Evolution of Glutamate Metabolism via GLUD2 Enhances Lactate-Dependent Synaptic Plasticity and Complex Cognition
Abstract
1. Introduction
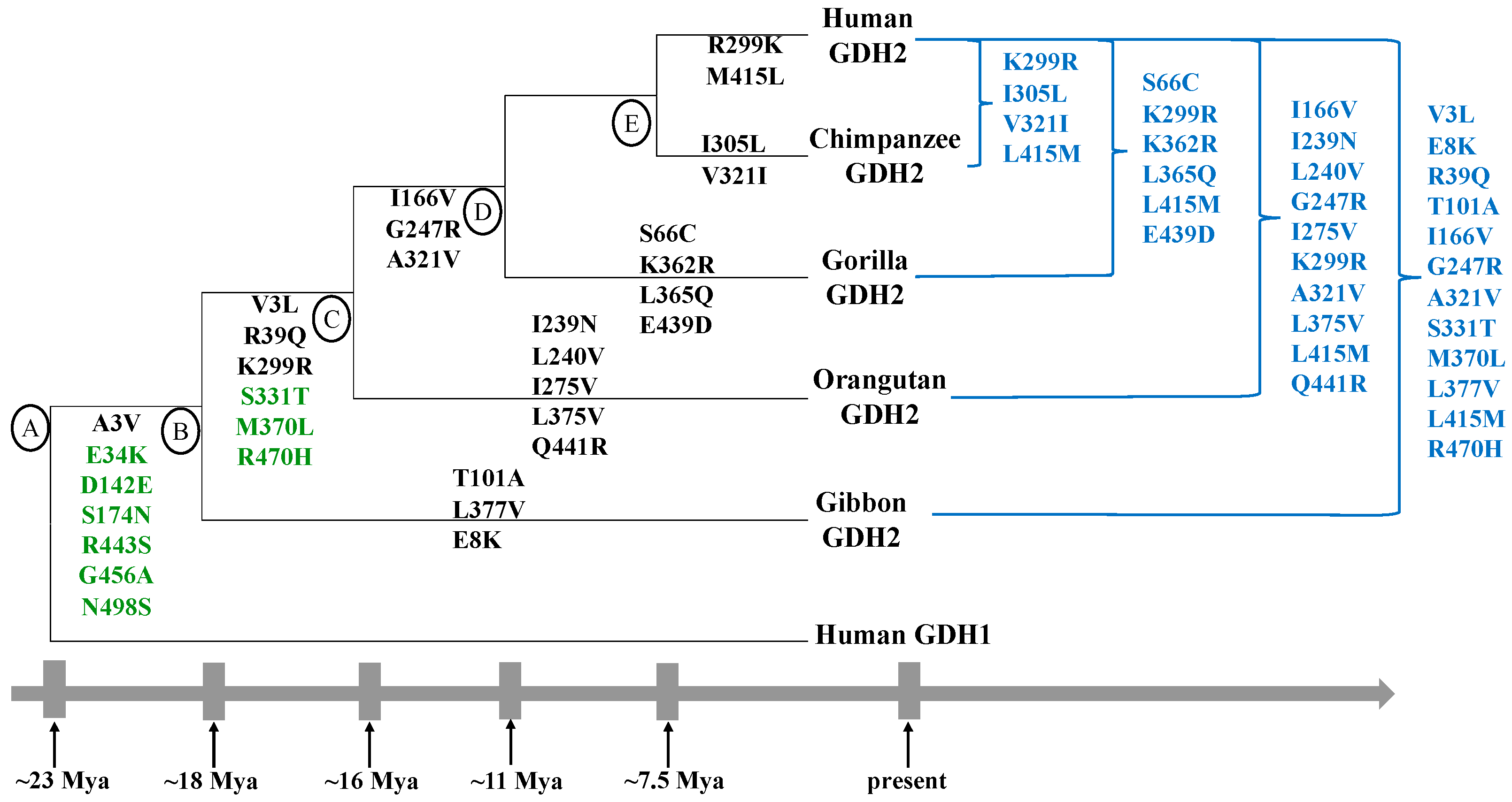
2. Emergence and Evolution of the GLUD2 Gene
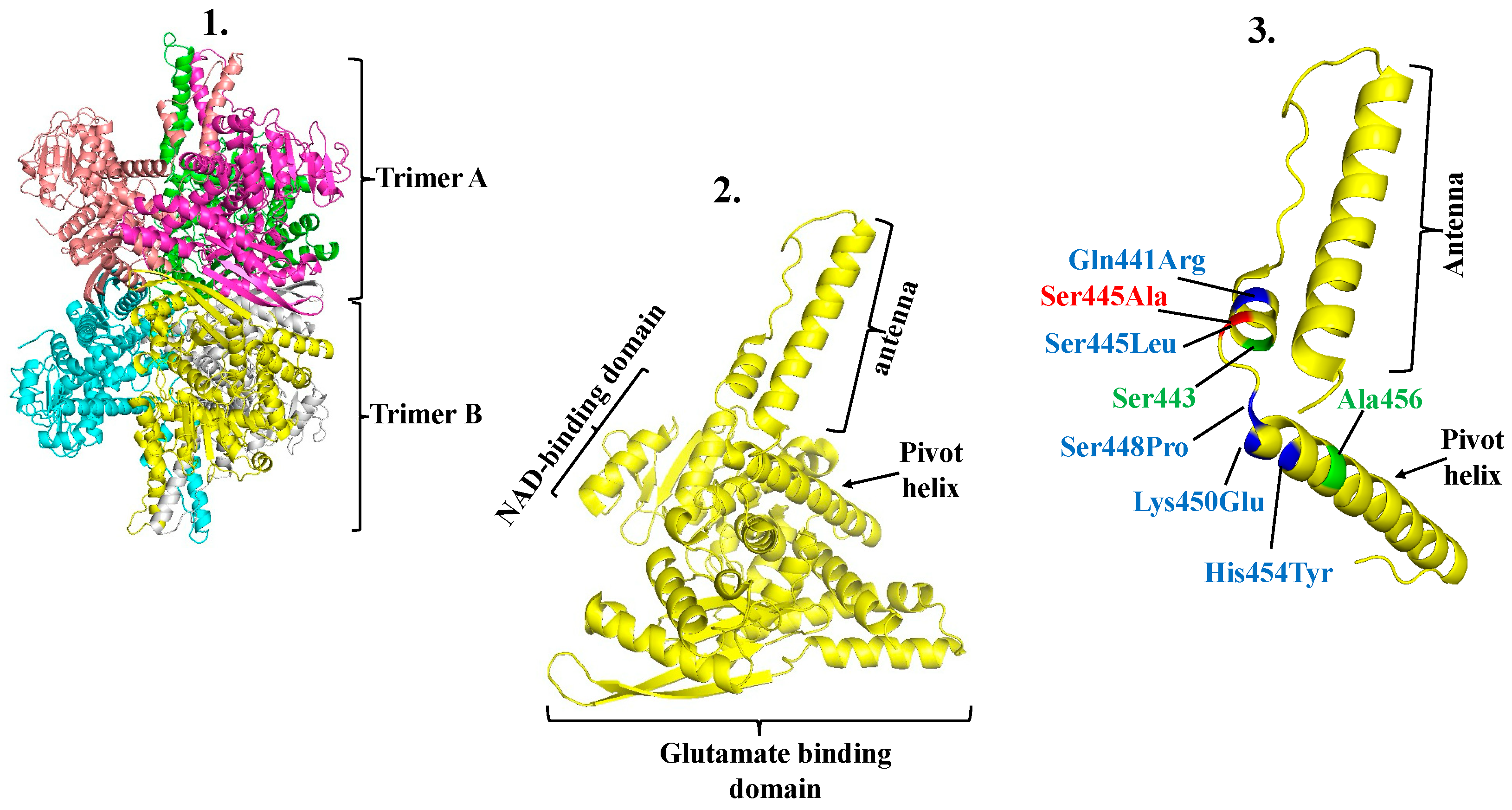
3. Functional and Structural Aspects of hGDH1 and hGDH2
4. Insights from the Study of Transgenic Mice Carrying the Human GLUD2 Gene
5. The Role of hGDH1/2 in Cellular Energetics: Clues from the Regulation of Glucose Homeostasis
6. The Potential Role of hGDH2 in Lactate Metabolism: Clues from Studies on Testicular Tissue
7. The Potential Role of GDH1/2 in Ammonia Metabolism: Clues from Studies on Renal Tissue
8. GLUD2 in Brain Biology
8.1. Studies on Human Central Nervous Tissue
8.2. Studies on Tg Mice’s Central Nervous System
8.3. GLUD2 Enhances Synaptic Plasticity (LTP) by Increasing NMDA Receptor Currents
8.4. GLUD2 Enhancement of Synaptic Plasticity Is Lactate-Mediated

8.5. GLUD2 Enhances Dendritic Spine Density/Synaptogenesis
8.6. GLUD2 Enhances Sensory Perception and Complex Cognition
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siletti, K.; Hodge, R.; Albiach, A.M.; Lee, K.W.; Ding, S.-L.; Hu, L.; Lönnerberg, P.; Bakken, T.; Casper, T.; Clark, M.; et al. Transcriptomic Diversity of Cell Types across the Adult Human Brain. Science 2023, 382, eadd7046. [Google Scholar] [CrossRef]
- Enard, W. The Molecular Basis of Human Brain Evolution. Curr. Biol. 2016, 26, R1109–R1117. [Google Scholar] [CrossRef] [PubMed]
- Weninger, A.; Arlotta, P. A Family Portrait of Human Brain Cells. Science 2023, 382, 168–169. [Google Scholar] [CrossRef] [PubMed]
- Vanderhaeghen, P.; Polleux, F. Developmental Mechanisms Underlying the Evolution of Human Cortical Circuits. Nat. Rev. Neurosci. 2023, 24, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Rochefort, N.L.; Konnerth, A. Dendritic Spines: From Structure to In Vivo Function. EMBO Rep. 2012, 13, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, S.; Stimpson, C.D.; Bauernfeind, A.L.; Schapiro, S.J.; Baze, W.B.; McArthur, M.J.; Bronson, E.; Hopkins, W.D.; Semendeferi, K.; Jacobs, B.; et al. Dendritic Morphology of Pyramidal Neurons in the Chimpanzee Neocortex: Regional Specializations and Comparison to Humans. Cereb. Cortex 2013, 23, 2429–2436. [Google Scholar] [CrossRef] [PubMed]
- Oberheim, N.A.; Wang, X.; Goldman, S.; Nedergaard, M. Astrocytic Complexity Distinguishes the Human Brain. Trends Neurosci. 2006, 29, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Jorstad, N.L.; Song, J.H.T.; Exposito-Alonso, D.; Suresh, H.; Castro-Pacheco, N.; Krienen, F.M.; Yanny, A.M.; Close, J.; Gelfand, E.; Long, B.; et al. Comparative Transcriptomics Reveals Human-Specific Cortical Features. Science 2023, 382. [Google Scholar] [CrossRef] [PubMed]
- Jorstad, N.L.; Close, J.; Johansen, N.; Yanny, A.M.; Barkan, E.R.; Travaglini, K.J.; Bertagnolli, D.; Campos, J.; Casper, T.; Crichton, K.; et al. Transcriptomic Cytoarchitecture Reveals Principles of Human Neocortex Organization. Science 2023, 382, eade9516. [Google Scholar] [CrossRef]
- Fu, X.; Giavalisco, P.; Liu, X.; Catchpole, G.; Fu, N.; Ning, Z.-B.; Guo, S.; Yan, Z.; Somel, M.; Pääbo, S.; et al. Rapid Metabolic Evolution in Human Prefrontal Cortex. Proc. Natl. Acad. Sci. USA 2011, 108, 6181–6186. [Google Scholar] [CrossRef]
