-
 Functional Precision in Pancreatic Cancer: Redefining Biomarkers with Patient-Derived Organoids
Functional Precision in Pancreatic Cancer: Redefining Biomarkers with Patient-Derived Organoids -
 Are Procoagulant Platelets an Emerging Therapeutic Target? A General Review with an Emphasis on Their Clinical Significance in Companion Animals
Are Procoagulant Platelets an Emerging Therapeutic Target? A General Review with an Emphasis on Their Clinical Significance in Companion Animals -
 MASLD Under the Microscope: How microRNAs and Microbiota Shape Hepatic Metabolic Disease Progression
MASLD Under the Microscope: How microRNAs and Microbiota Shape Hepatic Metabolic Disease Progression -
 Sequencing Anti-CD19 Therapies in Diffuse Large B-Cell Lymphoma: From Mechanistic Insights to Clinical Strategies
Sequencing Anti-CD19 Therapies in Diffuse Large B-Cell Lymphoma: From Mechanistic Insights to Clinical Strategies -
 Carbon Dots for Nucleic Acid-Based Diagnostics and Therapeutics: Focus on Oxidative DNA Damage
Carbon Dots for Nucleic Acid-Based Diagnostics and Therapeutics: Focus on Oxidative DNA Damage
Journal Description
International Journal of Molecular Sciences
International Journal of Molecular Sciences
is an international, peer-reviewed, open access journal providing an advanced forum for biochemistry, molecular and cell biology, molecular biophysics, molecular medicine, and all aspects of molecular research in chemistry, and is published semimonthly online by MDPI. The Australian Society of Plant Scientists (ASPS), Epigenetics Society, European Chitin Society (EUCHIS), Spanish Society for Cell Biology (SEBC) and others are affiliated with IJMS and their members receive a discount on the article processing charges.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, PMC, MEDLINE, Embase, CAPlus / SciFinder, and other databases.
- Journal Rank: JCR - Q1 (Biochemistry and Molecular Biology) / CiteScore - Q1 (Organic Chemistry)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 20.5 days after submission; acceptance to publication is undertaken in 2.6 days (median values for papers published in this journal in the first half of 2025).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
- Testimonials: See what our editors and authors say about IJMS.
- Companion journals for IJMS include: Biophysica, Stresses, Lymphatics and SynBio.
Impact Factor:
4.9 (2024);
5-Year Impact Factor:
5.7 (2024)
Latest Articles
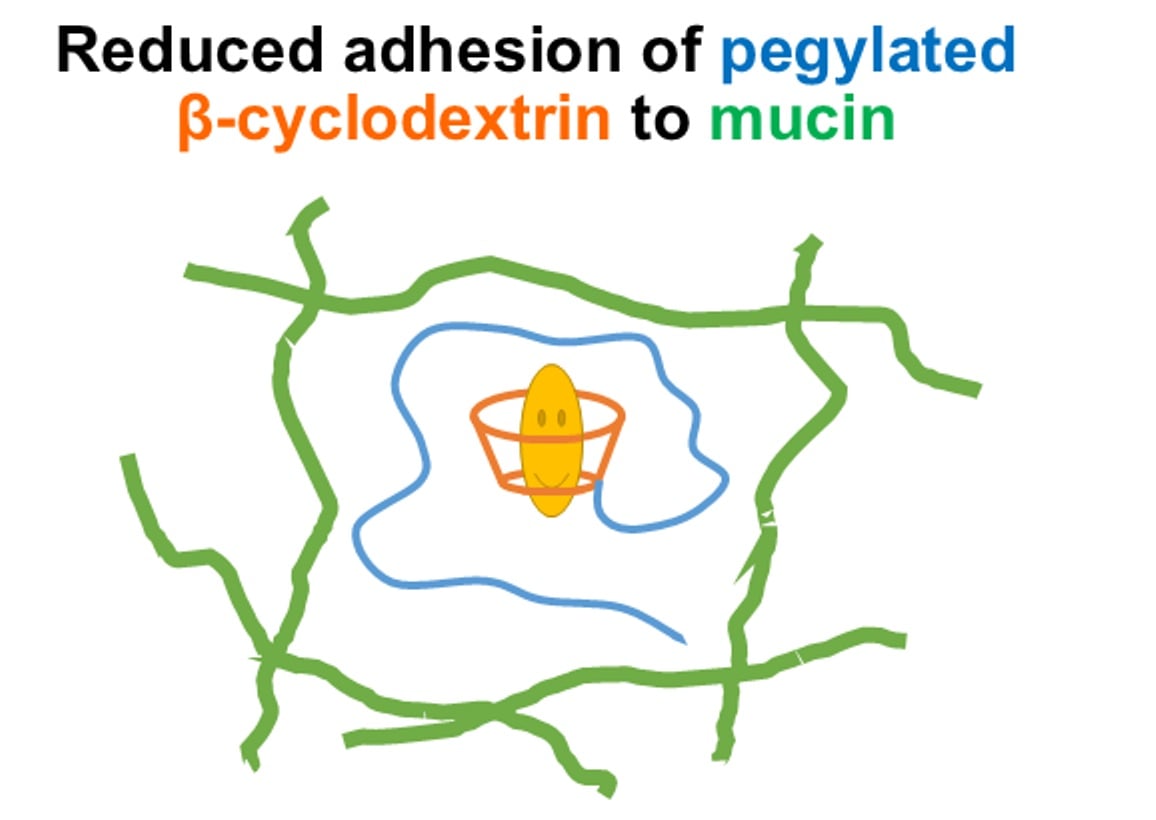
Mitigating Mucoadhesion of β–Cyclodextrins via PEGylation: Insights from 19F Diffusion NMR Analysis
Int. J. Mol. Sci. 2025, 26(23), 11690; https://doi.org/10.3390/ijms262311690 - 2 Dec 2025
Abstract
β–Cyclodextrin (β–CD)-based materials are widely used in drug delivery, yet their interactions with mucosal barriers remain insufficiently understood. Because the mucus layer coating epithelial surfaces can hinder drug transport, elucidating β–CD–mucin interactions is critical for optimizing cyclodextrin-based carriers. In this study, we examined
[...] Read more.
