70 and Calcitonin Receptor Protein in Extracellular Vesicles from Glioblastoma Multiforme: Biomarkers with Putative Roles in Carcinogenesis and Potential for Differentiating Tumor Types. Int. J. Mol. Sci. 2024, 26, 3415. [Google Scholar] [CrossRef]
- Alberti, G.; Campanella, C.; Paladino, L.; Porcasi, R.; Bavisotto, C.C.; Pitruzzella, A.; Graziano, F.; Florena, A.M.; Argo, A.; de Macario, E.C.; et al. The chaperone system in glioblastoma multiforme and derived cell lines: Diagnostic and mechanistic implications. Front. Biosci. 2022, 27, 97. [Google Scholar] [CrossRef]
- Graziano, F.; Iacopino, G.D.; Cammarata, G.; Scalia, G.; Campanella, C.; Giannone, A.G.; Porcasi, R.; Florena, A.M.; de Macario, E.C.; Macario, A.J.; et al. The Triad Hsp60-miRNAs-Extracellular Vesicles in Brain Tumors: Assessing Its Components for Understanding Tumorigenesis and Monitoring Patients. Appl. Sci. 2021, 11, 2867. [Google Scholar] [CrossRef]
- Vitale, A.M.; Santonocito, R.; Vergilio, G.; Marino Gammazza, A.; Campanella, C.; Conway de Macario, E.; Bucchieri, F.; Macario, A.J.; Caruso Bavisotto, C. Brain Tumor-Derived Extracellular Vesicles as Carriers of Disease Markers: Molecular Chaperones and MicroRNAs. Appl. Sci. 2020, 10, 6961. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Scalia, F.; Marino Gammazza, A.; Carlisi, D.; Bucchieri, F.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F.; Campanella, C. Extracellular Vesicle-Mediated Cell-Cell Communication in the Nervous System: Focus on Neurological Diseases. Int. J. Mol. Sci. 2019, 20, 434. [Google Scholar] [CrossRef]
- Alberti, G.; Sánchez-López, C.M.; Andres, A.; Santonocito, R.; Campanella, C.; Cappello, F.; Marcilla, A. Molecular Profile Study of Extracellular Vesicles for the Identification of Useful Small “Hit” in Cancer Diagnosis. Appl. Sci. 2021, 11, 10787. [Google Scholar] [CrossRef]
- Bellipanni, G.; Cappello, F.; Scalia, F.; Conway de Macario, E.; Macario, A.J.; Giordano, A. Zebrafish as a Model for the Study of Chaperonopathies. J. Cell. Physiol. 2016, 231, 2107–2114. [Google Scholar] [CrossRef] [PubMed]
- Scalia, F.; Marino Gammazza, A.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F. Myelin Pathology: Involvement of Molecular Chaperones and the Promise of Chaperonotherapy. Brain Sci. 2019, 9, 297. [Google Scholar] [CrossRef]
- Scalia, F.; Vitale, A.M.; Santonocito, R.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F. The Neurochaperonopathies: Anomalies of the Chaperone System with Pathogenic Effects in Neurodegenerative and Neuromuscular Disorders. Appl. Sci. 2021, 11, 898. [Google Scholar] [CrossRef]
- Shi, T.; Zhu, J.; Zhang, X.; Mao, X. The Role of Hypoxia and Cancer Stem Cells in Development of Glioblastoma. Cancers 2023, 15, 2613. [Google Scholar] [CrossRef]
- Saitoh, M. Involvement of partial EMT in cancer progression. J. Biochem. 2018, 164, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Lintz, M.; Muñoz, A.; Reinhart-King, C.A. The Mechanics of Single Cell and Collective Migration of Tumor Cells. J. Biomech. Eng. 2017, 139, 021005. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, B.; Lv, C.; Zhang, N.; Xing, K.; Wang, Z.; Lv, R.; Yu, M.; Xu, C.; Wang, Y. Single-cell RNA sequencing identifies critical transcription factors of tumor cell invasion induced by hypoxia microenvironment in glioblastoma. Theranostics 2023, 13, 3744–3760. [Google Scholar] [CrossRef] [PubMed]
- Dzhalilova, D.S.; Zolotova, N.A.; Mkhitarov, V.A.; Kosyreva, A.M.; Tsvetkov, I.S.; Khalansky, A.S.; Alekseeva, A.I.; Fatkhudinov, T.H.; Makarova, O.V. Morphological and molecular-biological features of glioblastoma progression in tolerant and susceptible to hypoxia Wistar rats. Sci. Rep. 2023, 13, 12694. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Semenza, G.L. A compendium of proteins that interact with HIF-1α. Exp. Cell Res. 2017, 356, 128–135. [Google Scholar] [CrossRef]
- Bar, E.E. Glioblastoma, cancer stem cells and hypoxia. Brain Pathol. 2011, 21, 119–129. [Google Scholar] [CrossRef]
- Krcek, R.; Matschke, V.; Theis, V.; Adamietz, I.A.; Bühler, H.; Theiss, C. Vascular Endothelial Growth Factor, Irradiation, and Axitinib Have Diverse Effects on Motility and Proliferation of Glioblastoma Multiforme Cells. Front. Oncol. 2017, 7, 182. [Google Scholar] [CrossRef]
- Schiffer, D.; Annovazzi, L.; Casalone, C.; Corona, C.; Mellai, M. Glioblastoma: Microenvironment and Niche Concept. Cancers 2018, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M. Vascular Endothelial Growth Factor (VEGF) and Its Receptor (VEGFR) Signaling in Angiogenesis: A Crucial Target for Anti- and Pro-Angiogenic Therapies. Genes Cancer 2011, 2, 1097–1105. [Google Scholar] [CrossRef]
- Xie, Y.; He, L.; Lugano, R.; Zhang, Y.; Cao, H.; He, Q.; Chao, M.; Liu, B.; Cao, Q.; Wang, J.; et al. Key molecular alterations in endothelial cells in human glioblastoma uncovered through single-cell RNA sequencing. JCI Insight 2021, 6, e150861. [Google Scholar] [CrossRef]
