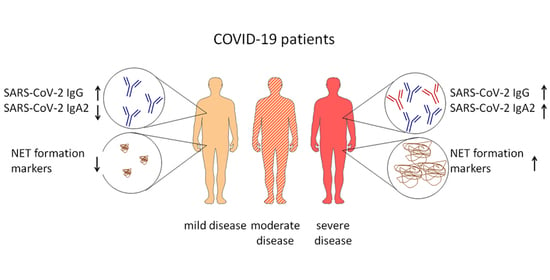
IgA2 Antibodies against SARS-CoV-2 Correlate with NET Formation and Fatal Outcome in Severely Diseased COVID-19 Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Healthy Controls
2.2. Measurement of Antibody Levels
2.3. Measurement of C-Reactive Protein (CRP), Extracellular DNA (ecDNA) and Neutrophil Elastase (NE) Activity
2.4. Statistical Analysis
3. Results
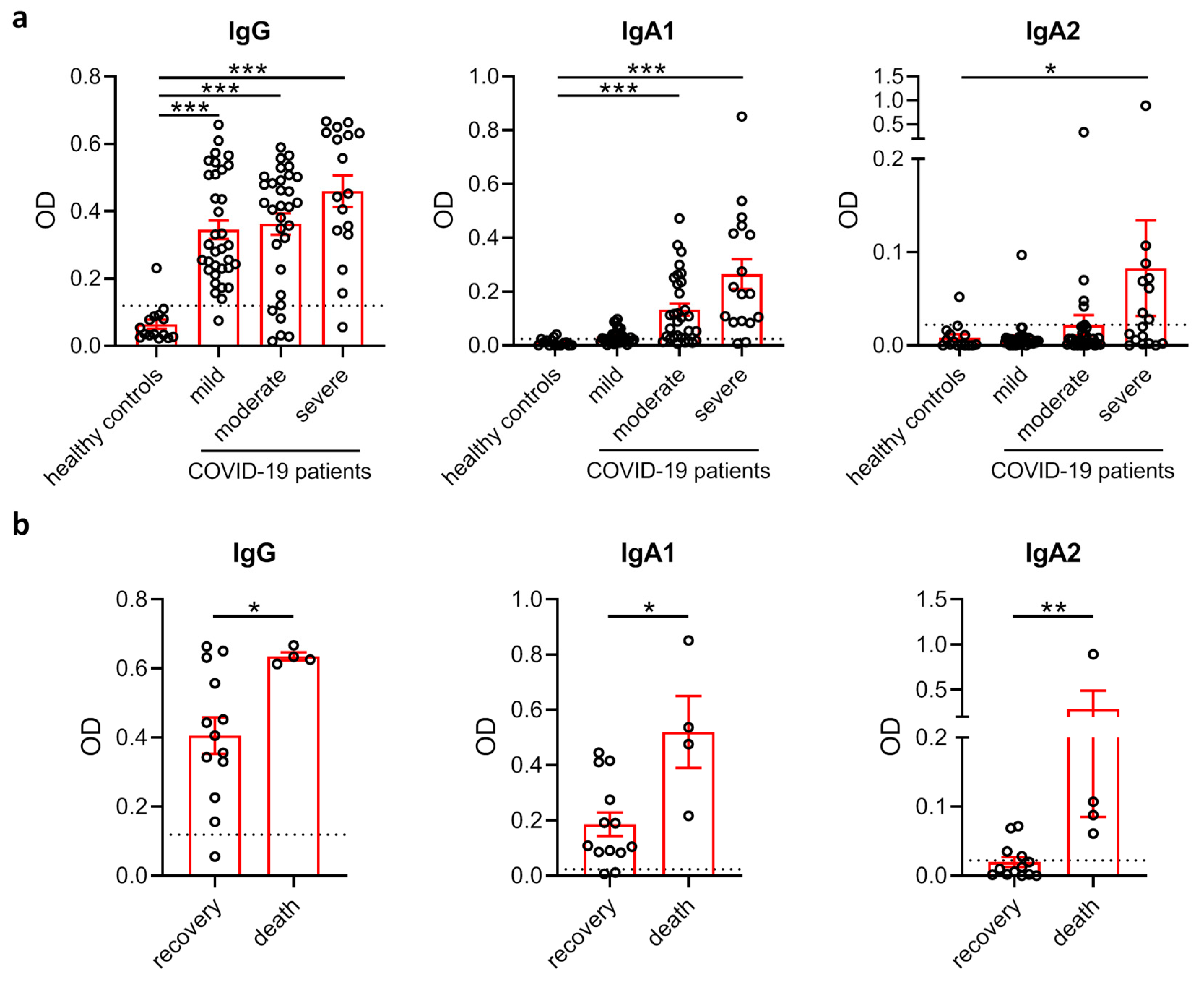