β–Cyclodextrin (β–CD)-based materials are widely used in drug delivery, yet their interactions with mucosal barriers remain insufficiently understood. Because the mucus layer coating epithelial surfaces can hinder drug transport, elucidating β–CD–mucin interactions is critical for optimizing cyclodextrin-based carriers. In this study, we examined whether PEGylation can attenuate the mucoadhesive behavior of β–CD. Monomethoxy poly(ethylene glycol)-modified β–CDs (MPEG–β–CDs) were evaluated using 19F self-diffusion NMR spectroscopy coupled with a kinetic diffusion model describing reversible binding to stationary substrates. Mucin hydrogels were prepared from bovine submaxillary mucin and served as a model mucus environment. Diffusion coefficients were extracted from the 19F NMR signals of 1-fluoroadamantane (1FA) molecules encapsulated within HP-β–CD or MPEG–β–CD cavities. The results demonstrate that PEGylation substantially reduces β–CD–mucin adhesion, with longer PEG chains (2000 Da) providing more effective steric shielding than shorter chains (500 Da). These findings indicate that PEGylation can protect β–CD-included drugs during transport across mucosal barriers by minimizing unwanted β–CD–mucin interactions.
Full article
(This article belongs to the Special Issue Research on Cyclodextrin)
►
Show Figures
Open AccessArticle
Transcriptomic Changes Underlying the Anti-Steatotic Effects of DHA Supplementation in Aged Obese Female Mice
by
Álvaro Pejenaute Martínez de Lizarrondo, Paula Martín-Climent, María Martínez-Rubio, Jesús Saborido-Gavilán, Neira Sáinz, Elisa Félix-Soriano, Miriam Samblas, Mónica Alfonso-Núñez, Elizabeth Guruceaga, M Pilar Lostao, Pedro González-Muniesa and María J Moreno-Aliaga
Int. J. Mol. Sci. 2025, 26(23), 11689; https://doi.org/10.3390/ijms262311689 - 2 Dec 2025
Abstract
The prevalence of metabolic dysfunction-associated steatotic liver disease (MASLD) increases with age and obesity. The aim of this study was to characterize the transcriptomic changes in liver underlying the beneficial effects of docosahexaenoic acid (DHA) supplementation on MASLD progression in aged obese female
[...] Read more.
The prevalence of metabolic dysfunction-associated steatotic liver disease (MASLD) increases with age and obesity. The aim of this study was to characterize the transcriptomic changes in liver underlying the beneficial effects of docosahexaenoic acid (DHA) supplementation on MASLD progression in aged obese female mice. Two-month-old C57BL/6J female mice were fed ad libitum with high fat saturated diet (HFD, 45%) for 4 months. These diet-induced obese (DIO) mice were divided into two groups: the DIO group that continued with HFD and the DIO + DHA mice fed up to 18 months with the HFD containing a DHA-rich concentrate. High quality RNA from liver was used to perform RNA sequencing analysis. Further functional and clustering analyses of differentially expressed genes (DEGs) in liver between treatments revealed that DHA regulated processes related to lipid metabolic processes. DHA treatment also affects the PPAR signaling pathway and the regulation of inflammatory response. These data suggest that long-term DHA supplementation ameliorates MASLD progression in aged obese female mice by promoting transcriptomic changes in genes related to lipid metabolism and inflammatory response.
Full article
(This article belongs to the Special Issue Cell Metabolism: Its Physiology, Dysfunction, and Related Metabolic Complications)
Open AccessArticle
Integrated Genomic–Metabolomic Analysis for Tri-Categorical Classification of Type 2 Diabetes Status in the Korean Ansan–Ansung Cohort
by
Junho Cha and Sungkyoung Choi
Int. J. Mol. Sci. 2025, 26(23), 11688; https://doi.org/10.3390/ijms262311688 - 2 Dec 2025
Abstract
Identifying high-risk individuals for type 2 diabetes (T2D), particularly during prediabetes (PD), remains challenging owing to its complex metabolic etiology. In this study, we aimed to develop and validate an integrative multi-omics model for the tri-categorical classification of T2D status (Normal Glucose Tolerance,
[...] Read more.
Identifying high-risk individuals for type 2 diabetes (T2D), particularly during prediabetes (PD), remains challenging owing to its complex metabolic etiology. In this study, we aimed to develop and validate an integrative multi-omics model for the tri-categorical classification of T2D status (Normal Glucose Tolerance, PD, and T2D) by combining genomic and metabolomic data from a Korean cohort. Based on cross-sectional data from 1819 participants in the Ansan–Ansung cohort, significant metabolites associated with glycemic traits and T2D status were identified using regression analysis. A metabolite-adjusted genome-wide association study (GWAS) was conducted to identify T2D-associated genetic variants. Finally, three nested prediction models (Clinical, Metabolite-Enriched, and integrated Multi-omics) were constructed using baseline-category logistic regression and evaluated using stratified five-fold cross-validation. Thirty-nine metabolites were identified as consistently associated with T2D status and related glycemic traits. GWAS identified 86 T2D-associated independent single-nucleotide polymorphisms (SNPs). The final integrated multi-omics model, combining clinical factors, 39 metabolites, and 86 SNPs, demonstrated strong predictive performance for classifying T2D status, achieving an area under the receiver operating characteristic curve (AUC) of 0.935, significantly improved over the clinical model (AUC = 0.695) and metabolite-enriched model (AUC = 0.874). It also outperformed previously established external models and represents an important step in our understanding of T2D status. Our findings thus demonstrate that integrating genomic and metabolomic data provides a useful framework for the tri-categorical classification of T2D status. This multi-omics approach significantly enhances risk stratification beyond that provided by clinical or single-omics data alone, thus offering valuable insights into the underlying pathophysiology in T2D with potential for shaping future T2D research and clinical practice.
