A Systematic Review of Genetic Polymorphisms Associated with Binge Eating Disorder
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Eligibility Criteria
2.3. Information Sources
2.4. Search
2.5. Study Selection
2.6. Data Collection Process
2.7. Data Items
2.8. Risk of Biases in Individual Studies
3. Results
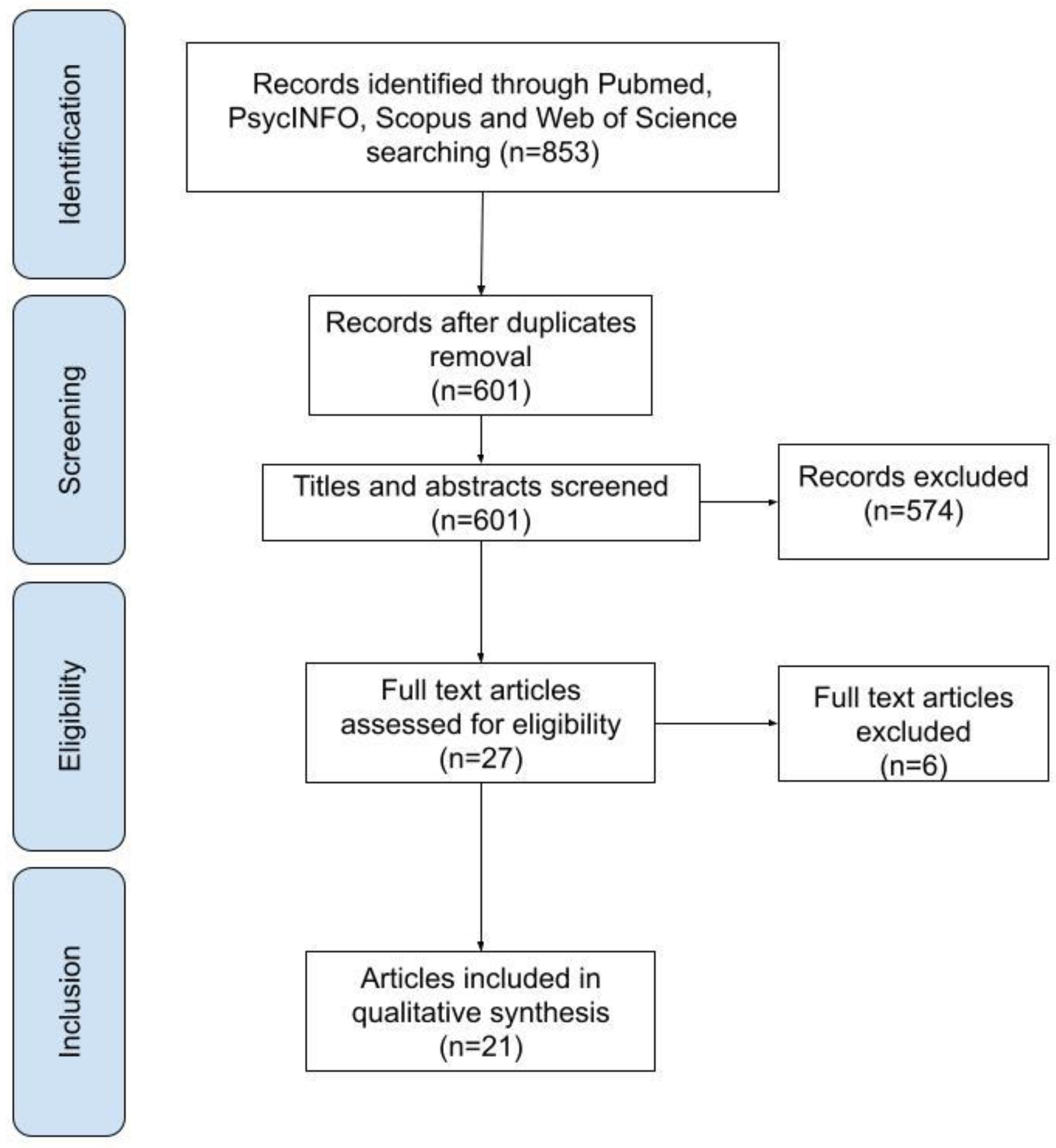
3.1. Study Selection
3.2. Study Characteristics
3.3. Risk of Bias within Studies
3.4. Results of Individual Studies
3.4.1. Serotonergic Genes
3.4.2. Dopaminergic Genes
3.4.3. Other Genes
4. Discussion
4.1. Summary of Evidence
4.2. Limitations
- The number of studies exploring associations between many genetic polymorphisms and BED is limited, and in some cases, the sample size used was small.