- Večeřa, J.; Procházková, J.; Šumberová, V.; Pánská, V.; Paculová, H.; Lánová, M.K.; Mašek, J.; Bohačiaková, D.; Andersson, E.R.; Pacherník, J. Hypoxia/Hif1α prevents premature neuronal differentiation of neural stem cells through the activation of Hes1. Stem Cell Res. 2020, 45, 101770. [Google Scholar] [CrossRef]
- Gustafsson, M.V.; Zheng, X.; Pereira, T.; Gradin, K.; Jin, S.; Lundkvist, J.; Ruas, J.L.; Poellinger, L.; Lendahl, U.; Bondesson, M. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell 2005, 9, 617–628. [Google Scholar] [CrossRef]
- Clausing, M.; William, D.; Preussler, M.; Biedermann, J.; Grützmann, K.; Richter, S.; Buchholz, F.; Temme, A.; Schröck, E.; Klink, B. Different Effects of RNAi-Mediated Downregulation or Chemical Inhibition of NAMPT in an Isogenic IDH Mutant and Wild-Type Glioma Cell Model. Int. J. Mol. Sci. 2022, 23, 5787. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, Y.; Shingu, T.; Feng, L.; Chen, Z.; Ogasawara, M.; Keating, M.J.; Kondo, S.; Huang, P. Metabolic alterations in highly tumorigenic glioblastoma cells: Preference for hypoxia and high dependency on glycolysis. J. Biol. Chem. 2011, 286, 32843–32853. [Google Scholar] [CrossRef]
- Saga, I.; Shibao, S.; Okubo, J.; Osuka, S.; Kobayashi, Y.; Yamada, S.; Fujita, S.; Urakami, K.; Kusuhara, M.; Yoshida, K.; et al. Integrated analysis identifies different metabolic signatures for tumor-initiating cells in a murine glioblastoma model. Neuro Oncol. 2014, 1, 1048–1056. [Google Scholar] [CrossRef]
- Sanzey, M.; Abdul Rahim, S.A.; Oudin, A.; Dirkse, A.; Kaoma, T.; Vallar, L.; Herold-Mende, C.; Bjerkvig, R.; Golebiewska, A.; Niclou, S.P. Comprehensive analysis of glycolytic enzymes as therapeutic targets in the treatment of glioblastoma. PLoS ONE 2015, 1, e0123544. [Google Scholar] [CrossRef] [PubMed]
- Marin-Valencia, I.; Cho, S.K.; Rakheja, D.; Hatanpaa, K.J.; Kapur, P.; Mashimo, T.; Jindal, A.; Vemireddy, V.; Good, L.B.; Raisanen, J.; et al. Glucose metabolism via the pentose phosphate pathway, glycolysis and Krebs cycle in an orthotopic mouse model of human brain tumors. NMR Biomed. 2012, 2, 1177–1186. [Google Scholar] [CrossRef]
- Erices, J.I.; Bizama, C.; Niechi, I.; Uribe, D.; Rosales, A.; Fabres, K.; Navarro-Martínez, G.; Torres, Á.; San Martín, R.; Roa, J.C.; et al. Glioblastoma Microenvironment and Invasiveness: New Insights and Therapeutic Targets. Int. J. Mol. Sci. 2023, 24, 7047. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lei, Q.; Yuan, B.; Liu, K.; Peng, L.; Xia, Z. A novel prognostic related lncRNA signature associated with amino acid metabolism in glioma. Front. Immunol. 2023, 14, 1014378. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, K.; Le Belle, J.E.; Muthukrishnan, S.D.; Sperry, J.; Condro, M.; Vlashi, E.; Pajonk, F.; Kornblum, H.I. Nicotinamide Adenine Dinucleotide Phosphate Oxidase Promotes Glioblastoma Radiation Resistance in a Phosphate and Tensin Homolog-Dependent Manner. Antioxid. Redox Signal. 2023, 39, 890–903. [Google Scholar] [CrossRef]
- Maraqah, H.H.; Abu-Asab, M.S.; Lee, H.S.; Aboud, O. Comparative survey of mitochondrial ultrastructure in IDH1-mutant astrocytoma and IDH1-wildtype glioblastoma (GBM). Ultrastruct. Pathol. 2023, 25, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.E.; Keatley, K.; Littlewood, D.T.; Meunier, B.; Holt, W.V.; An, Q.; Higgins, S.C.; Polyzoidis, S.; Stephenson, K.F.; Ashkan, K.; et al. Identification and functional prediction of mitochondrial complex III and IV mutations associated with glioblastoma. Neuro Oncol. 2015, 17, 942–952. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Lu, Y.; He, B.; Xie, T.; Yan, C.; Liu, T.; Wu, S.; Yeh, Y.; Li, Z.; Huang, W.; et al. Rab32 promotes glioblastoma migration and invasion via regulation of ERK/Drp1-mediated mitochondrial fission. Cell Death Dis. 2023, 14, 198. [Google Scholar] [CrossRef] [PubMed]
- Kulawiak, B.; Żochowska, M.; Bednarczyk, P.; Galuba, A.; Stroud, D.A.; Szewczyk, A. Loss of the large conductance calcium-activated potassium channel causes an increase in mitochondrial reactive oxygen species in glioblastoma cells. Pflugers Arch. 2023, 475, 1045–1060. [Google Scholar] [CrossRef] [PubMed]
- Wear, D.; Bhagirath, E.; Balachandar, A.; Vegh, C.; Pandey, S. Autophagy Inhibition via Hydroxychloroquine or 3-Methyladenine Enhances Chemotherapy-Induced Apoptosis in Neuro-Blastoma and Glioblastoma. Int. J. Mol. Sci. 2023, 24, 12052. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Oliva, R.; Domínguez-García, S.; Carrascal, L.; Abalos-Martínez, J.; Pardillo-Díaz, R.; Verástegui, C.; Castro, C.; Nunez-Abades, P.; Geribaldi-Doldán, N. Evolution of Experimental Models in the Study of Glioblastoma: Toward Finding Efficient Treatments. Front. Oncol. 2021, 10, 614295. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, H.B.; Blake, E.R.; Walder, A.S. A critique of the evidence for active host defence against cancer, based on personal studies of 27 murine tumours of spontaneous origin. Br. J. Cancer 1976, 33, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Peterson, D.L.; Sheridan, P.J.; Brown, W.E., Jr. Animal models for brain tumors: Historical perspectives and future directions. J. Neurosurg. 1994, 80, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Sughrue, M.E.; Yang, I.; Kane, A.J.; Rutkowski, M.J.; Fang, S.; James, C.D.; Parsa, A.T. Immunological considerations of modern animal models of malignant primary brain tumors. J. Transl. Med. 2009, 7, 84. [Google Scholar] [CrossRef]