- Grossman, L.I.; Schmidt, T.R.; Wildman, D.E.; Goodman, M. Molecular Evolution of Aerobic Energy Metabolism in Primates. Mol. Phylogenet Evol. 2001, 18, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Malenka, R.C.; Nicoll, R.A. NMDA-Receptor-Dependent Synaptic Plasticity: Multiple Forms and Mechanisms. Trends Neurosci. 1993, 16, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Engert, F.; Bonhoeffer, T. Dendritic Spine Changes Associated with Hippocampal Long-Term Synaptic Plasticity. Nature 1999, 399, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. How to Make an Ape Brain. Nat. Genet. 2004, 36, 1034–1036. [Google Scholar] [CrossRef] [PubMed]
- de Kock, C.P.J.; Feldmeyer, D. Shared and Divergent Principles of Synaptic Transmission between Cortical Excitatory Neurons in Rodent and Human Brain. Front. Synaptic Neurosci. 2023, 15, 1274383. [Google Scholar] [CrossRef] [PubMed]
- Cotney, J.; Leng, J.; Yin, J.; Reilly, S.K.; DeMare, L.E.; Emera, D.; Ayoub, A.E.; Rakic, P.; Noonan, J.P. The Evolution of Lineage-Specific Regulatory Activities in the Human Embryonic Limb. Cell 2013, 154, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Kronenberg, Z.N.; Fiddes, I.T.; Gordon, D.; Murali, S.; Cantsilieris, S.; Meyerson, O.S.; Underwood, J.G.; Nelson, B.J.; Chaisson, M.J.P.; Dougherty, M.L.; et al. High-Resolution Comparative Analysis of Great Ape Genomes. Science 2018, 360, eaar6343. [Google Scholar] [CrossRef] [PubMed]
- Muntané, G.; Horvath, J.E.; Hof, P.R.; Ely, J.J.; Hopkins, W.D.; Raghanti, M.A.; Lewandowski, A.H.; Wray, G.A.; Sherwood, C.C. Analysis of Synaptic Gene Expression in the Neocortex of Primates Reveals Evolutionary Changes in Glutamatergic Neurotransmission. Cereb. Cortex 2015, 25, 1596–1607. [Google Scholar] [CrossRef] [PubMed]
- Cáceres, M.; Lachuer, J.; Zapala, M.A.; Redmond, J.C.; Kudo, L.; Geschwind, D.H.; Lockhart, D.J.; Preuss, T.M.; Barlow, C. Elevated Gene Expression Levels Distinguish Human from Non-Human Primate Brains. Proc. Natl. Acad. Sci. USA 2003, 100, 13030–13035. [Google Scholar] [CrossRef]
- Spanaki, C.; Sidiropoulou, K.; Petraki, Z.; Diskos, K.; Konstantoudaki, X.; Volitaki, E.; Mylonaki, K.; Savvaki, M.; Plaitakis, A. Glutamate-Specific Gene Linked to Human Brain Evolution Enhances Synaptic Plasticity and Cognitive Processes. iScience 2024, 27, 108821. [Google Scholar] [CrossRef]
- Shashidharan, P.; Michaelidis, T.M.; Robakis, N.K.; Kresovali, A.; Papamatheakis, J.; Plaitakis, A. Novel Human Glutamate Dehydrogenase Expressed in Neural and Testicular Tissues and Encoded by an X-Linked Intronless Gene. J. Biol. Chem. 1994, 269, 16971–16976. [Google Scholar] [CrossRef] [PubMed]
- Burki, F.; Kaessmann, H. Birth and Adaptive Evolution of a Hominoid Gene That Supports High Neurotransmitter Flux. Nat. Genet. 2004, 36, 1061–1063. [Google Scholar] [CrossRef] [PubMed]
- Kaessmann, H.; Vinckenbosch, N.; Long, M. RNA-Based Gene Duplication: Mechanistic and Evolutionary Insights. Nat. Rev. Genet. 2009, 10, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Plaitakis, A.; Kalef-Ezra, E.; Kotzamani, D.; Zaganas, I.; Spanaki, C. The Glutamate Dehydrogenase Pathway and Its Roles in Cell and Tissue Biology in Health and Disease. Biology 2017, 6, 11. [Google Scholar] [CrossRef]