- The relationship between some polymorphisms and BED could also be influenced by gene–gene or gene–environment interactions.
- We could not pool data collected for a meta-analysis due to the heterogeneity of the genetic polymorphisms observed.
4.3. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Author | Reason for Exclusion | Comments |
|---|---|---|
| Castellini et al. (2017) [40] | Wrong sample | Not BED group, but only clinical populations with other eating disorders, such as BN and AN. |
| Gamero-Villarroel et al. (2015) [41] | Wrong sample | Not BED group; the study group included patients with binge eating behavior diagnosed with either BN or BED. |
| Genis-Mendoza et al. (2019) [42] | Wrong sample | Childhood, adolescence sample. |
| Hebebrand et al. (2004) [43] | Wrong sample | Childhood, adolescence sample. |
| Hersrud et al. (2009) [44] | Wrong sample | Not BED group. |
| Shinohara et al. (2004) [45] | Wrong sample | Not BED group, but only clinical populations with other eating disorders, such as BN and AN. |
References
- American Psychiatric Association. American Psychiatric Association, 2013. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; APA: Washington, DC, USA, 2013. [Google Scholar]
- Spitzer, R.L.; Yanovski, S.; Wadden, T.; Wing, R.; Marcus, M.D.; Stunkard, A.; Devlin, M.; Mitchell, J.; Hasin, D.; Horne, R.L. Binge Eating Disorder: Its Further Validation in a Multisite Study. Int. J. Eat. Disord. 1993. [Google Scholar] [CrossRef]
- Striegel-Moore, R.H.; Franko, D.L. Epidemiology of Binge Eating Disorder. Int. J. Eat. Disord. 2003, 34, S19–S29. [Google Scholar] [CrossRef] [Green Version]
- de Zwaan, M. Binge Eating Disorder and Obesity. Int. J. Obes. 2001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, J.D.; Tasca, G.A.; Little, J.; Chyurlia, L.; Ritchie, K.; Yeh, E.; Doucette, S.; Obregon, A.-M.; Bulman, D.E.; Doucet, E.; et al. Effects of Fat Mass and Obesity-Associated (FTO) Gene Polymorphisms on Binge Eating in Women with Binge-Eating Disorder: The Moderating Influence of Attachment Style. Nutrition 2019, 61, 208–212. [Google Scholar] [CrossRef]
- Bello, N.T.; Hajnal, A. Dopamine and Binge Eating Behaviors. Pharmacol. Biochem. Behav. 2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, C. The Epidemiology and Genetics of Binge Eating Disorder (BED). CNS Spectr. 2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naef, L.; Pitman, K.A.; Borgland, S.L. Mesolimbic Dopamine and Its Neuromodulators in Obesity and Binge Eating. CNS Spectr. 2015. [Google Scholar] [CrossRef]
- Monteleone, P.; Tortorella, A.; Castaldo, E.; Maj, M. Association of a Functional Serotonin Transporter Gene Polymorphism with Binge Eating Disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006, 141B, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Burnet, P.W.; Smith, K.A.; Cowen, P.J.; Fairburn, C.G.; Harrison, P.J. Allelic Variation of the 5-HT2C Receptor (HTR2C) in Bulimia Nervosa and Binge Eating Disorder. Psychiatr. Genet. 1999, 9, 101–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricca, V.; Nacmias, B.; Cellini, E.; Di Bernardo, M.; Rotella, C.M.; Sorbi, S. 5-HT2A Receptor Gene Polymorphism and Eating Disorders. Neurosci. Lett. 2002, 323, 105–108. [Google Scholar] [CrossRef]