- Seligman, A.M.; Shear, M.; Alexander, L. Studies in Carcinogenesis: VIII. Experimental Production of Brain Tumors in Mice with Methylcholanthrene. Am. J. Cancer 1939, 37, 364–395. [Google Scholar]
- Huse, J.T.; Holland, E.C. Genetically engineered mouse models of brain cancer and the promise of preclinical testing. Brain Pathol. 2009, 19, 132–143. [Google Scholar] [CrossRef]
- Grigore, F.N.; Yang, S.J.; Chen, C.C.; Koga, T. Pioneering models of pediatric brain tumors. Neoplasia 2023, 36, 100859. [Google Scholar] [CrossRef] [PubMed]
- Noorani, I. Genetically Engineered Mouse Models of Gliomas: Technological Developments for Translational Discoveries. Cancers 2019, 11, 1335. [Google Scholar] [CrossRef] [PubMed]
- Khaddour, K.; Johanns, T.M.; Ansstas, G. The Landscape of Novel Therapeutics and Challenges in Glioblastoma Multiforme: Contemporary State and Future Directions. Pharmaceuticals 2020, 13, 389. [Google Scholar] [CrossRef] [PubMed]
- Maes, W.; Van Gool, S.W. Experimental immunotherapy for malignant glioma: Lessons from two decades of research in the GL261 model. Cancer Immunol. Immunother. 2011, 60, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Sajjad, H.; Imtiaz, S.; Noor, T.; Siddiqui, Y.H.; Sajjad, A.; Zia, M. Cancer models in preclinical research: A chronicle review of advancement in effective cancer research. Anim. Model. Exp. Med. 2021, 4, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Letai, A.; Bhola, P.; Welm, A.L. Functional precision oncology: Testing tumors with drugs to identify vulnerabilities and novel combinations. Cancer Cell 2022, 40, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Guignard, F.; Zhao, D.; Liu, L.; Burns, D.K.; Mason, R.P.; Messing, A.; Parada, L.F. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell 2005, 8, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Uhrbom, L.; Dai, C.; Celestino, J.C.; Rosenblum, M.K.; Fuller, G.N.; Holland, E.C. Ink4a-Arf loss cooperates with KRas activation in astrocytes and neural progenitors to generate glioblastomas of various morphologies depending on activated Akt. Cancer Res. 2002, 62, 5551–5558. [Google Scholar]
- Alcantara Llaguno, S.; Chen, J.; Kwon, C.H.; Jackson, E.L.; Li, Y.; Burns, D.K.; Alvarez-Buylla, A.; Parada, L.F. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 2009, 15, 45–56. [Google Scholar] [CrossRef]
- Al-Sammarraie, N.; Ray, S.K. Applications of CRISPR-Cas9 Technology to Genome Editing in Glioblastoma Multiforme. Cells 2021, 10, 2342. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, K.; Wakimoto, H.; Iafrate, A.J.; Tanaka, S.; Loebel, F.; Lelic, N.; Wiederschain, D.; Bedel, O.; Deng, G.; Zhang, B.; et al. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell 2015, 28, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Szabo, E.; Schneider, H.; Seystahl, K.; Rushing, E.J.; Herting, F.; Weidner, K.M.; Weller, M. Autocrine VEGFR1 and VEGFR2 signaling promotes survival in human glioblastoma models in vitro and in vivo. Neuro Oncol. 2016, 18, 1242–1252. [Google Scholar] [CrossRef] [PubMed]
- Kaye, A.H.; Morstyn, G.; Gardner, I.; Pyke, K. Development of a xenograft glioma model in mouse brain. Cancer Res. 1986, 46, 1367–1373. [Google Scholar] [PubMed]
- Ponten, J. Neoplastic human glia cells in culture. In Human Tumor Cells In Vitro; Springer: Boston, MA, USA, 1975; pp. 175–185. [Google Scholar]
- Camphausen, K.; Purow, B.; Sproull, M.; Scott, T.; Ozawa, T.; Deen, D.F.; Tofilon, P.J. Orthotopic growth of human glioma cells quantitatively and qualitatively influences radiation-induced changes in gene expression. Cancer Res. 2005, 65, 10389–10393. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, J.E.; Kahng, J.Y.; Kim, S.H.; Park, J.S.; Yoon, S.J.; Um, J.Y.; Kim, W.K.; Lee, J.K.; Park, J.; et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 2018, 560, 243–247. [Google Scholar] [CrossRef]
- Rabotti, G.F.; Grove, A.S., Jr.; Sellers, R.L.; Anderson, W.R. Induction of multiple brain tumours (gliomata and leptomeningeal sarcomata) in dogs by Rous sarcoma virus. Nature 1966, 209, 884–886. [Google Scholar] [CrossRef] [PubMed]
- Paoloni, M.; Khanna, C. Translation of new cancer treatments from pet dogs to humans. Nat. Rev. Cancer 2008, 8, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N. Molecular pathology of malignant gliomas. Annu. Rev. Pathol. 2006, 1, 97–117. [Google Scholar] [CrossRef]