- Plaitakis, A.; Latsoudis, H.; Spanaki, C. The Human GLUD2 Glutamate Dehydrogenase and Its Regulation in Health and Disease. Neurochem. Int. 2011, 59, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Kanavouras, K.; Mastorodemos, V.; Borompokas, N.; Spanaki, C.; Plaitakis, A. Properties and Molecular Evolution of Human GLUD2 (Neural and Testicular Tissue-specific) Glutamate Dehydrogenase. J. Neurosci. Res. 2007, 85, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Zaganas, I.; Plaitakis, A. Single Amino Acid Substitution (G456A) in the Vicinity of the GTP Binding Domain of Human Housekeeping Glutamate Dehydrogenase Markedly Attenuates GTP Inhibition and Abolishes the Cooperative Behavior of the Enzyme. J. Biol. Chem. 2002, 277, 26422–26428. [Google Scholar] [CrossRef]
- Zaganas, I.; Spanaki, C.; Karpusas, M.; Plaitakis, A. Substitution of Ser for Arg-443 in the Regulatory Domain of Human Housekeeping (GLUD1) Glutamate Dehydrogenase Virtually Abolishes Basal Activity and Markedly Alters the Activation of the Enzyme by ADP and l-Leucine. J. Biol. Chem. 2002, 277, 46552–46558. [Google Scholar] [CrossRef]
- Choi, M.-M.; Kim, E.-A.; Yang, S.-J.; Choi, S.Y.; Cho, S.-W.; Huh, J.-W. Amino Acid Changes within Antenna Helix Are Responsible for Different Regulatory Preferences of Human Glutamate Dehydrogenase Isozymes. J. Biol. Chem. 2007, 282, 19510–19517. [Google Scholar] [CrossRef]
- Choi, M.-M.; Hwang, E.Y.; Kim, E.-A.; Huh, J.-W.; Cho, S.-W. Identification of Amino Acid Residues Responsible for Different GTP Preferences of Human Glutamate Dehydrogenase Isozymes. Biochem. Biophys. Res. Commun. 2008, 368, 742–747. [Google Scholar] [CrossRef]
- Yang, S.-J.; Huh, J.-W.; Hong, H.-N.; Kim, T.U.; Cho, S.-W. Important Role of Ser443 in Different Thermal Stability of Human Glutamate Dehydrogenase Isozymes 1. FEBS Lett. 2004, 562, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Rosso, L.; Marques, A.C.; Reichert, A.S.; Kaessmann, H. Mitochondrial Targeting Adaptation of the Hominoid-Specific Glutamate Dehydrogenase Driven by Positive Darwinian Selection. PLoS Genet. 2008, 4, e1000150. [Google Scholar] [CrossRef] [PubMed]
- Aleshina, Y.A.; Aleshin, V.A. Evolutionary Changes in Primate Glutamate Dehydrogenases 1 and 2 Influence the Protein Regulation by Ligands, Targeting and Posttranslational Modifications. Int. J. Mol. Sci. 2024, 25, 4341. [Google Scholar] [CrossRef]
- Litso, I.; Plaitakis, A.; Fadouloglou, V.E.; Providaki, M.; Kokkinidis, M.; Zaganas, I. Structural Evolution of Primate Glutamate Dehydrogenase 2 as Revealed by In Silico Predictions and Experimentally Determined Structures. Biomolecules 2023, 14, 22. [Google Scholar] [CrossRef] [PubMed]
- Almécija, S.; Hammond, A.S.; Thompson, N.E.; Pugh, K.D.; Moyà-Solà, S.; Alba, D.M. Fossil Apes and Human Evolution. Science 2021, 372, eabb4363. [Google Scholar] [CrossRef] [PubMed]
- Carbone, L.; Alan Harris, R.; Gnerre, S.; Veeramah, K.R.; Lorente-Galdos, B.; Huddleston, J.; Meyer, T.J.; Herrero, J.; Roos, C.; Aken, B.; et al. Gibbon Genome and the Fast Karyotype Evolution of Small Apes. Nature 2014, 513, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Langergraber, K.E.; Prüfer, K.; Rowney, C.; Boesch, C.; Crockford, C.; Fawcett, K.; Inoue, E.; Inoue-Muruyama, M.; Mitani, J.C.; Muller, M.N.; et al. Generation Times in Wild Chimpanzees and Gorillas Suggest Earlier Divergence Times in Great Ape and Human Evolution. Proc. Natl. Acad. Sci. USA 2012, 109, 15716–15721. [Google Scholar] [CrossRef]