- Ceccarini, M.R.; Tasegian, A.; Franzago, M.; Patria, F.F.; Albi, E.; Codini, M.; Conte, C.; Bertelli, M.; Dalla Ragione, L.; Stuppia, L.; et al. 5-HT2AR and BDNF Gene Variants in Eating Disorders Susceptibility. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2020, 183, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Dalley, J.W.; Roiser, J.P. Dopamine, Serotonin and Impulsivity. Neuroscience 2012. [Google Scholar] [CrossRef] [Green Version]
- Val-Laillet, D.; Aarts, E.; Weber, B.; Ferrari, M.; Quaresima, V.; Stoeckel, L.E.; Alonso-Alonso, M.; Audette, M.; Malbert, C.H.; Stice, E. Neuroimaging and Neuromodulation Approaches to Study Eating Behavior and Prevent and Treat Eating Disorders and Obesity. NeuroImage Clin. 2015. [Google Scholar] [CrossRef]
- Gonzalez, L.M.; Mota-Zamorano, S.; Garcia-Herraiz, A.; Lopez-Nevado, E.; Gervasini, G. Genetic Variants in Dopamine Pathways Affect Personality Dimensions Displayed by Patients with Eating Disorders. Eat. Weight Disord. Anorex. Bulim. Obes. 2019. [Google Scholar] [CrossRef]
- Davis, C.; Levitan, R.D.; Kaplan, A.S.; Carter, J.; Reid, C.; Curtis, C.; Patte, K.; Kennedy, J.L. Dopamine Transporter Gene (DAT1) Associated with Appetite Suppression to Methylphenidate in a Case-Control Study of Binge Eating Disorder. Neuropsychopharmacology 2007, 32, 2199–2206. [Google Scholar] [CrossRef] [PubMed]
- Leehr, E.J.; Schag, K.; Bruckmann, C.; Plewnia, C.; Zipfel, S.; Nieratschker, V.; Giel, K.E. A Putative Association of COMT Val(108/158)Met with Impulsivity in Binge Eating Disorder. Eur. Eat. Disord. Rev. 2016, 24, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Kindler, J.; Bailer, U.; de Zwaan, M.; Fuchs, K.; Leisch, F.; Grün, B.; Strnad, A.; Stojanovic, M.; Windisch, J.; Lennkh-Wolfsberg, C.; et al. No Association of the Neuropeptide Y (Leu7Pro) and Ghrelin Gene (Arg51Gln, Leu72Met, Gln90Leu) Single Nucleotide Polymorphisms with Eating Disorders. Nord. J. Psychiatry 2011, 65, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, P.; Tortorella, A.; Martiadis, V.; Di Filippo, C.; Canestrelli, B.; Maj, M. The CDNA 385C to A Missense Polymorphism of the Endocannabinoid Degrading Enzyme Fatty Acid Amide Hydrolase (FAAH) Is Associated with Overweight/Obesity but Not with Binge Eating Disorder in Overweight/Obese Women. Psychoneuroendocrinology 2008, 33, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Tortorella, A.; Monteleone, P.; del Giudice, E.M.; Cirillo, G.; Perrone, L.; Castaldo, E.; Maj, M. Melanocortin-4 Receptor Molecular Scanning and pro-Opiomelanocortin R236G Variant Screening in Binge Eating Disorder. Psychiatr. Genet. 2005, 15, 161. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, P.; Tortorella, A.; Docimo, L.; Maldonato, M.N.; Canestrelli, B.; De Luca, L.; Maj, M. Investigation of 3111T/C Polymorphism of the CLOCK Gene in Obese Individuals with or without Binge Eating Disorder: Association with Higher Body Mass Index. Neurosci. Lett. 2008, 435, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Altman, D.; Antes, G.; Atkins, D.; Barbour, V.; Barrowman, N.; Berlin, J.A.; et al. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, G.; Shea, B.; O’Connell, D.; Peterson, J.; Welch, V.; Losos, M.; Tugwell, P. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality If Nonrandomized Studies in Meta-Analyses. 2012. Available online: http//www.ohri.ca/programs/clinical_epidemiology/oxford.asp (accessed on 22 August 2020).