3.1. Comparison of SARS-CoV-2 Antibody Levels in COVID-19 Patients with Different Disease Severity and Outcome
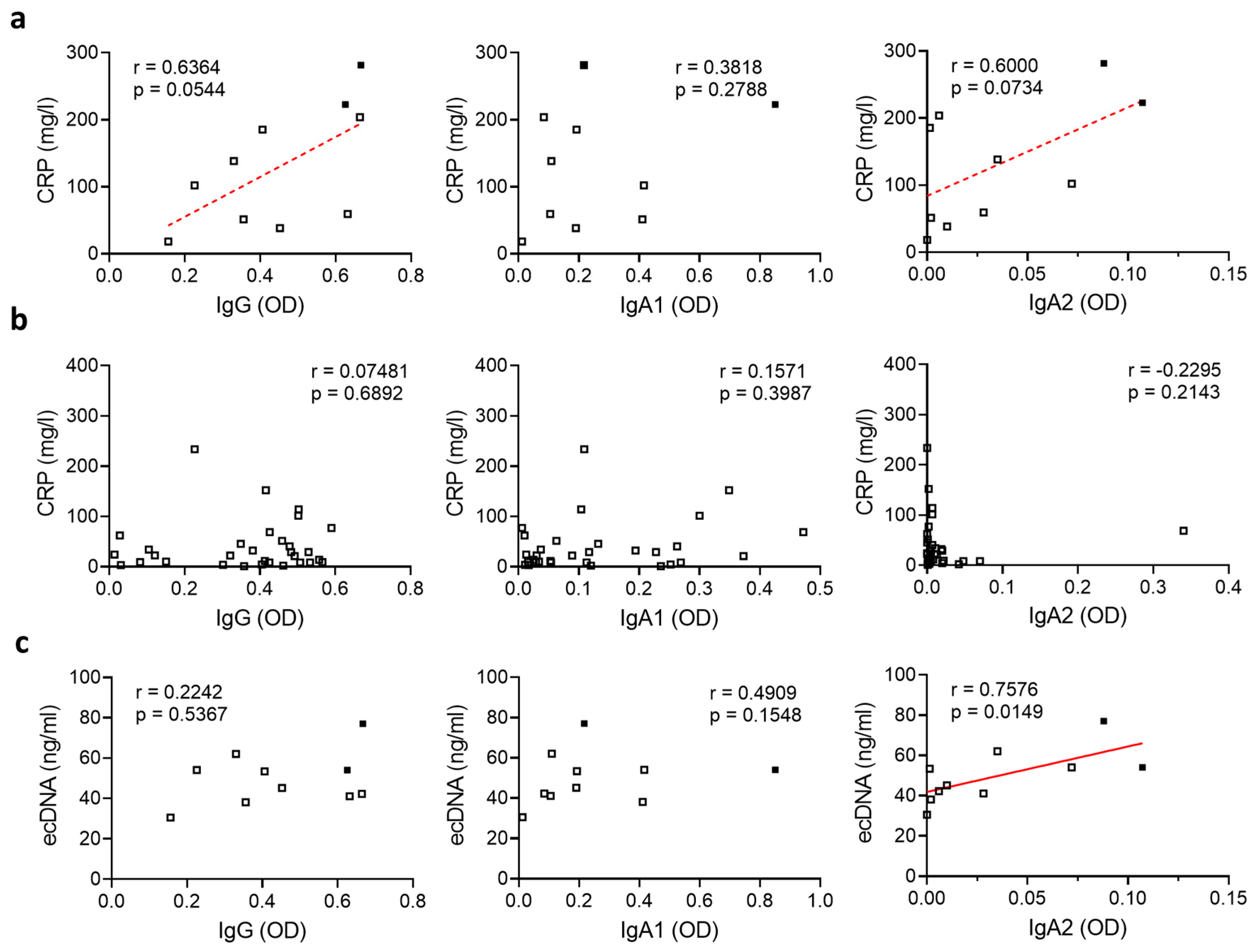
3.2. Relation of SARS-CoV-2-Specific Antibody Levels to Systemic Inflammation and NET Formation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Organization WHO. Coronavirus Disease 2019 (COVID-19): Situation Report. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed on 12 October 2020).