- Almécija, S.; Smaers, J.B.; Jungers, W.L. The Evolution of Human and Ape Hand Proportions. Nat. Commun. 2015, 6, 7717. [Google Scholar] [CrossRef]
- Moorjani, P.; Amorim, C.E.G.; Arndt, P.F.; Przeworski, M. Variation in the Molecular Clock of Primates. Proc. Natl. Acad. Sci. USA 2016, 113, 10607–10612. [Google Scholar] [CrossRef]
- Kalef-Ezra, E.; Kotzamani, D.; Zaganas, I.; Katrakili, N.; Plaitakis, A.; Tokatlidis, K. Import of a Major Mitochondrial Enzyme Depends on Synergy between Two Distinct Helices of Its Presequence. Biochem. J. 2016, 473, 2813–2829. [Google Scholar] [CrossRef]
- DeSilva, J.M.; Traniello, J.F.A.; Claxton, A.G.; Fannin, L.D. When and Why Did Human Brains Decrease in Size? A New Change-Point Analysis and Insights From Brain Evolution in Ants. Front. Ecol. Evol. 2021, 9, 742639. [Google Scholar] [CrossRef]
- Li, Q.; Guo, S.; Jiang, X.; Bryk, J.; Naumann, R.; Enard, W.; Tomita, M.; Sugimoto, M.; Khaitovich, P.; Pääbo, S. Mice Carrying a Human GLUD2 Gene Recapitulate Aspects of Human Transcriptome and Metabolome Development. Proc. Natl. Acad. Sci. USA 2016, 113, 5358–5363. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.J.; Schmidt, T.; Fang, J.; Wu, J.; Siuzdak, G.; Stanley, C.A. The Structure of Apo Human Glutamate Dehydrogenase Details Subunit Communication and Allostery. J. Mol. Biol. 2002, 318, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Dimovasili, C.; Fadouloglou, V.E.; Kefala, A.; Providaki, M.; Kotsifaki, D.; Kanavouras, K.; Sarrou, I.; Plaitakis, A.; Zaganas, I.; Kokkinidis, M. Crystal Structure of Glutamate Dehydrogenase 2, a Positively Selected Novel Human Enzyme Involved in Brain Biology and Cancer Pathophysiology. J. Neurochem. 2021, 157, 802–815. [Google Scholar] [CrossRef] [PubMed]
- Spanaki, C.; Zaganas, I.; Kleopa, K.A.; Plaitakis, A. Human GLUD2 Glutamate Dehydrogenase Is Expressed in Neural and Testicular Supporting Cells. J. Biol. Chem. 2010, 285, 16748–16756. [Google Scholar] [CrossRef] [PubMed]
- Spanaki, C.; Kotzamani, D.; Kleopa, K.; Plaitakis, A. Evolution of GLUD2 Glutamate Dehydrogenase Allows Expression in Human Cortical Neurons. Mol. Neurobiol. 2016, 53, 5140–5148. [Google Scholar] [CrossRef] [PubMed]
- Plaitakis, A.; Kotzamani, D.; Petraki, Z.; Delidaki, M.; Rinotas, V.; Zaganas, I.; Douni, E.; Sidiropoulou, K.; Spanaki, C. Transgenic Mice Carrying GLUD2 as a Tool for Studying the Expressional and the Functional Adaptation of This Positive Selected Gene in Human Brain Evolution. Neurochem. Res. 2019, 44, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Clayton, P.T.; Eaton, S.; Aynsley-Green, A.; Edginton, M.; Hussain, K.; Krywawych, S.; Datta, V.; Malingré, H.E.M.; Berger, R.; van den Berg, I.E.T. Hyperinsulinism in Short-Chain L-3-Hydroxyacyl-CoA Dehydrogenase Deficiency Reveals the Importance of β-Oxidation in Insulin Secretion. J. Clin. Investig. 2001, 108, 457–465. [Google Scholar] [CrossRef]
- Herrero-Yraola, A. Regulation of Glutamate Dehydrogenase by Reversible ADP-Ribosylation in Mitochondria. EMBO J. 2001, 20, 2404–2412. [Google Scholar] [CrossRef]
- Hoffpauir, Z.A.; Sherman, E.; Smith, T.J. Dissecting the Antenna in Human Glutamate Dehydrogenase: Understanding Its Role in Subunit Communication and Allosteric Regulation. Biochemistry 2019, 58, 4195–4206. [Google Scholar] [CrossRef]