Full article
(This article belongs to the Special Issue Genomics of Human Disease)
►▼
Show Figures
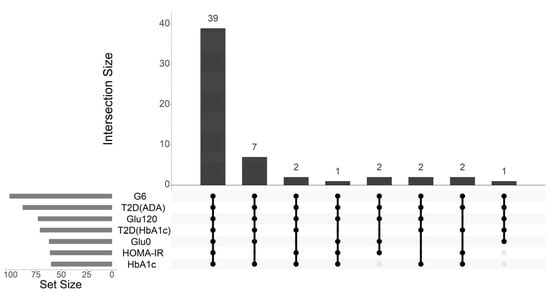
Figure 1
Open AccessArticle
Solid-Phase Synthesis Approaches and U-Rich RNA-Binding Activity of Homotrimer Nucleopeptide Containing Adenine Linked to L-azidohomoalanine Side Chain via 1,4-Linked-1,2,3-Triazole
by
Piotr Mucha, Małgorzata Pieszko, Irena Bylińska, Wiesław Wiczk, Jarosław Ruczyński and Piotr Rekowski
Int. J. Mol. Sci. 2025, 26(23), 11687; https://doi.org/10.3390/ijms262311687 - 2 Dec 2025
Abstract
Nucleopeptides (NPs) are unnatural hybrid polymers designed by coupling nucleobases to the side chains of amino acid residues within peptides. In this study, we present the synthesis of an Fmoc-protected nucleobase amino acid (NBA) monomer (Fmoc-1,4-TzlNBAA) with adenine attached to the
[...] Read more.
Nucleopeptides (NPs) are unnatural hybrid polymers designed by coupling nucleobases to the side chains of amino acid residues within peptides. In this study, we present the synthesis of an Fmoc-protected nucleobase amino acid (NBA) monomer (Fmoc-1,4-TzlNBAA) with adenine attached to the side chain of L-homoazidoalanine (Aha) through a 1,4-linked-1,2,3-triazole. The coupling was accomplished by a Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC) of Fmoc-Aha and N9-propargyladenine. Subsequently, a homotrinucleopeptide (HalTzlAAA) containing three 1,4-TzlNBAA residues was synthesized, using different solid-phase peptide synthesis (SPPS) approaches, and its ability to recognize U-rich motifs of RNAs involved in the HIV replication cycle was studied using circular dichroism (CD) and fluorescence spectroscopy. CD curves confirmed the binding of HalTzlAAA to U-rich motifs of the transactivation responsive element (TAR UUU RNA HIV-1) bulge and the anticodon stem–loop domain of human tRNALys3 (ASLLys3) by a decrease in the positive ellipticity band intensity around 265 nm during the complexation. 5′-(FAM(6))-labeled TAR UUU and hASLLys3 were used for fluorescence anisotropy binding studies. Fluorescence data revealed that HalTzlAAA bound TAR’s UUU bulge with a moderate affinity (Kd ≈ 38 µM), whereas the ASLLys3 UUUU-containing loop sequence was recognized with 2.5 times lower affinity (with Kd ≈ 75 µM). Both the standard SPPS method and its variants, which involved the attachment of adenine to the L-Aha side chain using the click reaction during the synthesis on the resin or after the nucleopeptide cleavage, were characterized by a similar efficiency and yield. The CD and fluorescence results demonstrated that HalTzlAAA recognized the U-rich sequences of the RNAs with moderate and varied affinities. It is likely that both the hydrogen bonds associated with the complementarity of the interacting sequences and the conformational aspects associated with the high conformational dynamics of U-rich motifs are important in the recognition process. The nucleopeptide represents a new class of RNA binders and may be a promising scaffold for the development of new antiviral drugs.
Full article
(This article belongs to the Section Molecular Biology)
►▼
Show Figures

Figure 1
Open AccessReview
Plasma Extracellular Vesicles as Liquid Biopsies for Glioblastoma: Biomarkers, Subpopulation Enrichment, and Clinical Translation
by
Abudumijiti Aibaidula, Ali Gharibi Loron, Samantha M. Bouchal, Megan M. J. Bauman, Hyo Bin You, Fabrice Lucien and Ian F. Parney
Int. J. Mol. Sci. 2025, 26(23), 11686; https://doi.org/10.3390/ijms262311686 - 2 Dec 2025
Abstract
Glioblastoma (GBM), the most common primary malignant brain tumor in adults, has a median survival of 14–15 months despite aggressive treatment. Monitoring relies on MRI, but differentiating tumor progression from pseudo-progression or radiation necrosis remains difficult. Plasma extracellular vesicles (EVs) are emerging as
[...] Read more.
Glioblastoma (GBM), the most common primary malignant brain tumor in adults, has a median survival of 14–15 months despite aggressive treatment. Monitoring relies on MRI, but differentiating tumor progression from pseudo-progression or radiation necrosis remains difficult. Plasma extracellular vesicles (EVs) are emerging as promising non-invasive biomarkers due to their molecular cargos and accessibility. This review evaluates studies that specifically isolated plasma EVs for molecular profiling in GBM diagnosis and monitoring. Biomarkers (miRNA, RNA, DNA, proteins), EV characterization methods, and advancements in enriching tumor-derived EV subpopulations and assessing their diagnostic and prognostic potential are highlighted. Plasma EVs carry diverse cargos, including miRNAs (e.g., miR-21, miR-15b-3p), mRNAs (e.g., EGFRvIII), circRNAs, and proteins (e.g., CD44, GFAP). Composite molecular signatures have achieved sensitivities of 87–100% and specificities of 73–100% for GBM diagnosis. Tumor-derived EVs, enriched using techniques like SEC-CD44 immunoprecipitation, microfluidic platforms, or 5-ALA-induced PpIX fluorescence, enhance biomarker detection. Non-tumor-derived EVs may also reflect GBM’s systemic effects. Challenges include EV heterogeneity, non-EV contamination, and variable biomarker expression across studies. Plasma-EV-based liquid biopsies offer significant potential for GBM monitoring, with advanced enrichment methods improving tumor-specific biomarker detection. Standardizing isolation protocols and validating biomarkers in larger cohorts are critical for clinical translation.