- Palmeira, L.; Cunha, M.; Padez, C.; Alvarez, M.; Pinto-Gouveia, J.; Manco, L. Association Study of Variants in Genes FTO, SLC6A4, DRD2, BDNF and GHRL with Binge Eating Disorder (BED) in Portuguese Women. Psychiatry Res. 2019, 273, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.; Levitan, R.D.; Kaplan, A.S.; Carter, J.; Reid, C.; Curtis, C.; Patte, K.; Hwang, R.; Kennedy, J.L. Reward Sensitivity and the D2 Dopamine Receptor Gene: A Case-Control Study of Binge Eating Disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 2008, 32, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.A.; Levitan, R.D.; Reid, C.; Carter, J.C.; Kaplan, A.S.; Patte, K.A.; King, N.; Curtis, C.; Kennedy, J.L. Dopamine for “Wanting” and Opioids for “Liking”: A Comparison of Obese Adults with and without Binge Eating. Obesity 2009, 17, 1220–1225. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.; Levitan, R.D.; Yilmaz, Z.; Kaplan, A.S.; Carter, J.C.; Kennedy, J.L. Binge Eating Disorder and the Dopamine D2 Receptor: Genotypes and Sub-Phenotypes. Prog. Neuropsychopharmacol. Biol. Psychiatry 2012, 38, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Palacios, A.; Canto, P.; Tejeda, M.E.; Stephano, S.; Lujan, H.; Garcia-Garcia, E.; Rojano-Mejia, D.; Mendez, J.P. Complete Sequence of the ANKK1 Gene in Mexican-Mestizo Individuals with Obesity, with or without Binge Eating Disorder. Eur. Psychiatry 2018, 54, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Gervasini, G.; Gonzalez, L.M.; Mota-Zamorano, S.; Gamero-Villarroel, C.; Carrillo, J.A.; Flores, I.; Garcia-Herraiz, A. Association of COMT Val158Met Polymorphism with Psychopathological Symptoms in Patients with Eating Disorders. Curr. Mol. Med. 2018, 18, 65–70. [Google Scholar] [CrossRef]
- Cellini, E.; Castellini, G.; Ricca, V.; Bagnoli, S.; Tedde, A.; Rotella, C.M.; Faravelli, C.; Sorbi, S.; Nacmias, B. Glucocorticoid Receptor Gene Polymorphisms in Italian Patients with Eating Disorders and Obesity. Psychiatr. Genet. 2010, 20, 282–288. [Google Scholar] [CrossRef]
- Monteleone, P.; Zanardini, R.; Tortorella, A.; Gennarelli, M.; Castaldo, E.; Canestrelli, B.; Maj, M. The 196G/A (Val66met) Polymorphism of the BDNF Gene Is Significantly Associated with Binge Eating Behavior in Women with Bulimia Nervosa or Binge Eating Disorder. Neurosci. Lett. 2006, 406, 133–137. [Google Scholar] [CrossRef]
- Monteleone, P.; Tortorella, A.; Castaldo, E.; Di Filippo, C.; Maj, M. The Leu72Met Polymorphism of the Ghrelin Gene Is Significantly Associated with Binge Eating Disorder. Psychiatr. Genet. 2007, 17, 13–16. [Google Scholar] [CrossRef]
- Blundell, J.E. Serotonin Manipulations and the Structure of Feeding Behaviour. Appetite 1986. [Google Scholar] [CrossRef]
- Noble, E.P. Addiction and Its Reward Process through Polymorphisms of the D2 Dopamine Receptor Gene: A Review. Eur. Psychiatry 2000. [Google Scholar] [CrossRef]
- Dubertret, C.; Gouya, L.; Hanoun, N.; Deybach, J.C.; Adès, J.; Hamon, M.; Gorwood, P. The 3′ Region of the DRD2 Gene Is Involved in Genetic Susceptibility to Schizophrenia. Schizophr. Res. 2004. [Google Scholar] [CrossRef]
- Neville, M.J.; Johnstone, E.C.; Walton, R.T. Identification and Characterization of ANKK1: A Novel Kinase Gene Closely Linked to DRD2 on Chromosome Band 11q23.1. Hum. Mutat. 2004. [Google Scholar] [CrossRef] [PubMed]
- Koo-Loeb, J.H.; Costello, N.; Light, K.C.; Girdler, S.S. Women with Eating Disorder Tendencies Display Altered Cardiovascular, Neuroendocrine, and Psychosocial Profiles. Psychosom. Med. 2000. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.D.; Marcus, M.D. Eating Behavior Following Stress in Women with and without Bulimic Symptoms. Ann. Behav. Med. 1997. [Google Scholar] [CrossRef] [PubMed]