- Goyal, P.; Choi, J.J.; Pinheiro, L.C.; Schenck, E.J.; Chen, R.; Jabri, A.; Satlin, M.J.; Campion, T.R.; Nahid, M.; Ringel, J.B.; et al. Clinical Characteristics of Covid-19 in New York City. N. Engl. J. Med. 2020, 382, 2372–2374. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Morales, A.J.; Cardona-Ospina, J.A.; Gutiérrez-Ocampo, E.; Villamizar-Peña, R.; Holguin-Rivera, Y.; Escalera-Antezana, J.P.; Alvarado-Arnez, L.E.; Bonilla-Aldana, D.K.; Franco-Paredes, C.; Henao-Martinez, A.F.; et al. Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Med. Infect. Dis. 2020, 34, 101623. [Google Scholar] [CrossRef]
- Fourati, S.; Hue, S.; Pawlotsky, J.M.; Mekontso-Dessap, A.; de Prost, N. SARS-CoV-2 viral loads and serum IgA/IgG immune responses in critically ill COVID-19 patients. Intensive Care Med. 2020, 46, 1781–1783. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020, 71, 2027–2034. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Zeng, W.; He, H.; Zhao, D.; Jiang, D.; Zhou, P.; Cheng, L.; Li, Y.; Ma, X.; Jin, T. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020, 17, 773–775. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Z.; Poh, C.M.; Renia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef]
- Hoepel, W.; Chen, H.J.; Allahverdiyeva, S.; Manz, X.; Aman, J.; Bonta, P.; Brouwer, P.; de Taeye, S.; Caniels, T.; van der Straten, K.; et al. Anti-SARS-CoV-2 IgG from severely ill COVID-19 patients promotes macrophage hyper-inflammatory responses. bioRxiv 2020. [Google Scholar] [CrossRef]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A.; Borczuk, A.; Cools-Lartigue, J.; Crawford, J.M.; Daßler-Plenker, J.; Guerci, P.; Huynh, C.; Knight, J.S.; et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5. [Google Scholar] [CrossRef] [Green Version]
- Leppkes, M.; Knopf, J.; Naschberger, E.; Lindemann, A.; Singh, J.; Herrmann, I.; Stürzl, M.; Staats, L.; Mahajan, A.; Schauer, C.; et al. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine 2020, 58, 102925. [Google Scholar] [CrossRef] [PubMed]
- Middleton, E.A.; He, X.Y.; Denorme, F.; Campbell, R.A.; Ng, D.; Salvatore, S.P.; Mostyka, M.; Baxter-Stoltzfus, A.; Borczuk, A.C.; Loda, M.; et al. Neutrophil Extracellular Traps (NETs) Contribute to Immunothrombosis in COVID-19 Acute Respiratory Distress Syndrome. Blood 2020. [Google Scholar] [CrossRef] [PubMed]
- Nicolai, L.; Leunig, A.; Brambs, S.; Kaiser, R.; Weinberger, T.; Weigand, M.; Muenchhoff, M.; Hellmuth, J.C.; Ledderose, S.; Schulz, H.; et al. Immunothrombotic Dysregulation in COVID-19 Pneumonia Is Associated With Respiratory Failure and Coagulopathy. Circulation 2020, 142, 1176–1189. [Google Scholar] [CrossRef] [PubMed]
- Skendros, P.; Mitsios, A.; Chrysanthopoulou, A.; Mastellos, D.C.; Metallidis, S.; Rafailidis, P.; Ntinopoulou, M.; Sertaridou, E.; Tsironidou, V.; Tsigalou, C.; et al. Complement and tissue factor-enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Investig. 2020, 130, 6151–6157. [Google Scholar] [CrossRef]
- Arcanjo, A.; Logullo, J.; Menezes, C.C.B.; Giangiarulo, T.C.D.S.C.; Dos Reis, M.C.; De Castro, G.M.M.; Fontes, Y.D.S.; Todeschini, A.R.; Freire-De-Lima, L.; Decoté-Ricardo, D.; et al. The emerging role of neutrophil extracellular traps in severe acute respiratory syndrome coronavirus 2 (COVID-19). Sci. Rep. 2020, 10, 19630. [Google Scholar] [CrossRef]