Full article
(This article belongs to the Section Molecular Oncology)
Open AccessArticle
Benchmarking Molecular Mutation Operators for Evolutionary Drug Design
by
Raúl Acosta Murillo, Patricio Adrián Zapata-Morin and José Carlos Ortiz-Bayliss
Int. J. Mol. Sci. 2025, 26(23), 11685; https://doi.org/10.3390/ijms262311685 - 2 Dec 2025
Abstract
This study investigates and compares different molecular mutation strategies to optimize their application as genetic algorithm operators in drug design. We evaluated five distinct mutation methods—Graph-Based Genetic Algorithm, Graph-Based Generative Model, SmilesClickChem, SELFIES Token, and SMILES Token Mutation—by assessing their computational efficiency, validity,
[...] Read more.
This study investigates and compares different molecular mutation strategies to optimize their application as genetic algorithm operators in drug design. We evaluated five distinct mutation methods—Graph-Based Genetic Algorithm, Graph-Based Generative Model, SmilesClickChem, SELFIES Token, and SMILES Token Mutation—by assessing their computational efficiency, validity, and impact on molecular complexity and structural conservation. Our results reveal that the Graph-Based Genetic Algorithm achieves the highest molecular validity (96.5%) while maintaining computational efficiency, making it ideal for rapid iterative drug discovery. SmilesClickChem and Graph-Based Generative Model tend to increase molecular complexity, whereas SF-T simplifies molecular structures, suggesting different applications in lead optimization. Additionally, we analyzed mutation-induced changes in pIC50 potency and found that SELFIES Token caused the most substantial shifts in bioactivity, particularly in SRC-targeted molecules. These findings underscore the importance of selecting the appropriate mutation strategy to balance validity, structural diversity, and computational cost in AI-driven drug design. Our insights help refine evolutionary algorithms for molecular generation and optimize candidate selection in early-stage drug discovery.
Full article
(This article belongs to the Special Issue Advances in Computer-Aided Drug Design Strategies)
►▼
Show Figures
Graphical abstract
Open AccessArticle
Targeting Pan-Cancer Stemness: Core Regulatory lncRNAs as Novel Therapeutic Vulnerabilities
by
Shengcheng Deng, Yufan Yang, Dapeng Gao, Jiajun Gao and Yuanyan Xiong
Int. J. Mol. Sci. 2025, 26(23), 11684; https://doi.org/10.3390/ijms262311684 - 2 Dec 2025
Abstract
Tumor stemness represents a key biological process that drives tumor progression and therapeutic resistance across various cancer types. To systematically elucidate the regulatory roles of long non-coding RNAs (lncRNAs) in this process, we integrated bulk transcriptomic data from The Cancer Genome Atlas (TCGA)
[...] Read more.
Tumor stemness represents a key biological process that drives tumor progression and therapeutic resistance across various cancer types. To systematically elucidate the regulatory roles of long non-coding RNAs (lncRNAs) in this process, we integrated bulk transcriptomic data from The Cancer Genome Atlas (TCGA) with publicly available pan-cancer single-cell transcriptomic atlases. Using machine-learning-based stemness metrics, we successfully quantified stemness features and identified unique lncRNA gene sets for each cancer type at the bulk data level. The high-stemness subtype exhibited enhanced proliferation, an immunosuppressive microenvironment, and profound metabolic reprogramming. Based on these findings, we constructed a robust prognostic model with remarkable predictive performance across multiple cancer types. At the single-cell resolution, we reconstructed the dynamic trajectory of stemness evolution, uncovering distinctive metabolic and cell-communication patterns within cancer stem cells (CSCs). This multi-scale analysis consistently nominated a core set of regulatory lncRNAs, including NEAT1 and MALAT1. Our work not only nominates potential targets for stemness-directed therapy but also provides a comprehensive framework for understanding lncRNA-driven mechanisms of cancer aggressiveness and resistance.
Full article
(This article belongs to the Section Molecular Informatics)
Open AccessArticle
Cell Suspension of the Tree Fern Cyathea smithii (J.D. Hooker) and Its Metabolic Potential During Cell Growth: Preliminary Studies
by
Jan J. Rybczyński, Łukasz Marczak, Katarzyna Skórkowska-Telichowska, Maciej Stobiecki, Jan Szopa and Anna Mikuła
Int. J. Mol. Sci. 2025, 26(23), 11683; https://doi.org/10.3390/ijms262311683 - 2 Dec 2025
Abstract
The purpose of this study was to present a chemical analysis of the metabolome of cell aggregates of the tree fern Cyathea smithii (J.D. Hooker) cell suspension culture. The LC/MS and GC/MS techniques were used for identification of metabolites. The kinetics of fresh
[...] Read more.
The purpose of this study was to present a chemical analysis of the metabolome of cell aggregates of the tree fern Cyathea smithii (J.D. Hooker) cell suspension culture. The LC/MS and GC/MS techniques were used for identification of metabolites. The kinetics of fresh weight, dry weight, and ash content showed 3.5-fold increases during 15-day-long culture. The analysis demonstrated high metabolic activity of cultured cells. In total, 160 metabolites from primary and secondary metabolism and almost 2000 compounds of unknown identity were identified. Three flavonoids—the chalcone isookanin [(2S)-2-(3,4-dihydroxyphenyl)-7,8-dihydroxy-2,3-dihydrochromen-4-one], a methoxy derivative of the flavone gardenin B (5-Hydroxy-2-(4-methoxyphenyl)-6,7,8-trimethoxy-4H-1-benzopyran-4-one), and the isoflavone tectoridin (4′,5-Dihydro-6-methoxy-7-(O-glucoside)isoflavone)—had not been previously detected in the cell culture of C. smithii. Principal component analysis revealed five distinct groups of samples; groups 4 and 5 showed the greatest similarity and corresponded to cultures on days 12 and 15, respectively. The number of differentiating compounds was 75, indicated by a heatmap showing positive and negative correlations between the days of culture. The studies described in this paper are crucial for further identification of metabolites and establishing the relationship between the metabolic composition of tree fern cells in culture and their biological activity, assessed by physiological parameters. By determining the relationship between the chemical composition of cells and their growth from culture initiation to senescence, we will provide a more complete picture of the potential for environmental factors to regulate this relationship. Based on previous studies, environmental stimuli such as electromagnetic fields or light of different wavelengths can result in altered growth physiology and cell mass, as well as metabolite diversification and accumulation. The research results presented in this paper provide a foundation for further studies aimed at predicting and regulating the productivity of C. smithii cells in suspension culture and elucidating the significance of tree fern-derived metabolic products in human cell biology, particularly in thyroid cells.