- Kyrou, I.; Tsigos, C. Stress Mechanisms and Metabolic Complications. Horm. Metab. Res. 2007, 39, 430–438. [Google Scholar] [CrossRef]
- Castellini, G.; Franzago, M.; Bagnoli, S.; Lelli, L.; Balsamo, M.; Mancini, M.; Nacmias, B.; Ricca, V.; Sorbi, S.; Antonucci, I.; et al. Fat Mass and Obesity-Associated Gene (FTO) Is Associated to Eating Disorders Susceptibility and Moderates the Expression of Psycho-Pathological Traits. PLoS ONE 2017, 12, e0173560. [Google Scholar] [CrossRef] [PubMed]
- Gamero-Villarroel, C.; Rodriguez-Lopez, R.; Jimenez, M.; Carrillo, J.A.; Garcia-Herraiz, A.; Albuquerque, D.; Flores, I.; Gervasini, G. Melanocortin-4 Receptor Gene Variants Are Not Associated with Binge-Eating Behavior in Nonobese Patients with Eating Disorders. Psychiatr. Genet. 2015, 25, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Genis-Mendoza, A.D.; Ruiz-Ramos, D.; López-Narvaez, M.L.; Tovilla-Zárate, C.A.; Rosa García, A.; Cortes Meda, G.; Martinez-Magaña, J.J.; González-Castro, T.B.; Juárez-Rojop, I.E.; Nicolini, H. Genetic Association Analysis of 5-HTR2A Gene Variants in Eating Disorders in a Mexican Population. Brain Behav. 2019, 9. [Google Scholar] [CrossRef] [Green Version]
- Hebebrand, J.; Geller, F.; Dempfle, A.; Heinzel-Gutenbrunner, M.; Raab, M.; Gerber, G.; Wermter, A.-K.; Horro, F.F.; Blundell, J.; Schafer, H.; et al. Binge-Eating Episodes Are Not Characteristic of Carriers of Melanocortin-4 Receptor Gene Mutations. Mol. Psychiatry 2004, 9, 796–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hersrud, S.L.; Stoltenberg, S.F. Epistatic Interaction between COMT and DAT1 Genes on Eating Behavior: A Pilot Study. Eat. Behav. 2009, 10, 131–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinohara, M.; Mizushima, H.; Hirano, M.; Shioe, K.; Nakazawa, M.; Hiejima, Y.; Ono, Y.; Kanba, S. Eating Disorders with Binge-Eating Behaviour Are Associated with the s Allele of the 3′-UTR VNTR Polymorphism of the Dopamine Transporter Gene. J. Psychiatry Neurosci. 2004, 29, 134–137. [Google Scholar] [PubMed]
| Article Characteristics | Study Populations | Candidates Genes and Polymorphisms Associated | Methodology | Results | NOS Score | |||
|---|---|---|---|---|---|---|---|---|
| Author(s) and (year) | Groups (N) | Sex: M, F; Age: years or Mean (sd) | Diagnostic criteria of BED | Gene(s) | Polymorphism(s) | Study design | Main findings | |
| Cameron et al. (2019) [5] | BED with overweight or obesity (73) Overweight or obese without BED (55) Normal weight (50) | F = 73; 44.2 (11.2) F = 55; 46.1 (11.9) F = 50; 43.8 (11.9) | DSM-IV | FTO (fat mass and obesity associated) | 5 SNPs: rs9939609, rs8050136, rs3751812, rs1421085 and rs1121980. | Case-control | There were no significant between-group differences for frequencies of FTO allele, nor were there any significant anthropometric associations. | 8 |
| Monteleone et al. (2006) [9] | BED with obesity (77) CTRs (61) | F = 77; NA F = 61; NA | DSM-IV | 5HTT (5HT transporter gene) | 5HTTLPR | Case-control | Statistical analysis showed that both the LL genotype and the L allele of the 5HTTLPR were significantly more frequent in BED subjects. Moreover, the L allele was associated with a moderate but significant risk to develop BED. | 6 |
| Burnet et al. (1999) [10] | BN (40) BED (21) CTRs (92) | F = 40; 16–35 F = 21; 16–35 F = 92; age-matched | DSM-IV | 5-HT2C (serotonin 2C receptor) | Serine (ser) is substituted for cysteine (cys) at codon 23 (cys23ser) | Case-control | Genotype and allele frequencies were entirely unaltered in both case groups, compared to screened healthy controls from the same population. | 8 |