- Veras, F.P.; Pontelli, M.; Silva, C.; Toller-Kawahisa, J.; de Lima, M.; Nascimento, D.; Schneider, A.; Caetite, D.; Rosales, R.; Colon, D.; et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Aleyd, E.; Al, M.; Tuk, C.W.; van der Laken, C.J.; van Egmond, M. IgA Complexes in Plasma and Synovial Fluid of Patients with Rheumatoid Arthritis Induce Neutrophil Extracellular Traps via FcalphaRI. J. Immunol. 2016, 197, 4552–4559. [Google Scholar] [CrossRef]
- Brandsma, A.M.; Bondza, S.; Evers, M.; Koutstaal, R.; Nederend, M.; Jansen, J.H.M.; Rösner, T.; Valerius, T.; Leusen, J.H.; Broeke, T.T. Potent Fc Receptor Signaling by IgA Leads to Superior Killing of Cancer Cells by Neutrophils Compared to IgG. Front. Immunol. 2019, 10, 704. [Google Scholar] [CrossRef] [Green Version]
- Valerius, T.; Stockmeyer, B.; van Spriel, A.B.; Graziano, R.F.; van den Herik-Oudijk, I.E.; Repp, R.; Deo, Y.M.; Lund, J.; Kalden, J.R.; Gramatzki, M.; et al. FcalphaRI (CD89) as a novel trigger molecule for bispecific antibody therapy. Blood 1997, 90, 4485–4492. [Google Scholar] [CrossRef]
- Hansen, I.S.; Baeten, D.L.P.; den Dunnen, J. The inflammatory function of human IgA. Cell. Mol. Life Sci. 2019, 76, 1041–1055. [Google Scholar] [CrossRef] [Green Version]
- Steffen, U.; Koeleman, C.A.; Sokolova, M.V.; Bang, H.; Kleyer, A.; Rech, J.; Unterweger, H.; Schicht, M.; Garreis, F.; Hahn, J.; et al. IgA subclasses have different effector functions associated with distinct glycosylation profiles. Nat. Commun. 2020, 11, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dogan, M.; Kozhaya, L.; Placek, L.; Gunter, C.L.; Yigit, M.; Hardy, R.; Plassmeyer, M.; Coatney, P.; Lillard, K.; Bukhari, Z.; et al. Novel SARS-CoV-2 specific antibody and neutralization assays reveal wide range of humoral immune response during COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Graham, N.R.; Whitaker, A.N.; A Strother, C.; Miles, A.K.; Grier, D.; McElvany, B.D.; A Bruce, E.; E Poynter, M.; Pierce, K.K.; Kirkpatrick, B.D.; et al. Kinetics and isotype assessment of antibodies targeting the spike protein receptor-binding domain of severe acute respiratory syndrome-coronavirus-2 in COVID-19 patients as a function of age, biological sex and disease severity. Clin. Transl. Immunol. 2020, 9, e1189. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Chen, H.; Xue, M.; Huang, H.; Zheng, P.; Luo, W.; Liang, X.; Sun, B.; Zhong, N. Characteristics and roles of SARS-CoV-2 specific antibodies in patients with different severities of COVID-19. Clin. Exp. Immunol. 2020, 202, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Ali, O.H.; Bomze, D.; Risch, L.; Brugger, S.D.; Paprotny, M.; Weber, M.; Thiel, S.; Kern, L.; Albrich, W.C.; Kohler, P.; et al. Severe COVID-19 is associated with elevated serum IgA and antiphospholipid IgA-antibodies. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Horns, F.; Vollmers, C.; Croote, D.; Mackey, S.F.; Swan, G.E.; Dekker, C.L.; Davis, M.M.; Quake, S.R. Lineage tracing of human B cells reveals the in vivo landscape of human antibody class switching. eLife 2016, 5. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Wong, B.H.L.; Chan, K.-H.; Chu, C.-M.; Tsoi, H.-W.; Huang, Y.; Peiris, J.S.M.; Yuen, K.-Y. Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin. Diagn. Lab. Immunol. 2004, 11, 665–668. [Google Scholar] [CrossRef] [Green Version]