Full article
(This article belongs to the Special Issue Molecular Approach to Fern Development)
►▼
Show Figures
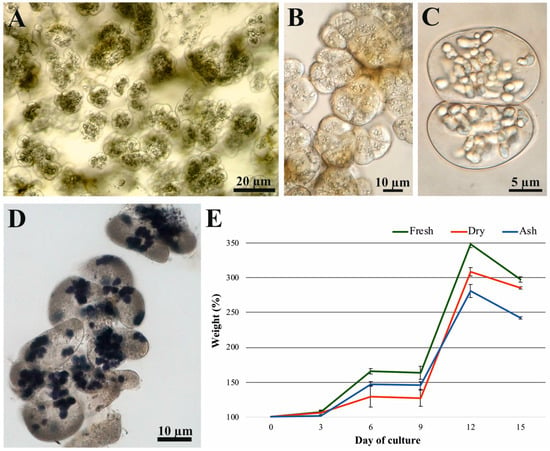
Figure 1
Open AccessReview
Exploring Cyclodextrin Complexes of Lipophilic Antioxidants: Benefits and Challenges in Nutraceutical Development
by
Mario Jug, Kristina Radić, Laura Nižić Nodilo, Emerik Galić, Tea Petković, Marina Jurić, Nikolina Golub, Ivanka Jerić and Dubravka Vitali Čepo
Int. J. Mol. Sci. 2025, 26(23), 11682; https://doi.org/10.3390/ijms262311682 - 2 Dec 2025
Abstract
Antioxidants are essential bioactive compounds widely recognized for their health benefits in preventing oxidative stress-related diseases. However, many lipophilic antioxidants suffer from poor aqueous solubility, low chemical stability, and limited bioavailability, restricting their application in food, nutraceutical, and pharmaceutical industries. Cyclodextrins (CDs), a
[...] Read more.
Antioxidants are essential bioactive compounds widely recognized for their health benefits in preventing oxidative stress-related diseases. However, many lipophilic antioxidants suffer from poor aqueous solubility, low chemical stability, and limited bioavailability, restricting their application in food, nutraceutical, and pharmaceutical industries. Cyclodextrins (CDs), a class of cyclic oligosaccharides with a hydrophilic exterior and lipophilic interior, present an effective strategy to encapsulate and deliver these compounds by improving their solubility, stability, and therapeutic efficacy. This review critically examines the structural features and derivatives of cyclodextrins relevant for antioxidant encapsulation, mechanisms and thermodynamics of inclusion complex formation, and advanced characterization techniques. It evaluates the influence of CD encapsulation on the oral bioavailability and antioxidant activity of various lipophilic antioxidants supported by recent in vitro and in vivo studies. Moreover, sustainable preparation methods for CD complexes are discussed alongside safety and regulatory considerations. The comprehensive synthesis of current knowledge contributes to guiding the rational design and development of CD-based antioxidant nutraceuticals, addressing formulation challenges while promoting efficacy and consumer safety.
Full article
(This article belongs to the Special Issue Research on Cyclodextrin: Properties and Biomedical Applications)
Open AccessArticle
The Immunostimulatory Effect of MIL-101(Al)-NH2 In Vivo and Its Potential to Overcome Bacterial Resistance to Penicillin Enhanced by Hypericin-Induced Photodynamic Therapy
by
Mariana Máčajová, Ľuboš Ambro, Majlinda Meta, Ľuboš Zauška, Terézia Gulyásová, Boris Bilčík, Ivan Čavarga, Gabriela Zelenková, Erik Sedlák, Miroslav Almáši and Veronika Huntošová
Int. J. Mol. Sci. 2025, 26(23), 11681; https://doi.org/10.3390/ijms262311681 - 2 Dec 2025
Abstract
The increasing prevalence of multidrug-resistant bacteria necessitates alternative therapeutic strategies that combine antimicrobial efficacy with immunomodulatory properties. Here, we report the immunostimulatory activity and antibacterial potential of the amino-functionalized metal–organic framework MIL-101(Al)-NH2 as a carrier for penicillin (PEN) and hypericin (Hyp), a
[...] Read more.
The increasing prevalence of multidrug-resistant bacteria necessitates alternative therapeutic strategies that combine antimicrobial efficacy with immunomodulatory properties. Here, we report the immunostimulatory activity and antibacterial potential of the amino-functionalized metal–organic framework MIL-101(Al)-NH2 as a carrier for penicillin (PEN) and hypericin (Hyp), a photodynamically active compound. Structural and physicochemical characterization confirmed successful encapsulation of PEN, Hyp, and their combination within MIL-101(Al)-NH2, with distinct effects on porosity, release kinetics, and thermal stability. Drug release studies revealed rapid Hyp liberation triggered by serum components, whereas PEN exhibited a biphasic, diffusion-controlled profile. Using a quail chorioallantoic membrane (CAM) model, we demonstrated that MIL-101(Al)-NH2 enhances interferon-α expression, indicating intrinsic immunostimulatory activity, and that Hyp-loaded systems promote angiogenic responses. In a bacterial infection CAM model, MIL-101(Al)-NH2 carriers loaded with Hyp or Hyp/PEN induced immunomodulatory changes and, upon photodynamic activation, inhibited bacterial growth. While Gram-negative Escherichia coli remained resistant, Gram-positive Staphylococcus epidermidis was effectively suppressed by photodynamic therapy (PDT), and Hyp/PEN co-delivery overcame bacterial resistance to PEN. These results highlight MIL-101(Al)-NH2 as a multifunctional nanoplatform with immunostimulatory capacity and PDT-enhanced antibacterial activity, offering a promising strategy to combat antibiotic resistance and infections associated with medical implants.