- Mastorodemos, V.; Kanavouras, K.; Sundaram, S.; Providaki, M.; Petraki, Z.; Kokkinidis, M.; Zaganas, I.; Logothetis, D.E.; Plaitakis, A. Side-chain Interactions in the Regulatory Domain of Human Glutamate Dehydrogenase Determine Basal Activity and Regulation. J. Neurochem. 2015, 133, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Stanley, C.A. Two Genetic Forms of Hyperinsulinemic Hypoglycemia Caused by Dysregulation of Glutamate Dehydrogenase. Neurochem. Int. 2011, 59, 465–472. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stanley, C.A. Regulation of Glutamate Metabolism and Insulin Secretion by Glutamate Dehydrogenase in Hypoglycemic Children. Am. J. Clin. Nutr. 2009, 90, 862S–866S. [Google Scholar] [CrossRef] [PubMed]
- Kanavouras, K.; Borompokas, N.; Latsoudis, H.; Stagourakis, A.; Zaganas, I.; Plaitakis, A. Mutations in Human GLUD2 Glutamate Dehydrogenase Affecting Basal Activity and Regulation. J. Neurochem. 2009, 109, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Plaitakis, A.; Latsoudis, H.; Kanavouras, K.; Ritz, B.; Bronstein, J.M.; Skoula, I.; Mastorodemos, V.; Papapetropoulos, S.; Borompokas, N.; Zaganas, I.; et al. Gain-of-Function Variant in GLUD2 Glutamate Dehydrogenase Modifies Parkinson’s Disease Onset. Eur. J. Hum. Genet. 2010, 18, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gong, J.; Ding, L.; Zhang, Z.; Pan, X.; Chen, X.; Guo, W.; Zhang, X.; Yang, X.; Peng, G.; et al. Functional Validation of a Human GLUD2 Variant in a Murine Model of Parkinson’s Disease. Cell Death Dis. 2020, 11, 897. [Google Scholar] [CrossRef]
- Smith, E.L.; Austin, B.M.; Blumenthal, K.M.; Nyc, J.F. The Enzymes; Boyer, P.D., Ed.; Academic Press: New York, NY, USA, 1975. [Google Scholar]
- Bahi-Buisson, N.; Roze, E.; Dionisi, C.; Escande, F.; Valayannopoulos, V.; Feillet, F.; Heinrichs, C. Neurological Aspects of Hyperinsulinism–Hyperammonaemia Syndrome. Dev. Med. Child. Neurol. 2008, 50, 945–949. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.F.; Cember, A.T.J.; Matschinsky, F.M. Glutamate Dehydrogenase: Role in Regulating Metabolism and Insulin Release in Pancreatic β-Cells. J. Appl. Physiol. 2018, 125, 419–428. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A Single–Cell Type Transcriptomics Map of Human Tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]
- Brauchi, S.; Rauch, M.C.; Alfaro, I.E.; Cea, C.; Concha, I.I.; Benos, D.J.; Reyes, J.G. Kinetics, Molecular Basis, and Differentiation of L-lactate Transport in Spermatogenic Cells. Am. J. Physiol. Cell Physiol. 2005, 288, C523–C534. [Google Scholar] [CrossRef] [PubMed]
- Ni, F.-D.; Hao, S.-L.; Yang, W.-X. Multiple Signaling Pathways in Sertoli Cells: Recent Findings in Spermatogenesis. Cell Death Dis. 2019, 10, 541. [Google Scholar] [CrossRef] [PubMed]
- Matos, B.; Publicover, S.J.; Castro, L.F.C.; Esteves, P.J.; Fardilha, M. Brain and Testis: More Alike than Previously Thought? Open Biol. 2021, 11, 200322. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Stern, S.A.; Bozdagi, O.; Huntley, G.W.; Walker, R.H.; Magistretti, P.J.; Alberini, C.M. Astrocyte-Neuron Lactate Transport Is Required for Long-Term Memory Formation. Cell 2011, 144, 810–823. [Google Scholar] [CrossRef] [PubMed]