- Snyder, S.A.; Dewhirst, M.W.; Hauck, M.L. The role of hypoxia in canine cancer. Vet. Comp. Oncol. 2008, 6, 213–223. [Google Scholar] [CrossRef]
- Hubbard, M.E.; Arnold, S.; Bin Zahid, A.; Mc Pheeters, M.; Gerard O’Sullivan, M.; Tabaran, A.F.; Hunt, M.A.; Pluhar, G.E. Naturally Occurring Canine Glioma as a Model for Novel Therapeutics. Cancer Investig. 2018, 36, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Herranz, C.; Fernández, F.; Martín-Ibáñez, R.; Blasco, E.; Crespo, E.; De la Fuente, C.; Añor, S.; Rabanal, R.M.; Canals, J.M.; Pumarola, M. Spontaneously Arising Canine Glioma as a Potential Model for Human Glioma. J. Comp. Pathol. 2016, 154, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.B.; Anderson, K.J.; Boudreau, C.E.; Martinez-Ledesma, E.; Kocakavuk, E.; Johnson, K.C.; Barthel, F.P.; Varn, F.S.; Kassab, C.; Ling, X.; et al. Comparative Molecular Life History of Spontaneous Canine and Human Gliomas. Cancer Cell 2020, 37, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Ingram, M.; Buckwalter, J.G.; Jacques, D.B.; Freshwater, D.B.; Abts, R.M.; Techy, G.B.; Miyagi, K.; Shelden, C.H.; Rand, R.W.; English, L.W. Immunotherapy for recurrent malignant glioma: An interim report on survival. Neurol. Res. 1990, 12, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Stoica, G.; Lungu, G.; Martini-Stoica, H.; Waghela, S.; Levine, J.; Smith, R. Identification of cancer stem cells in dog glioblastoma. Vet. Pathol. 2009, 46, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Higgins, R.J.; Dickinson, P.J.; LeCouteur, R.A.; Bollen, A.W.; Wang, H.; Wang, H.; Corely, L.J.; Moore, L.M.; Zang, W.; Fuller, G.N. Spontaneous canine gliomas: Overexpression of EGFR, PDGFRalpha and IGFBP2 demonstrated by tissue microarray immunophenotyping. J. Neurooncol. 2010, 98, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, P.J.; LeCouteur, R.A.; Higgins, R.J.; Bringas, J.R.; Larson, R.F.; Yamashita, Y.; Krauze, M.T.; Forsayeth, J.; Noble, C.O.; Drummond, D.C.; et al. Canine spontaneous glioma: A translational model system for convection-enhanced delivery. Neuro Oncol. 2010, 2, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Bentley, R.T.; Ahmed, A.U.; Yanke, A.B.; Cohen-Gadol, A.A.; Dey, M. Dogs are man’s best friend: In sickness and in health. Neuro Oncol. 2017, 19, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Kimmelman, J.; Nalbantoglu, J. Faithful companions: A proposal for neurooncology trials in pet dogs. Cancer Res. 2007, 67, 4541–4544. [Google Scholar] [CrossRef]
- Schook, L.B.; Collares, T.V.; Darfour-Oduro, K.A.; De, A.K.; Rund, L.A.; Schachtschneider, K.M.; Seixas, F.K. Unraveling the swine genome: Implications for human health. Annu. Rev. Anim. Biosci. 2015, 3, 219–244. [Google Scholar] [CrossRef]
- Ruvinsky, A.; Rothschild, M.F.; Larson, G.; Gongora, J. Systematics and evolution of the pig. In The Genetics of the Pig, 2nd ed.; Rothschild, M.F., Ruvinsky, A., Eds.; CABI: Wallingford, UK, 2011; pp. 1–13. [Google Scholar]
- Lind, N.M.; Moustgaard, A.; Jelsing, J.; Vajta, G.; Cumming, P.; Hansen, A.K. The use of pigs in neuroscience: Modeling brain disorders. Neurosci. Biobehav. Rev. 2007, 31, 728–751. [Google Scholar] [CrossRef]
- Sauleau, P.; Lapouble, E.; Val-Laillet, D.; Malbert, C.H. The pig model in brain imaging and neurosurgery. Animal 2009, 3, 1138–1151. [Google Scholar] [CrossRef]
- Selek, L.; Seigneuret, E.; Nugue, G.; Wion, D.; Nissou, M.F.; Salon, C.; Seurin, M.J.; Carozzo, C.; Ponce, F.; Roger, T.; et al. Imaging and histological characterization of a human brain xenograft in pig: The first induced glioma model in a large animal. J. Neurosci. Methods 2014, 221, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Khoshnevis, M.; Carozzo, C.; Bonnefont-Rebeix, C.; Belluco, S.; Leveneur, O.; Chuzel, T.; Pillet-Michelland, E.; Dreyfus, M.; Roger, T.; Berger, F.; et al. Development of induced glioblastoma by implantation of a human xenograft in Yucatan minipig as a large animal model. J. Neurosci. Methods 2017, 282, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Boettcher, A.N.; Schachtschneider, K.M.; Schook, L.B.; Tuggle, C.K. Swine models for translational oncological research: An evolving landscape and regulatory considerations. Mamm. Genome 2022, 33, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.; Gibbs, R.A. Comparative primate genomics: Emerging patterns of genome content and dynamics. Nat. Rev. Genet. 2014, 15, 347–359. [Google Scholar] [CrossRef]
- Foster, C.; Sheng, W.A.; Heed, T.; Ben Hamed, S. The macaque ventral intraparietal area has expanded into three homologue human parietal areas. Prog. Neurobiol. 2022, 209, 102185. [Google Scholar] [CrossRef]
- Van Essen, D.C.; Donahue, C.J.; Coalson, T.S.; Kennedy, H.; Hayashi, T.; Glasser, M.F. Cerebral cortical folding, parcellation, and connectivity in humans, nonhuman primates, and mice. Proc. Natl. Acad. Sci. USA 2019, 116, 26173–26180. [Google Scholar] [CrossRef]