| Ricca et al. (2002) [11] | BED (54) Obese non-BED (132) | NA; 39.5 (13.76) NA; 43.3 (12.75) | DSM-IV | 5-HT2A (serotonin 2A receptor gene) | −1438 G/A | Case-control | No significant differences were found between obese BED and obese non-BED individuals, suggesting that this polymorphism does not genetically distinguish these two phenotypes. | 6 |
| Ceccarini et al. (2020) [12] | AN (311) BN (115) BE (130) CTRs (355) | M = 5, F = 306; 22.4 (± 8.9) M = 5, F = 110; 26.3 (± 9.4) M = 15, F = 115; 37.8 (± 18) M = 38, F = 317; 27.7 (± 7.9) | DSM-V | 5-HT2AR (serotonin 2A receptor) BDNF (brain-derived neurotrophic factor) | 1 SNP: rs6311 (−1438A/G) in the promoter region of 5-HT2AR 1 SNP: rs6265 (196C/T) in the coding region (Val66Met) of BDNF | Case-control | There was association between the rs6311 SNP of the 5-HT2AR gene and AN, but not with BN and BE. The Val66Met SNP in the coding region of BDNF showed a strong association with both AN and BE patients. Their data show that Met/Met genotype is present in 7.1% of AN, 6.1% of BN, and 9.2% of BE against only 2% of CTRs. 6 | 6 |
| Gonzalez et al. (2019) [15] | AN (210) BN (80) BED (34) | F = 210; NA F = 80; NA F = 34; NA | DSM-5 | DAT 1 (dopamine transporter) DRD2 (dopamine D2 receptor) DRD3 (dopamine D3 receptor) | 1 SNP: VNTR (rs28363170) 1 SNP: Taq1A (rs1800497) 1 SNP: Ser9Gly (rs6280) | Case-only | There were no differences between AN, BN, and BED with regard to the distribution of the different genotypes. | 3 |
| Davis et al. (2007) [16] | BED (32) CTRs (46) | F = 84.4%; 33.7 (± 6.5) F = 97.8%; 32.9 (± 7.0) | DSM-IV | DAT1 (dopamine transporter) | 3′-UTR VNTR | Case-Control | At baseline (the placebo condition), there was no significant group difference in DAT1 genotype frequencies. BED subjects with at least one copy of the 9-repeat allele showed a significant suppression of appetite in response to methylphenidate compared with controls with this allele, or to subjects with the 10/10 genotype (irrespective of diagnosis) whose drug response was indistinguishable from placebo. | 7 |
| Leehr et al. (2016) [17] | BED+ (BED with obesity) (21) OBED- (Obese controls without BED) (23) NWC (Normal-weight healthy controls) (25) | F = 21; 31.0 (12.3) F = 23; 31.7 (11.2) F = 25; 31.4 (10.9) | DSM-IV | COMT (catechol-O-methyltransferase) | Val(108/158)Met | Case-control | In the BED+ group, COMT Met/Met homozygous individuals showed stronger deficits in inhibitory control. | 6 |
| Kindler et al. (2011) [18] | AN (46) BN (30) BED (38) CTRs (164) | M = 1; F = 45; 30.4 M = 1; F = 29; 26.6 M = 1; F = 37; 39.3 F = 164; 32.2 | DSM-IV | prepro-NPY gene (prepro neuropeptide Y) prepro-GHRL (preproghrelin) | 1 SNP: Leu7Pro 3 SNPs: Arg51Gln, Leu72Met and Gln90Leu | Case-control | No difference was seen in the genotype distribution between patients with EDs and controls in any of the four SNPs. The analysis of the diagnostic subgroups AN, BN, and BED also did not reveal any significant differences compared with controls. | 7 |
| Monteleone et al. (2008) [19] | BED with obesity/ overweight (115) Obese without BED (74) Normal weight CTRs (110) | F = 115; 34.8 (11.1) F = 74; 37.5 (12.7) F = 110; 27.3 (6.8) | DSM-IV | FAAH (fatty acid amide hydrolase) | 1 SNP: cDNA 385C to A | Case-control | Compared to healthy controls, the whole group of overweight/obese BED and non-BED patients had a significantly higher frequency of the CA genotype and the A allele of the FAAH gene cDNA 385C to A SNP. Moreover, the SNP resulted significantly correlated to the presence of overweight/obesity, but not to the occurrence of BED | 7 |