- Matricardi, P.M.; Dal Negro, R.W.; Nisini, R. The first, holistic immunological model of COVID-19: Implications for prevention, diagnosis, and public health measures. Pediatr. Allergy Immunol. 2020. [Google Scholar] [CrossRef]
- Paces, J.; Strizova, Z.; Smrz, D.; Cerny, J. COVID-19 and the immune system. Physiol. Res. 2020, 69, 379–388. [Google Scholar] [CrossRef]
- Karsten, C.M.; Kohl, J. The immunoglobulin, IgG Fc receptor and complement triangle in autoimmune diseases. Immunobiology 2012, 217, 1067–1079. [Google Scholar] [CrossRef]
- Toubi, E.; Vadasz, Z. Innate immune-responses and their role in driving autoimmunity. Autoimmun. Rev. 2019, 18, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Ravetch, J.V. Functional diversification of IgGs through Fc glycosylation. J. Clin. Investig. 2019, 129, 3492–3498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Xu, S.; Yu, M.; Wang, K.; Tao, Y.; Zhou, Y.; Shi, J.; Zhou, M.; Wu, B.; Yang, Z.; et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J. Allergy Clin. Immunol. 2020, 146, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Iyer, A.S.; Jones, F.K.; Nodoushani, A.; Kelly, M.; Becker, M.; Slater, D.; Mills, R.; Teng, E.; Kamruzzaman, M.; Garcia-Beltran, W.F.; et al. Dynamics and significance of the antibody response to SARS-CoV-2 infection. medRxiv 2020. [Google Scholar] [CrossRef]
| Healthy Controls (N = 15) | Mild Disease (N = 34) | Moderate Disease * (N = 31) | Severe Disease ** (N = 17) | |
|---|---|---|---|---|
| Age (ys): median (range) | 34 (23–63) | 39 (19–65) | 68 (34–96) | 68 (37–78) |
| Male, N (%) | 5 (33%) | 28 (82%) | 17 (55%) | 13 (76%) |
| Female, N (%) | 10 (67%) | 6 (18%) | 14 (45%) | 4 (24%) |
| CAD, N (%) | n.a. | 0 (0%) | 8 (26%) | 5 (29%) |
| Hypertension, N (%) | n.a. | 1 (3%) | 18 (58%) | 13 (76%) |
| Diabetes mellitus, N (%) | n.a. | 2 (6%) | 8 (26%) | 6 (35%) |
| COPD, N (%) | n.a. | 0 (0%) | 1 (3%) | 4 (24%) |
| Days in hospital: median (range) | 0 (0–0) | 0 (0–14) | 8 (1–64) | 31 (2–50) |
| Death, N (%) *** | 0 (0%) | 0 (0%) | 0 (0%) | 4 (24%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Staats, L.A.N.; Pfeiffer, H.; Knopf, J.; Lindemann, A.; Fürst, J.; Kremer, A.E.; Hackstein, H.; Neurath, M.F.; Muñoz, L.E.; Achenbach, S.; et al. IgA2 Antibodies against SARS-CoV-2 Correlate with NET Formation and Fatal Outcome in Severely Diseased COVID-19 Patients. Cells 2020, 9, 2676. https://doi.org/10.3390/cells9122676
Staats LAN, Pfeiffer H, Knopf J, Lindemann A, Fürst J, Kremer AE, Hackstein H, Neurath MF, Muñoz LE, Achenbach S, et al. IgA2 Antibodies against SARS-CoV-2 Correlate with NET Formation and Fatal Outcome in Severely Diseased COVID-19 Patients. Cells. 2020; 9(12):2676. https://doi.org/10.3390/cells9122676
Chicago/Turabian StyleStaats, Léonie A. N., Hella Pfeiffer, Jasmin Knopf, Aylin Lindemann, Julia Fürst, Andreas E. Kremer, Holger Hackstein, Markus F. Neurath, Luis E. Muñoz, Susanne Achenbach, and et al. 2020. "IgA2 Antibodies against SARS-CoV-2 Correlate with NET Formation and Fatal Outcome in Severely Diseased COVID-19 Patients" Cells 9, no. 12: 2676. https://doi.org/10.3390/cells9122676