- Lonser, R.R.; Walbridge, S.; Vortmeyer, A.O.; Pack, S.D.; Nguyen, T.T.; Gogate, N.; Olson, J.J.; Akbasak, A.; Bobo, R.H.; Goffman, T.; et al. Induction of glioblastoma multiforme in nonhuman primates after therapeutic doses of fractionated whole-brain radiation therapy. J. Neurosurg. 2002, 9, 1378–1389. [Google Scholar] [CrossRef]
- Kanthaswamy, S.; Reader, R.; Tarara, R.; Oslund, K.; Allen, M.; Ng, J.; Grinberg, C.; Hyde, D.; Glenn, D.G.; Lerche, N. Large scale pedigree analysis leads to evidence for founder effects of Hypertrophic Cardiomyopathy in Rhesus Macaques (Macaca mulatta). J. Med. Primatol. 2014, 4, 288–291. [Google Scholar] [CrossRef]
- Comuzzie, A.G.; Cole, S.A.; Martin, L.; Carey, K.D.; Mahaney, M.C.; Blangero, J.; VandeBerg, J.L. The baboon as a nonhuman primate model for the study of the genetics of obesity. Obes. Res. 2003, 11, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Roelfsema, P.R.; Treue, S. Basic neuroscience research with nonhuman primates: A small but indispensable component of biomedical research. Neuron 2014, 82, 1200–1204. [Google Scholar] [CrossRef]
- Dray, B.K.; Raveendran, M.; Harris, R.A.; Benavides, F.; Gray, S.B.; Perez, C.J.; McArthur, M.J.; Williams, L.E.; Baze, W.B.; Doddapaneni, H.; et al. Mismatch repair gene mutations lead to lynch syndrome colorectal cancer in rhesus macaques. Genes Cancer 2018, 9, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Huszthy, P.C.; Daphu, I.; Niclou, S.P.; Stieber, D.; Nigro, J.M.; Sakariassen, P.Ø.; Miletic, H.; Thorsen, F.; Bjerkvig, R. In vivo models of primary brain tumors: Pitfalls and perspectives. Neuro Oncol. 2012, 14, 979–993. [Google Scholar] [CrossRef]
- Lowenstine, L.J. Neoplasms and Proliferative Disorders in Nonhuman Primates. In Primates; Benirschke, K., Ed.; Springer; New York, NY, USA, 1986; pp. 781–814.
- Kim, T.; Song, B.; Lee, I.S. Drosophila Glia: Models for Human Neurodevelopmental and Neurodegenerative Disorders. Int. J. Mol. Sci. 2020, 21, 4859. [Google Scholar] [CrossRef]
- Reiter, L.T.; Bier, E. Using Drosophila melanogaster to uncover human disease gene function and potential drug target proteins. Expert. Opin. Ther. Targets 2002, 6, 387–399. [Google Scholar]
- Wilson, C.W.; Chuang, P.T. Mechanism and evolution of cytosolic Hedgehog signal transduction. Development 2010, 137, 2079–2094. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Neufeld, T.P.; Pan, D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev. Biol. 2000, 221, 404–418. [Google Scholar] [CrossRef] [PubMed]
- Gateff, E.; Schneiderman, H.A. Neoplasms in mutant and cultured wild-tupe tissues of Drosophila. Natl. Cancer Inst. Monogr. 1969, 31, 365–397. [Google Scholar]
- Gateff, E. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science 1978, 200, 1448–1459. [Google Scholar] [CrossRef]
- Stork, T.; Bernardos, R.; Freeman, M.R. Analysis of glial cell development and function in Drosophila. Cold Spring Harb. Protoc. 2012, 2012, 1–17. [Google Scholar] [CrossRef]
- Read, R.D.; Cavenee, W.K.; Furnari, F.B.; Thomas, J.B. A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 2009, 5, e1000374. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.S.; Read, R.D. Drosophila melanogaster as a Model System for Human Glioblastomas. Adv. Exp. Med. Biol. 2019, 1167, 207–224. [Google Scholar] [PubMed]
- Saborio, J.G.; Young, E.E.; Chen, A.S.; Read, R.D. A protocol to use Drosophila melanogaster larvae to model human glioblastoma. STAR Protoc. 2022, 3, 101609. [Google Scholar] [CrossRef]
- Morgan, T.H. Sex limited inheritance in drosophila. Science 1910, 32, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Read, R.D. Drosophila melanogaster as a model system for human brain cancers. Glia 2011, 59, 1364–1376. [Google Scholar] [CrossRef]
- Bertrand, M.; Szeremeta, F.; Hervouet-Coste, N.; Sarou-Kanian, V.; Landon, C.; Morisset-Lopez, S.; Decoville, M. An adult Drosophila glioma model to highlight metabolic dysfunctions and evaluate the role of the serotonin 5-HT7 receptor as a potential therapeutic target. FASEB J. 2023, 37, e23230. [Google Scholar] [CrossRef]
- Jeibmann, A.; Paulus, W. Drosophila melanogaster as a model organism of brain diseases. Int. J. Mol. Sci. 2009, 10, 407–440. [Google Scholar] [CrossRef]
- Hughes, T.T.; Allen, A.L.; Bardin, J.E.; Christian, M.N.; Daimon, K.; Dozier, K.D.; Hansen, C.L.; Holcomb, L.M.; Ahlander, J. Drosophila as a genetic model for studying pathogenic human viruses. Virology 2012, 423, 1–5. [Google Scholar] [CrossRef]
- Zhao, S.; Huang, J.; Ye, J. A fresh look at zebrafish from the perspective of cancer research. J. Exp. Clin. Cancer Res. 2015, 34, 80. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.H.; Chua, H.L.; Gong, Z.; Lam, T.J.; Sin, Y.M. Development and maturation of the immune system in zebrafish, Danio rerio: A gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 2004, 28, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Guo, S. Using zebrafish to assess the impact of drugs on neural development and function. Expert. Opin. Drug Discov. 2009, 4, 715–726. [Google Scholar] [CrossRef] [PubMed]