| Tortorella et al. (2005) [20] | BED with Obesity (48) BED without obesity (10) CTRs (40) | F = 48; 18–58 F = 10; 18–58 NA; 18–39 | DSM-IV | MC4R (melanocortin-4 receptor gene) | mutational scanning | Case-control | Two polymorphisms, Val103Ile and Ile251Leu of the MC4R gene were detected in BED patients. No variants of the MC4R gene were observed in the 40 controls. | 5 |
| Monteleone et al. (2008) [21] | Overweight/obese patients wit3h and/or without BED (192) CTRs (92) | Obese with/without BED: M = 28; F = 164; NA M = 14; F = 79; NA | DSM-IV | CLOCK (Circadian locomotor output cycles kaput) | 1 SNP: 3111T/C | Case-control | Genotype and allele frequencies did not significantly differ between normal-weight controls and overweight/obese patients with and/or without BED. | 6 |
| Palmeira et al. (2019) [24] | Overweight (24) Obese (69, of which 31 with BED) CTRs (62) | F = 24; 41.52 F = 69; 41.52 F = 62; 42.73 | DSM-5 | BDNF (brain-derived neurotrophic factor) GHRL (ghrelin gene) SLC6A4/5-HTT (serotonin transporter gene) DRD2 (dopamine D2 receptor) FTO (fat mass obesity-associate d gene) | 2 SNPs: rs16917237 and rs6265 (Val66Met) 2 SNPs: rs696217 (Leu72met) and rs4684677(Gln9 0Leu) 5-HTTLPR 1 SNP: rs1800497 (Taq1A) 1 SNP: rs9939609 | Case-Control | No significant associations were found between polymorphisms and BED. Of interest, a markedly lower frequency of the FTO rs9939609 obesity risk A allele was found in BED patients (0.290) in relation to the control group (0.402). Contrasting with anorexia nervosa and bulimia nervosa, their data suggest that rs9939609 A allele has no potential role in BED. | 8 |
| Davis et al. (2008) [25] | BED (56) Obese CTRs (51) Normal-weight CTRs (59) | M = 12, F = 44; 34.84 (6.41) M = 12; F = 39; 36.29 (6.34) M = 7; F = 52; 33.49 (7.53) | DSM-IV | DRD2 (dopamine D2 receptor) | 6 SNPs: Taq1A, −141C Ins/Del, −241 A/G, Taq1D, C957T, and rs4648317 | Case-Control | Among the six SNPs related to DRD2, the findings for the Taq1A are perhaps of greatest interest given its considerable links to addictions in general, and obesity more specifically. BED and obese participants reported greater reward sensitivity compared to those with normal weight. The Taq1A genotype had a moderating influence on this relationship. Higher reward sensitivity was only observed in BED and obese subjects who carried the A1 allele. | 8 |
| Davis et al. (2009) [26] | BED with obesity (66) Obese CTRs (70) | M = 13, F = 53; 34.7 (6.5) M = 17, F = 53 37.0 (6.7) | DSM-IV | DRD2 (dopamine D2 receptor) OPRM1 (mu-opioid receptor) | 3 SNPs: taq1A, −141c Ins/del, C957T 1 SNP: A118G | Case-Control | Their results show that a larger proportion of the obese control group carried the rare A1 allele whereas for the latter comparison, a greater proportion of the BED group carried the rare G allele. It can be seen that in the genotype combination characterized by A1− and G +, 80% are BED participants whereas only 20% are obese controls. The opposite pattern pertains for the genotype group carrying A1+ and G−, where about 65% were obese controls and about 35% were obese binge eaters. | 8 |
| Davis et al. (2012) [27] | BED with obesity (79) Obese without BED (151) | M = 12, F = 67; 38.6 (7.2) M = 47, F = 104; 38.7 (7.1) | DSM-IV | DRD2 (dopamine D2 receptor) ANKK1 (D2 receptor gene) | 4 SNPs: rs1799732 (−141C ins/del), rs6277 (C957T), rs2283265 and rs12364283 1 SNP: rs1800497 (Taq1A) | Case-Control | Compared to weight-matched controls, BED was significantly related to the rs1800497 (were in the A2/A2 genotype group) and rs6277 (were homozygous for the T allele) genotypes that reflect enhanced dopamine neurotransmission. BED participants were also less likely to carry the minor T allele of rs2283265. | 8 |