- Sonnewald, U.; Westergaard, N.; Petersen, S.B.; Unsgård, G.; Schousboe, A. Metabolism of [U -13 C]Glutamate in Astrocytes Studied by 13 C NMR Spectroscopy: Incorporation of More Label into Lactate than into Glutamine Demonstrates the Importance of the Tricarboxylic Acid Cycle. J. Neurochem. 1993, 61, 1179–1182. [Google Scholar] [CrossRef] [PubMed]
- Sonnewald, U. Glutamate Synthesis Has to Be Matched by Its Degradation—Where Do All the Carbons Go? J. Neurochem. 2014, 131, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Olsen, G.M.; Sonnewald, U. Glutamate: Where Does It Come from and Where Does It Go? Neurochem. Int. 2015, 88, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Nissim, I. Newer Aspects of Glutamine/Glutamate Metabolism: The Role of Acute PH Changes. Am. J. Physiol. Ren. Physiol. 1999, 277, F493–F497. [Google Scholar] [CrossRef] [PubMed]
- Treberg, J.R.; Clow, K.A.; Greene, K.A.; Brosnan, M.E.; Brosnan, J.T. Systemic Activation of Glutamate Dehydrogenase Increases Renal Ammoniagenesis: Implications for the Hyperinsulinism/Hyperammonemia Syndrome. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E1219–E1225. [Google Scholar] [CrossRef]
- Romanov, V.; Whyard, T.; Bonala, R.; Johnson, F.; Grollman, A. Glutamate Dehydrogenase Requirement for Apoptosis Induced by Aristolochic Acid in Renal Tubular Epithelial Cells. Apoptosis 2011, 16, 1217–1228. [Google Scholar] [CrossRef]
- Voss, C.M.; Arildsen, L.; Nissen, J.D.; Waagepetersen, H.S.; Schousboe, A.; Maechler, P.; Ott, P.; Vilstrup, H.; Walls, A.B. Glutamate Dehydrogenase Is Important for Ammonia Fixation and Amino Acid Homeostasis in Brain During Hyperammonemia. Front. Neurosci. 2021, 15, 646291. [Google Scholar] [CrossRef] [PubMed]
- Drews, L.; Zimmermann, M.; Westhoff, P.; Brilhaus, D.; Poss, R.E.; Bergmann, L.; Wiek, C.; Brenneisen, P.; Piekorz, R.P.; Mettler-Altmann, T.; et al. Ammonia Inhibits Energy Metabolism in Astrocytes in a Rapid and Glutamate Dehydrogenase 2-Dependent Manner. Dis. Model. Mech. 2020, 13, dmm047134. [Google Scholar] [CrossRef]
- Huang, F.; Luo, X.; Ou, Y.; Gao, Z.; Tang, Q.; Chu, Z.; Zhu, X.; He, Y. Control of Histone Demethylation by Nuclear-Localized α-Ketoglutarate Dehydrogenase. Science 2023, 381, eadf8822. [Google Scholar] [CrossRef]
- Bonvento, G.; Bolaños, J.P. Astrocyte-Neuron Metabolic Cooperation Shapes Brain Activity. Cell Metab. 2021, 33, 1546–1564. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.V.; Schousboe, A. Glial Glutamine Homeostasis in Health and Disease. Neurochem. Res. 2023, 48, 1100–1128. [Google Scholar] [CrossRef] [PubMed]
- Lehre, K.; Levy, L.; Ottersen, O.; Storm-Mathisen, J.; Danbolt, N. Differential Expression of Two Glial Glutamate Transporters in the Rat Brain: Quantitative and Immunocytochemical Observations. J. Neurosci. 1995, 15, 1835–1853. [Google Scholar] [CrossRef] [PubMed]
- Danbolt, N.C.; Furness, D.N.; Zhou, Y. Neuronal vs. Glial Glutamate Uptake: Resolving the Conundrum. Neurochem. Int. 2016, 98, 29–45. [Google Scholar] [CrossRef]
- Chen, W.; Mahadomrongkul, V.; Berger, U.V.; Bassan, M.; DeSilva, T.; Tanaka, K.; Irwin, N.; Aoki, C.; Rosenberg, P.A. The Glutamate Transporter GLT1a Is Expressed in Excitatory Axon Terminals of Mature Hippocampal Neurons. J. Neurosci. 2004, 24, 1136–1148. [Google Scholar] [CrossRef]
- Danbolt, N.C. Glutamate Uptake. Prog. Neurobiol. 2001, 65, 1–105. [Google Scholar] [CrossRef]