- White, R.M. Genomic Approaches to Zebrafish Cancer. Adv. Exp. Med. Biol. 2016, 916, 125–145. [Google Scholar] [PubMed]
- Costa, B.; Estrada, M.F.; Mendes, R.V.; Fior, R. Zebrafish Avatars towards Personalized Medicine-A Comparative Review between Avatar Models. Cells 2020, 9, 293. [Google Scholar] [CrossRef]
- Hason, M.; Bartůněk, P. Zebrafish Models of Cancer-New Insights on Modeling Human Cancer in a Non-Mammalian Vertebrate. Genes 2019, 10, 935. [Google Scholar] [CrossRef]
- Teame, T.; Zhang, Z.; Ran, C.; Zhang, H.; Yang, Y.; Ding, Q.; Xie, M.; Gao, C.; Ye, Y.; Duan, M.; et al. The use of zebrafish (Danio rerio) as biomedical models. Anim. Front. 2019, 9, 68–77. [Google Scholar] [CrossRef]
- Rudzinska-Radecka, M.; Janczewski, Ł.; Gajda, A.; Godlewska, M.; Chmielewska-Krzesinska, M.; Wasowicz, K.; Podlasz, P. The Anti-Tumoral Potential of Phosphonate Analog of Sulforaphane in Zebrafish Xenograft Model. Cells 2021, 10, 3219. [Google Scholar] [CrossRef] [PubMed]
- Caja, L.; Dadras, M.S.; Mezheyeuski, A.; Rodrigues-Junior, D.M.; Liu, S.; Webb, A.T.; Gomez-Puerto, M.C.; Ten Dijke, P.; Heldin, C.H.; Moustakas, A. The protein kinase LKB1 promotes self-renewal and blocks invasiveness in glioblastoma. J. Cell Physiol. 2022, 237, 743–762. [Google Scholar] [CrossRef]
- Umans, R.A.; Ten Kate, M.; Pollock, C.; Sontheimer, H. Fishing for Contact: Modeling Perivascular Glioma Invasion in the Zebrafish Brain. ACS Pharmacol. Transl. Sci. 2020, 4, 1295–1305. [Google Scholar] [CrossRef]
- Gamble, J.T.; Reed-Harris, Y.; Barton, C.L.; La Du, J.; Tanguay, R.; Greenwood, J.A. Quantification of glioblastoma progression in zebrafish xenografts: Adhesion to laminin alpha 5 promotes glioblastoma microtumor formation and inhibits cell invasion. Biochem. Biophys. Res. Commun. 2018, 506, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.; Ye, Z.; Xiao, C.; Zhong, J.; Lancman, J.J.; Chen, X.; Pan, X.; Yang, Y.; Zhou, L.; Wang, X.; et al. Clinically relevant orthotopic xenograft models of patient-derived glioblastoma in zebrafish. Dis. Model. Mech. 2022, 15, dmm049109. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, X.P.; Wang, H.X.; Shen, Q.Y.; Li, X.P.; Wen, L.; Qin, X.J.; Jia, Q.L.; Kung, H.F.; Peng, Y. VEGF induces angiogenesis in a zebrafish embryo glioma model established by transplantation of human glioma cells. Oncol. Rep. 2012, 28, 937–942. [Google Scholar] [CrossRef] [PubMed]
- Almstedt, E.; Rosén, E.; Gloger, M.; Stockgard, R.; Hekmati, N.; Koltowska, K.; Krona, C.; Nelander, S. Real-time evaluation of glioblastoma growth in patient-specific zebrafish xenografts. Neuro Oncol. 2022, 24, 726–738. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.J.; Tsai, J.C.; Tseng, Y.T.; Wu, M.S.; Liu, W.S.; Lam, H.I.; Yu, J.H.; Nozell, S.E.; Benveniste, E.N. Small G protein Rac GTPases regulate the maintenance of glioblastoma stem-like cells in vitro and in vivo. Oncotarget 2017, 8, 18031–18049. [Google Scholar] [CrossRef]
- Pudelko, L.; Edwards, S.; Balan, M.; Nyqvist, D.; Al-Saadi, J.; Dittmer, J.; Almlöf, I.; Helleday, T.; Bräutigam, L. An orthotopic glioblastoma animal model suitable for high-throughput screenings. Neuro Oncol. 2018, 20, 1475–1484. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, L.; Astell, K.R.; Velikova, G.; Sieger, D. A Zebrafish Live Imaging Model Reveals Differential Responses of Microglia Toward Glioblastoma Cells In Vivo. Zebrafish 2016, 13, 523–534. [Google Scholar] [CrossRef]
- Wilms, C.; Lepka, K.; Häberlein, F.; Edwards, S.; Felsberg, J.; Pudelko, L.; Lindenberg, T.T.; Poschmann, G.; Qin, N.; Volbracht, K.; et al. Glutaredoxin 2 promotes SP-1-dependent CSPG4 transcription and migration of wound healing NG2 glia and glioma cells: Enzymatic Taoism. Redox Biol. 2022, 49, 102221. [Google Scholar] [CrossRef]
- Berghmans, S.; Jette, C.; Langenau, D.; Hsu, K.; Stewart, R.; Look, T.; Kanki, J.P. Making waves in cancer research: New models in the zebrafish. Biotechniques 2005, 39, 227–237. [Google Scholar] [CrossRef]
- Peglion, F.; Coumailleau, F.; Etienne-Manneville, S. Live Imaging of Microtubule Dynamics in Glioblastoma Cells Invading the Zebrafish Brain. J. Vis. Exp. 2022, 185, e64093. [Google Scholar]
- Kim, C.H.; Ueshima, E.; Muraoka, O.; Tanaka, H.; Yeo, S.Y.; Huh, T.L.; Miki, N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 1996, 216, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Rampazzo, E.; Persano, L.; Pistollato, F.; Moro, E.; Frasson, C.; Porazzi, P.; Della Puppa, A.; Bresolin, S.; Battilana, G.; Indraccolo, S.; et al. Wnt activation promotes neuronal differentiation of glioblastoma. Cell Death Dis. 2013, 4, e500. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.A.; Freundlich, T.; Weissman, T.A.; Schoppik, D.; Wang, X.C.; Zimmerman, S.; Ciruna, B.; Sanes, J.R.; Lichtman, J.W.; Schier, A.F. Zebrabow: Multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 2013, 140, 2835–2846. [Google Scholar] [CrossRef] [PubMed]