| Palacios et al. (2018) [28] | Obese with BED (25) Obese without BED (25) CTRs (100) | M = 9; F = 16; 27–53 M = 12, F = 13; 30–52 M = 38; F = 62; 21–38 | DSM-5 | ANKK1 | 8 SNPs: Two polymorphisms in exon 2 (rs17115439 and rs4938013), one in exon 5 (rs7118900), one in exon 6 (rs11604671), and four in exon 8 (rs4938016, rs2734849, rs2734848 and rs1800497). | Case-control | After ANKK1 sequencing we did not find any mutations; however, rs1800497 (also known as Taq1A) in exon 8, showed an association with BED. Remarkably, for this study, the number of individuals for this polymorphism and additive model was sufficient to derive strong statistical power. | 8 |
| Gervasini et al. (2018) [29] | AN (199) BN (74) BED (30) | F = 199; 16.94 (± 4.58) F = 74; 17.76 (± 4.98) F = 30; 20.18 (± 7.38) | DSM-5 | COMT (catechol-O-methyltransferase) | 1 SNP: Val158Met | Case-only | The distribution of the different genotypes was similar in the three diagnosis groups and was comparable to the 1000 genomes database. | 3 |
| Cellini et al. (2010) [30] | AN (118) BN (108) BED (62) Obese non-BED (177) CTRs (107) | F = 94.4%; 19.21 (± 3.05) F = 98.2%; 20.64 (± 6.17) F = 87.3%; 23.50 (± 8.69) F = 80.2%; 24.00 (± 7.07) NA; 27.3 (± 1.5) | DSM-IV | GR (Glucocorticoids receptor) | 4 SNPs: exon 9-b (rs6198), ER22/23EK (rs6189–6190), N363S (rs56149945) and the intronic BclI (rs41423247) | Case-Control | The results highlight two major findings. First, a significant association between BMI and GR genotypes was identified for the SNP rs56149945 (N363S) in obese patients and in all the ED groups, independent of eating psychopathology. Moreover, a significant association between the rs6198 polymorphism and the binge eating symptoms was detected. | 7 |
| Monteleone et al. (2006) [31] | BED (84) BN (126) CTRs (121) | F= 84; NA F= 126; NA F= 121; NA | DSM-IV | BDNF (brain-derived neurotrophic factor) | 1 SNP: 196 G/A | Case-control | No significant differences were found in the frequencies of the 196G/A variants of the BDNF gene among patients with BN or BED and healthy controls. | 6 |
| Monteleone et al. (2007) [32] | BED (90) CTRs (119) | F = 90; NA F = 119; NA | DSM-IV | GHRL (ghrelin gene) | 2 SNP: G152A (Arg51Gln) and C214A (Leu72Met) | Case-control | Statistical analyses showed that the Leu72Met ghrelin gene variant was significantly more frequent in BED patients and was associated with a moderate, but significant risk to develop binge eating disorder. No significant difference, instead, emerged in the distribution of Arg51Gln ghrelin gene variant between BED patients and healthy controls. | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manfredi, L.; Accoto, A.; Couyoumdjian, A.; Conversi, D. A Systematic Review of Genetic Polymorphisms Associated with Binge Eating Disorder. Nutrients 2021, 13, 848. https://doi.org/10.3390/nu13030848
Manfredi L, Accoto A, Couyoumdjian A, Conversi D. A Systematic Review of Genetic Polymorphisms Associated with Binge Eating Disorder. Nutrients. 2021; 13(3):848. https://doi.org/10.3390/nu13030848
Chicago/Turabian StyleManfredi, Lucia, Alessandra Accoto, Alessandro Couyoumdjian, and David Conversi. 2021. "A Systematic Review of Genetic Polymorphisms Associated with Binge Eating Disorder" Nutrients 13, no. 3: 848. https://doi.org/10.3390/nu13030848