Full article
(This article belongs to the Special Issue New Molecular Insights into Antimicrobial Photo-Treatments)
Open AccessArticle
PRSS38 Is a Novel Sperm Serine Protease Involved in Human and Mouse Fertilization
by
Ania Antonella Manjon, Gustavo Luis Verón, Rosario Vitale, Georgina Stegmayer, Fernanda Gonzalez Echeverria-Raffo, Lydie Lane and Mónica Hebe Vazquez-Levin
Int. J. Mol. Sci. 2025, 26(23), 11680; https://doi.org/10.3390/ijms262311680 - 2 Dec 2025
Abstract
Sperm proteases are involved in several gamete interaction events leading to fertilization. This report presents a detailed analysis of the expression and localization of serine protease PRSS38 in human and in mouse spermatozoa and its involvement in fertilization-related events, using bioinformatics, cellular, biochemical,
[...] Read more.
Sperm proteases are involved in several gamete interaction events leading to fertilization. This report presents a detailed analysis of the expression and localization of serine protease PRSS38 in human and in mouse spermatozoa and its involvement in fertilization-related events, using bioinformatics, cellular, biochemical, molecular, and functional approaches. Bioinformatics analyses included genomics and data analysis, prediction of protein subcellular localization and post-translational modifications, Self-Organizing Maps (SOMs) unsupervised training with other serine proteases, protein modeling (AlphaFold), and genetic variant analysis. For cellular, biochemical, and functional studies, human semen samples were obtained from healthy normozoospermic volunteers, and cauda epididymal sperm were collected from adult Balb-c/C57 mice. PRSS38 presence was detected in human and mouse sperm protein extracts by Western immunoblotting. Sperm PRSS38 subcellular localization was determined by fluorescence immunocytochemistry. Human sperm–oocyte interaction events were assessed by means of the mouse Cumulus Penetration Assay (CPA) using mouse COCs, the Human Hemizona Assay (HZA), and the ZP-free hamster egg Sperm Penetration Assay (SPA). Mouse sperm–oocyte interactions were evaluated by means of in vitro fertilization (IVF) with COCs and denuded oocytes. PRSS38 is proposed to be a GPI-anchored serine protease (active site: His-100, Asp-150, and Ser-245) based on bioinformatics analyses. Using commercial antibodies, protein forms of the expected Mr (human: 31 kDa; mouse: 32 and 24 kDa) were specifically immunodetected in protein sperm extracts. Immunocytochemical analysis revealed a specific PRSS38 signal in the human sperm acrosomal region, equatorial segment, and flagellum. Mouse sperm PRSS38 was immunolocalized in the equatorial segment and hook. Human sperm preincubation with specific antibodies resulted in inhibition (p < 0.05) of CPA, HZA, and SPA. Mouse sperm preincubation with PRSS38 antibodies impaired (p < 0.05) homologous IVF using COCs and denuded oocytes. Genetic variants affecting residues involved in the GPI anchor and the catalytic triad were found in individuals from the general population whose PRSS38 protease function could be altered. This study provides, for the first time, an integrated analysis of PRSS38 in human and mouse sperm, contributing to our understanding of mammalian fertilization and male infertility.
Full article
(This article belongs to the Special Issue The Molecular Life of Sperm: New Horizons in Male Infertility)
Open AccessReview
Interferon-Based Therapeutics in Cancer Therapy: Past, Present, and Future
by
Kristina Vorona, Anastasia Ryapolova, Olesya Sokolova, Alexander Karabelsky, Roman Ivanov, Vasiliy Reshetnikov and Ekaterina Minskaia
Int. J. Mol. Sci. 2025, 26(23), 11679; https://doi.org/10.3390/ijms262311679 - 2 Dec 2025
Abstract
Interferons (IFNs) are well-known immunostimulants involved in both innate and adaptive immune responses. These multifunctional proteins mediate an early antiviral response and have pronounced immunomodulatory and antiproliferative properties. Due to their potency, IFNs have been used not only in the treatment of viral
[...] Read more.
Interferons (IFNs) are well-known immunostimulants involved in both innate and adaptive immune responses. These multifunctional proteins mediate an early antiviral response and have pronounced immunomodulatory and antiproliferative properties. Due to their potency, IFNs have been used not only in the treatment of viral infections but also various other diseases. However, the use of IFNs in antitumor therapy has been limited by the frequent severe side effects, which reduced their appeal for the treatment of cancer. In this review, we focused on current data on recombinant IFNs used for anticancer therapy, as well as the development of promising IFN-based gene therapy approaches, with a focus on their safety and therapeutic efficacy. We also highlighted various types of IFNs and their application niches in the treatment of not only cancers in combination therapy but also of certain rare diseases. Taken together, this review improves our understanding of the existing IFN applications in cancer therapy, the disadvantages of using IFNs, and possible approaches for their improvement.
Full article
(This article belongs to the Special Issue Novel Therapeutic Targets in Cancers: 4th Edition)
Open AccessArticle
Intensity-Based Estimation of Monomeric Brightness for Fluorescent Proteins
by
Michael R. Stoneman, Sanam Bista, Thomas D. Killeen, Ionel Popa and Valerică Raicu
Int. J. Mol. Sci. 2025, 26(23), 11678; https://doi.org/10.3390/ijms262311678 - 2 Dec 2025
Abstract
Fluorescence fluctuation spectroscopy (FFS) techniques rely on determination of monomeric molecular brightness, i.e., the fluorescence intensity of a single, non-aggregated fluorophore, as a critical reference for estimating protein oligomer size. By comparing measured molecular brightness of fluorescently labeled proteins of interest to this
[...] Read more.
Fluorescence fluctuation spectroscopy (FFS) techniques rely on determination of monomeric molecular brightness, i.e., the fluorescence intensity of a single, non-aggregated fluorophore, as a critical reference for estimating protein oligomer size. By comparing measured molecular brightness of fluorescently labeled proteins of interest to this monomeric brightness benchmark, FFS enables inference of oligomerization states. However, widely used fluorescent proteins often exhibit self-association, compromising monomeric brightness calibration and introducing errors in brightness-derived oligomer-size estimates. This study compares two strategies for determining monomeric brightness: the conventional fluctuation-based method and a more recently proposed average-intensity-based alternative. The comparison uses two model fluorophores, a fluorescent protein (mCitrine) and the small-molecule dye Janelia Fluor 525 (JF525) conjugated to HaloTag. Our results show strong agreement between intensity- and fluctuation-derived brightness values only in the minimally aggregating JF525–HaloTag benchmark. In contrast, in the mCitrine samples, where aggregation is more prevalent, only the intensity-based method maintains a consistent estimate across sample preparation, while the fluctuation-based method overestimates brightness when aggregation effects become pronounced. This robustness makes the intensity-based approach a valuable cross-check for monomeric brightness calibration. Our results support a combined strategy, using both methods to improve the reliability of monomeric brightness calibration and protein oligomerization analysis in FFS.