- Vittori, M.; Motaln, H.; Turnšek, T.L. The study of glioma by xenotransplantation in zebrafish early life stages. J. Histochem. Cytochem. 2015, 63, 749–761. [Google Scholar] [CrossRef] [PubMed]
- Gamble, J.T.; Elson, D.J.; Greenwood, J.A.; Tanguay, R.L.; Kolluri, S.K. The Zebrafish Xenograft Models for Investigating Cancer and Cancer Therapeutics. Biology 2021, 10, 252. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Patron, L.A.; Agudelo-Dueñas, N.; Madrid-Wolff, J.; Venegas, J.A.; González, J.M.; Forero-Shelton, M.; Akle, V. Xenotransplantation of Human glioblastoma in Zebrafish larvae: In vivo imaging and proliferation assessment. Biol. Open 2019, 8, bio043257. [Google Scholar] [CrossRef] [PubMed]
- Breznik, B.; Motaln, H.; Vittori, M.; Rotter, A.; Lah Turnšek, T. Mesenchymal stem cells differentially affect the invasion of distinct glioblastoma cell lines. Oncotarget 2017, 8, 25482–25499. [Google Scholar] [CrossRef] [PubMed]
- Zeng, A.; Ye, T.; Cao, D.; Huang, X.; Yang, Y.; Chen, X.; Xie, Y.; Yao, S.; Zhao, C. Identify a Blood-Brain Barrier Penetrating Drug-TNB using Zebrafish Orthotopic Glioblastoma Xenograft Model. Sci. Rep. 2017, 7, 14372. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Cui, W.; Gu, A.; Xu, C.; Yu, S.C.; Li, T.T.; Cui, Y.H.; Zhang, X.; Bian, X.W. A novel zebrafish xenotransplantation model for study of glioma stem cell invasion. PLoS ONE 2013, 8, e61801. [Google Scholar] [CrossRef]
- Finotto, L.; Cole, B.; Giese, W.; Baumann, E.; Claeys, A.; Vanmechelen, M.; Decraene, B.; Derweduwe, M.; Dubroja Lakic, N.; Shankar, G.; et al. Single-cell profiling and zebrafish avatars reveal LGALS1 as immunomodulating target in glioblastoma. EMBO Mol. Med. 2023, 15, e18144. [Google Scholar] [CrossRef]
- Welker, A.M.; Jaros, B.D.; An, M.; Beattie, C.E. Changes in tumor cell heterogeneity after chemotherapy treatment in a xenograft model of glioblastoma. Neuroscience 2017, 356, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Banasavadi-Siddegowda, Y.K.; Welker, A.M.; An, M.; Yang, X.; Zhou, W.; Shi, G.; Imitola, J.; Li, C.; Hsu, S.; Wang, J.; et al. PRMT5 as a druggable target for glioblastoma therapy. Neuro Oncol. 2018, 20, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Haldi, M.; Ton, C.; Seng, W.L.; McGrath, P. Human melanoma cells transplanted into zebrafish proliferate, migrate, produce melanin, form masses and stimulate angiogenesis in zebrafish. Angiogenesis 2006, 9, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, Y.; Yao, T.; Jia, R. Benefits of Zebrafish Xenograft Models in Cancer Research. Front. Cell Dev. Biol. 2021, 9, 616551. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, Y.; Gao, Y.; Zhu, Z.; Zeng, X.; Liang, W.; Sun, S.; Chen, X.; Wang, H. Radiated glioblastoma cell-derived exosomal circ_0012381 induce M2 polarization of microglia to promote the growth of glioblastoma by CCL2/CCR2 axis. J. Transl. Med. 2022, 20, 388. [Google Scholar] [CrossRef] [PubMed]
- Canella, A.; Welker, A.M.; Yoo, J.Y.; Xu, J.; Abas, F.S.; Kesanakurti, D.; Nagarajan, P.; Beattie, C.E.; Sulman, E.P.; Liu, J.; et al. Efficacy of Onalespib, a Long-Acting Second-Generation HSP90 Inhibitor, as a Single Agent and in Combination with Temozolomide against Malignant Gliomas. Clin. Cancer Res. 2017, 23, 6215–6226. [Google Scholar] [CrossRef] [PubMed]
- Welker, A.M.; Jaros, B.D.; Puduvalli, V.K.; Imitola, J.; Kaur, B.; Beattie, C.E. Correction: Standardized orthotopic xenografts in zebrafish reveal glioma cell-line-specific characteristics and tumor cell heterogeneity. Dis. Model. Mech. 2016, 9, 1063–1065. [Google Scholar] [CrossRef] [PubMed]
- Wehmas, L.C.; Tanguay, R.L.; Punnoose, A.; Greenwood, J.A. Developing a Novel Embryo-Larval Zebrafish Xenograft Assay to Prioritize Human Glioblastoma Therapeutics. Zebrafish 2016, 13, 317–329. [Google Scholar] [CrossRef]
- Engebraaten, O.; Hjortland, G.O.; Hirschberg, H.; Fodstad, O. Growth of precultured human glioma specimens in nude rat brain. J. Neurosurg. 1999, 90, 125–132. [Google Scholar] [CrossRef]
- Miyai, M.; Tomita, H.; Soeda, A.; Yano, H.; Iwama, T.; Hara, A. Current trends in mouse models of glioblastoma. J. Neurooncol. 2017, 135, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Caruso Bavisotto, C.; Graziano, F.; Rappa, F.; Marino Gammazza, A.; Logozzi, M.; Fais, S.; Maugeri, R.; Bucchieri, F.; Conway de Macario, E.; Macario, A.J.L.; et al. Exosomal Chaperones and miRNAs in Gliomagenesis: State-of-Art and Theranostics Perspectives. Int. J. Mol. Sci. 2018, 19, 2626. [Google Scholar] [CrossRef] [PubMed]
- Strand, A.D.; Aragaki, A.K.; Baquet, Z.C.; Hodges, A.; Cunningham, P.; Holmans, P.; Jones, K.R.; Jones, L.; Kooperberg, C.; Olson, J.M. Conservation of regional gene expression in mouse and human brain. PLoS Genet. 2007, 3, e59. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Sohrabi, A.; Seidlits, S.K. Integrating the glioblastoma microenvironment into engineered experimental models. Future Sci. OA 2017, 3, FSO189. [Google Scholar] [CrossRef]