- McKenna, M.C. The Glutamate-glutamine Cycle Is Not Stoichiometric: Fates of Glutamate in Brain. J. Neurosci. Res. 2007, 85, 3347–3358. [Google Scholar] [CrossRef]
- McKenna, M.C.; Sonnewald, U.; Huang, X.; Stevenson, J.; Zielke, H.R. Exogenous Glutamate Concentration Regulates the Metabolic Fate of Glutamate in Astrocytes. J. Neurochem. 1996, 66, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Nissen, J.D.; Pajęcka, K.; Stridh, M.H.; Skytt, D.M.; Waagepetersen, H.S. Dysfunctional TCA-Cycle Metabolism in Glutamate Dehydrogenase Deficient Astrocytes. Glia 2015, 63, 2313–2326. [Google Scholar] [CrossRef] [PubMed]
- Westergaard, N.; Drejer, J.; Schousboe, A.; Sonnewald, U. Evaluation of the Importance of Transamination versus Deamination in Astrocytic Metabolism of [U-13C] Glutamate. Glia 1996, 17, 160–168. [Google Scholar] [CrossRef]
- Yu, A.C.; Schousboec, A.; Hertz, L. Metabolic Fate of 14C-Labeled Glutamate in Astrocytes in Primary Cultures. J. Neurochem. 1982, 39, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Sonnewald, U.; White, L.R.; Ødegård, E.; Westergaard, N.; Bakken, I.J.; Aasly, J.; Unsgård, G.; Schousboe, A. MRS Study of Glutamate Metabolism in Cultured Neurons/Glia. Neurochem. Res. 1996, 21, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Schousboe, A.; Westergaard, N.; Waagepetersen, H.S.; Larsson, O.M.; Bakken, I.J.; Sonnewald, U. Trafficking between Glia and Neurons of TCA Cycle Intermediates and Related Metabolites. Glia 1997, 21, 99–105. [Google Scholar] [CrossRef]
- Farinelli, S.E.; Nicklas, W.J. Glutamate Metabolism in Rat Cortical Astrocyte Cultures. J. Neurochem. 1992, 58, 1905–1915. [Google Scholar] [CrossRef] [PubMed]
- Hohnholt, M.C.; Andersen, V.H.; Andersen, J.V.; Christensen, S.K.; Karaca, M.; Maechler, P.; Waagepetersen, H.S. Glutamate Dehydrogenase Is Essential to Sustain Neuronal Oxidative Energy Metabolism during Stimulation. J. Cereb. Blood Flow. Metab. 2018, 38, 1754–1768. [Google Scholar] [CrossRef]
- Jourdain, P.; Rothenfusser, K.; Ben-Adiba, C.; Allaman, I.; Marquet, P.; Magistretti, P.J. Dual Action of L-Lactate on the Activity of NR2B-Containing NMDA Receptors: From Potentiation to Neuroprotection. Sci. Rep. 2018, 8, 13472. [Google Scholar] [CrossRef]
- Yang, L.; Yang, Q.; Zhang, K.; Li, Y.-J.; Wu, Y.-M.; Liu, S.-B.; Zheng, L.-H.; Zhao, M.-G. Neuroprotective Effects of Daphnetin against NMDA Receptor-Mediated Excitotoxicity. Molecules 2014, 19, 14542–14555. [Google Scholar] [CrossRef]
- Magistretti, P.J.; Allaman, I. Lactate in the Brain: From Metabolic End-Product to Signalling Molecule. Nat. Rev. Neurosci. 2018, 19, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Descalzi, G.; Gao, V.; Steinman, M.Q.; Suzuki, A.; Alberini, C.M. Lactate from Astrocytes Fuels Learning-Induced MRNA Translation in Excitatory and Inhibitory Neurons. Commun. Biol. 2019, 2, 247. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.S.; Dankovich, T.M.; Mandad, S.; Rammner, B.; Jähne, S.; Salimi, V.; Koerbs, C.; Leibrandt, R.; Urlaub, H.; Schikorski, T.; et al. A Large-Scale Nanoscopy and Biochemistry Analysis of Postsynaptic Dendritic Spines. Nat. Neurosci. 2021, 24, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Rilling, J.K.; Glasser, M.F.; Preuss, T.M.; Ma, X.; Zhao, T.; Hu, X.; Behrens, T.E.J. The Evolution of the Arcuate Fasciculus Revealed with Comparative DTI. Nat. Neurosci. 2008, 11, 426–428. [Google Scholar] [CrossRef] [PubMed]