Full article
(This article belongs to the Section Molecular Biophysics)
►▼
Show Figures
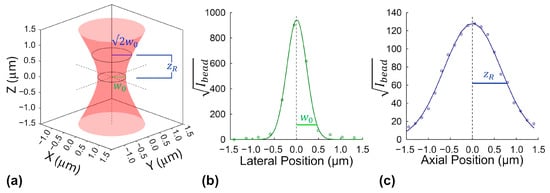
Figure 1
Open AccessArticle
Integrative Assessment of Glycyrrhiza uralensis Extract in Cosmetics Using HPLC Analysis, Network Pharmacology, and Computational Threshold of Toxicological Concern-Based Safety Evaluation
by
Hiyoung Kim, Kihoon Park, Young Bong Kim and Minjee Kim
Int. J. Mol. Sci. 2025, 26(23), 11677; https://doi.org/10.3390/ijms262311677 - 2 Dec 2025
Abstract
Licorice (Glycyrrhiza uralensis) contains bioactive flavonoids and saponins, primarily liquiritin and glycyrrhizin, which exhibit pharmacological activities but also potential dose-dependent toxicity. This study aimed to establish an integrative workflow combining analytical chemistry, network pharmacology, and computational toxicology to evaluate the skin-related safety of
[...] Read more.
Licorice (Glycyrrhiza uralensis) contains bioactive flavonoids and saponins, primarily liquiritin and glycyrrhizin, which exhibit pharmacological activities but also potential dose-dependent toxicity. This study aimed to establish an integrative workflow combining analytical chemistry, network pharmacology, and computational toxicology to evaluate the skin-related safety of these compounds. High-performance liquid chromatography (HPLC) was employed to quantify liquiritin and glycyrrhizin in licorice extract. Network pharmacology and molecular docking analyses were conducted to identify core toxicity-related targets. In silico toxicity and threshold of toxicological concern (TTC) assessments were performed using VEGA and database-driven prediction models to estimate dermal exposure risk. Liquiritin and glycyrrhizin were identified as major constituents of G. uralensis. Network analysis revealed three key targets—EGFR, STAT3, and SRC—linked to skin sensitivity and toxicological pathways, including TRP channel regulation and EGFR signaling. Molecular docking showed strong binding affinities to SRC. The threshold of toxicological concern evaluation indicated that liquiritin exposure remained below safety thresholds, while glycyrrhizin slightly exceeded but remained within acceptable limits. The proposed HPLC–network pharmacology–TTC workflow provides a novel, non-animal approach for early-stage cosmetic safety screening. Both compounds demonstrate acceptable safety margins, supporting their controlled use in dermal formulations.
Full article
(This article belongs to the Special Issue New Insights into Network Pharmacology)
►▼
Show Figures
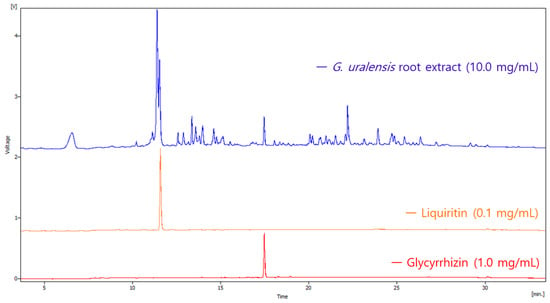
Figure 1
Open AccessCase Report
Novel NUTM1 Fusions in Relapsed Acute Myeloid Leukemia: Expanding the Genetic and Clinical Landscape
by
Parastou Tizro, Lisa Chang, Amandeep Salhotra, Javier Arias-Stella, Milhan Telatar, Vanina Tomasian, Karl Gaal, Joo Song, Lorinda Soma, Sandra Fuentes, Lino Garcia, Fei Fei, Anamaria Munteanu, Guido Marcucci and Michelle Afkhami
Int. J. Mol. Sci. 2025, 26(23), 11676; https://doi.org/10.3390/ijms262311676 - 2 Dec 2025
Abstract
Gene fusions involving NUTM1 have been increasingly recognized in hematologic malignancies, though their role in acute myeloid leukemia (AML) remains poorly understood. We retrospectively analyzed 565 unique AML patients with reported fusion results who underwent comprehensive next-generation sequencing (NGS) between March 2022 and
[...] Read more.
Gene fusions involving NUTM1 have been increasingly recognized in hematologic malignancies, though their role in acute myeloid leukemia (AML) remains poorly understood. We retrospectively analyzed 565 unique AML patients with reported fusion results who underwent comprehensive next-generation sequencing (NGS) between March 2022 and December 2023. Among them, three novel in-frame NUTM1 fusion transcripts, LARP1::NUTM1, ARHGAP15::NUTM1, and GABPB1::NUTM1, were identified in three relapsed or refractory AML cases, all with monocytic differentiation. Ancillary studies included flow cytometry, cytogenetics, FISH, and comprehensive mutational profiling. All three patients eventually relapsed and succumbed to their disease, despite initial responses in one case. Each case also harbored co-occurring mutations associated with adverse prognosis, such as BCOR, ASXL1, and RUNX1. These findings suggest NUTM1 fusions in AML could represent a distinct molecular subset with potentially poor prognosis, warranting further functional and clinical investigation to clarify their biological and therapeutic significance.