| Tumor Model | The Origin of the Tumor | Advantages | Disadvantages | References |
|---|---|---|---|---|
| Carcinogen-induced tumor model | Tumor induced by carcinogens | - Used to evaluate efficacy and toxicity of anticancer agents - Study of resistance and response biomarkers | - High animal mortality rate - Location and number of lesions are not uniform among individuals | [59] |
| Syngeneic tumor model | Transplanted mouse tumor cells | - Simple system able to recapitulate host immunity - Easily reproducible - Easy to manipulate | - It does not faithfully represent the tumor microenvironment - Reduced genetic heterogeneity of cells compared to the native tumor | [60] |
| Genetically engineered and viral-vector-mediated transduction model | De novo formed tumor induced by introduced mutations | - Models used to identify detailed information about the sequence of events underlying genetic alterations that occur in response to specific mutations - Adapted for the study of the microenvironment in tumor biology - Models suitable for preclinical and therapeutic studies | - Models often not representative of the genetic changes involved in GBM in humans - They do not faithfully reflect the intra-tumoral genetic and phenotypic heterogeneities of GBM therapeutic studies because of the beginning of the tumor reproducibility failure | [61,62,63] |
| Xenograft model of GBM (heterotopic) | Patient-derived tumor | - Models suitable for testing the effectiveness of drugs - Genetically stable | - An immunocompromised mouse is required to develop this model - It does not allow for testing of immunomodulatory therapies - It does not reproduce the original niche | [64] |
| Xenograft model of GBM (orthotopic) | Patient-derived tumor | - Models suitable for testing the effectiveness of drugs - Genetically stable - Models capable of maintaining the original tumor architecture and histological characteristics of the human tumor of origin | - An immunocompromised mouse is required to develop this model - It does not allow for testing of immunomodulatory therapies - It does not reproduce the original niche | [64] |
| Characteristics | Zebrafish Model | Rodent Models (e.g., Mice, Rats) | Non-Human Primate Models | Refs. |
|---|---|---|---|---|
| Genetics and Manipulation | Well-characterized genome, relatively simple genetic manipulation via CRISPR/Cas9. | Extensive genetic tools available, including transgenic and knockout technologies. | Closer genetic similarity to humans, enabling translational research but with higher technical demands. | [126,166] |
| Size and Accessibility | Small size, easy tissue observation and access for in vivo microscopy. | Larger size, variable accessibility depending on tumor location, and invasive procedures required. | Similar size to humans, facilitating surgical techniques and imaging studies, but with ethical and logistical challenges. | [95,124] |
| Technical Drawbacks | Lack of some genes conserved in humans. | Potential tumor heterogeneity due to different genetic backgrounds. | Ethical considerations, higher costs, and longer timelines for experiments. | [127,167] |
| Life Cycle and Development | Rapid life cycle and embryonic transparency facilitate tumor development studies. | Longer life span, enabling longitudinal studies and recapitulation of disease progression. | Longer life span, closer developmental timeline to humans, allowing for investigation of aging-related factors. | [95,123] |
| Costs and Time | Relatively low in terms of cost and time for model creation and maintenance. | Moderate costs for model creation and maintenance, varying depending on genetic manipulations. | Higher costs due to housing, care, and ethical considerations; longer timelines for experiments. | [106,123] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alberti, G.; Amico, M.D.; Caruso Bavisotto, C.; Rappa, F.; Marino Gammazza, A.; Bucchieri, F.; Cappello, F.; Scalia, F.; Szychlinska, M.A. Speeding up Glioblastoma Cancer Research: Highlighting the Zebrafish Xenograft Model. Int. J. Mol. Sci. 2024, 25, 5394. https://doi.org/10.3390/ijms25105394
Alberti G, Amico MD, Caruso Bavisotto C, Rappa F, Marino Gammazza A, Bucchieri F, Cappello F, Scalia F, Szychlinska MA. Speeding up Glioblastoma Cancer Research: Highlighting the Zebrafish Xenograft Model. International Journal of Molecular Sciences. 2024; 25(10):5394. https://doi.org/10.3390/ijms25105394
Chicago/Turabian StyleAlberti, Giusi, Maria Denise Amico, Celeste Caruso Bavisotto, Francesca Rappa, Antonella Marino Gammazza, Fabio Bucchieri, Francesco Cappello, Federica Scalia, and Marta Anna Szychlinska. 2024. "Speeding up Glioblastoma Cancer Research: Highlighting the Zebrafish Xenograft Model" International Journal of Molecular Sciences 25, no. 10: 5394. https://doi.org/10.3390/ijms25105394