- Sierpowska, J.; Bryant, K.L.; Janssen, N.; Blazquez Freches, G.; Römkens, M.; Mangnus, M.; Mars, R.B.; Piai, V. Comparing Human and Chimpanzee Temporal Lobe Neuroanatomy Reveals Modifications to Human Language Hubs beyond the Frontotemporal Arcuate Fasciculus. Proc. Natl. Acad. Sci. USA 2022, 119, e2118295119. [Google Scholar] [CrossRef] [PubMed]
- Ragozzino, M.E. The Contribution of the Medial Prefrontal Cortex, Orbitofrontal Cortex, and Dorsomedial Striatum to Behavioral Flexibility. Ann. N. Y. Acad. Sci. 2007, 1121, 355–375. [Google Scholar] [CrossRef] [PubMed]
- Heisler, J.M.; Morales, J.; Donegan, J.J.; Jett, J.D.; Redus, L.; O’Connor, J.C. The Attentional Set Shifting Task: A Measure of Cognitive Flexibility in Mice. J. Vis. Exp. 2015, 96, e51944. [Google Scholar] [CrossRef] [PubMed]
- Bissonette, G.B.; Powell, E.M.; Roesch, M.R. Neural Structures Underlying Set-Shifting: Roles of Medial Prefrontal Cortex and Anterior Cingulate Cortex. Behav. Brain Res. 2013, 250, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Maren, S.; Phan, K.L.; Liberzon, I. The Contextual Brain: Implications for Fear Conditioning, Extinction and Psychopathology. Nat. Rev. Neurosci. 2013, 14, 417–428. [Google Scholar] [CrossRef]
- Fanselow, M.S.; Poulos, A.M. The Neuroscience of Mammalian Associative Learning. Annu. Rev. Psychol. 2005, 56, 207–234. [Google Scholar] [CrossRef]
- Garelick, M.G.; Storm, D.R. The Relationship between Memory Retrieval and Memory Extinction. Proc. Natl. Acad. Sci. USA 2005, 102, 9091–9092. [Google Scholar] [CrossRef] [PubMed]
- Nash, A.; Aumann, T.D.; Pigoni, M.; Lichtenthaler, S.F.; Takeshima, H.; Munro, K.M.; Gunnersen, J.M. Lack of Sez6 Family Proteins Impairs Motor Functions, Short-Term Memory, and Cognitive Flexibility and Alters Dendritic Spine Properties. Cereb. Cortex 2020, 30, 2167–2184. [Google Scholar] [CrossRef] [PubMed]
- Festa, L.K.; Irollo, E.; Platt, B.J.; Tian, Y.; Floresco, S.; Meucci, O. CXCL12-Induced Rescue of Cortical Dendritic Spines and Cognitive Flexibility. Elife 2020, 9, e49717. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plaitakis, A.; Sidiropoulou, K.; Kotzamani, D.; Litso, I.; Zaganas, I.; Spanaki, C. Evolution of Glutamate Metabolism via GLUD2 Enhances Lactate-Dependent Synaptic Plasticity and Complex Cognition. Int. J. Mol. Sci. 2024, 25, 5297. https://doi.org/10.3390/ijms25105297
Plaitakis A, Sidiropoulou K, Kotzamani D, Litso I, Zaganas I, Spanaki C. Evolution of Glutamate Metabolism via GLUD2 Enhances Lactate-Dependent Synaptic Plasticity and Complex Cognition. International Journal of Molecular Sciences. 2024; 25(10):5297. https://doi.org/10.3390/ijms25105297
Chicago/Turabian StylePlaitakis, Andreas, Kyriaki Sidiropoulou, Dimitra Kotzamani, Ionela Litso, Ioannis Zaganas, and Cleanthe Spanaki. 2024. "Evolution of Glutamate Metabolism via GLUD2 Enhances Lactate-Dependent Synaptic Plasticity and Complex Cognition" International Journal of Molecular Sciences 25, no. 10: 5297. https://doi.org/10.3390/ijms25105297
APA StylePlaitakis, A., Sidiropoulou, K., Kotzamani, D., Litso, I., Zaganas, I., & Spanaki, C. (2024). Evolution of Glutamate Metabolism via GLUD2 Enhances Lactate-Dependent Synaptic Plasticity and Complex Cognition. International Journal of Molecular Sciences, 25(10), 5297. https://doi.org/10.3390/ijms25105297