Full article
(This article belongs to the Special Issue Immunotherapy Versus Immune Modulation of Leukemia)
►▼
Show Figures
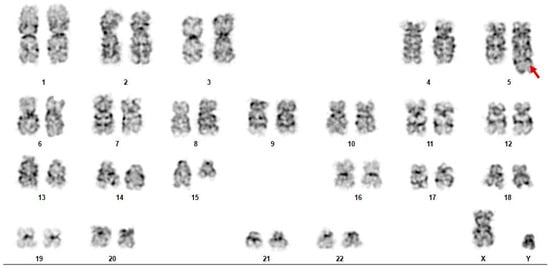
Figure 1
Open AccessArticle
Respiratory Delivery of Highly Conserved Antiviral siRNAs Suppress SARS-CoV-2 Infection
by
Yuan Zhang, Matt D. Johansen, Scott Ledger, Stuart Turville, Pall Thordarson, Philip M. Hansbro, Anthony D. Kelleher and Chantelle L. Ahlenstiel
Int. J. Mol. Sci. 2025, 26(23), 11675; https://doi.org/10.3390/ijms262311675 - 2 Dec 2025
Abstract
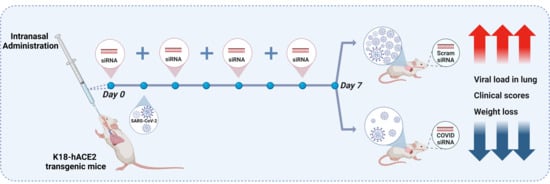
COVID-19 has resulted in over 777 million confirmed cases and more than 7 million deaths globally. While vaccination offers protection for individuals with a functional immune system, immunocompromised populations will not generate sufficient responses, highlighting the critical need for new antiviral treatments. Here
[...] Read more.
COVID-19 has resulted in over 777 million confirmed cases and more than 7 million deaths globally. While vaccination offers protection for individuals with a functional immune system, immunocompromised populations will not generate sufficient responses, highlighting the critical need for new antiviral treatments. Here we evaluated four highly conserved anti-COVID siRNAs targeting the ORF1a-Nsp1, Membrane, and Nucleocapsid regions by identifying their antiviral efficacy in vitro and investigated the direct delivery of naked siRNAs to the respiratory tract of mice via intranasal instillation to provide proof-of-concept evidence of their in vivo antiviral activity. Dose-response analysis of siRNAs revealed a range of IC50 0.02 nM to 0.9 nM. Intranasal administration of naked anti-COVID siRNA-18 in a K18-hACE2 transgenic SARS-CoV-2 mouse model was capable of reducing viral mRNA levels and disease severity. While anti-COVID siRNA-30 induced modest interferon-stimulated gene expression in vitro and immune cell infiltration in vivo, these effects were markedly reduced by 2′-O-methyl-AS456 chemical modification, which preserved antiviral efficacy against SARS-CoV-2 while minimizing off-target immune activation. These results demonstrate the feasibility of direct respiratory siRNA administration for in vivo viral suppression and highlight the benefit of using conserved target sequences and chemical modification to enhance therapeutic safety and efficacy.
Full article
(This article belongs to the Special Issue The Role of SARS-CoV-2 in Immunomodulation—Post-Pandemic Reflection on the Role of Viruses in Human Immunology)
►▼
Show Figures
Graphical abstract
Open AccessArticle
Heterogeneity of Primary Ciliary Dyskinesia Gene Variants: A Genetic Database Analysis in Russia
by
Elena I. Kondratyeva, Sergey N. Avdeev, Tatiana A. Kyian, Oksana P. Ryzhkova, Yuliya L. Melyanovskaya, Victoria V. Zabnenkova, Maria V. Bulakh, Zamira M. Merzhoeva, Artem V. Bukhonin, Natalia V. Trushenko, Baina B. Lavginova, Daria O. Zhukova and Sergey I. Kutsev
Int. J. Mol. Sci. 2025, 26(23), 11674; https://doi.org/10.3390/ijms262311674 - 2 Dec 2025
Abstract
Primary ciliary dyskinesia (PCD) is a rare hereditary disorder belonging to the group of ciliopathies, with autosomal recessive, autosomal dominant, and, less frequently, X-linked inheritance patterns. The aim of this study was to investigate the genetic heterogeneity of the Russian population of PCD
[...] Read more.
Primary ciliary dyskinesia (PCD) is a rare hereditary disorder belonging to the group of ciliopathies, with autosomal recessive, autosomal dominant, and, less frequently, X-linked inheritance patterns. The aim of this study was to investigate the genetic heterogeneity of the Russian population of PCD patients based on national registry data. The study included patients with PCD confirmed by molecular genetic testing. Quantitative data were analyzed using non-parametric statistical methods. Differences were considered statistically significant at p < 0.05. The study included 109 patients with PCD. Molecular genetic testing identified pathogenic variants in 29 autosomal recessive genes. The analysis of pathogenic variant distribution in the Russian PCD cohort revealed the highest number of changes in the DNAH5 and DNAH11 genes. 26 genetic variants in 13 genes were identified for the first time in the Russian population. Variants in the DNAH5 gene were significantly more frequent in Kartagener’s syndrome (KS) patients (32/55%) compared to those without KS (11/21.5%) (χ2 = 12.8; p = 0.0004; OR = 4.48). Preliminary data indicate that the frequency spectrum of DNAH5 and DNAH11 genes in Russian patients is similar to international trends. Additionally, there is an accumulation of pathogenic variants in the DNAH5, DNAH11, CCDC39, and CFAP300 genes.
Full article
(This article belongs to the Special Issue Molecular Mechanisms Underlying the Pathogenesis of Genetic Diseases)
►▼
Show Figures
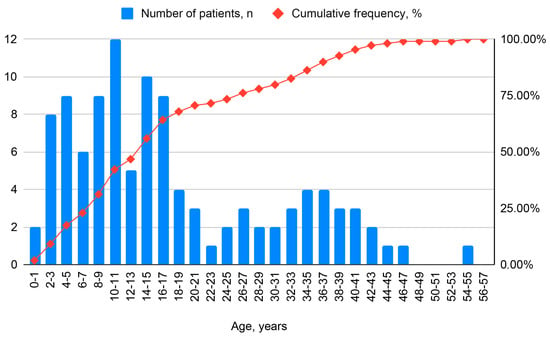
Figure 1
Open AccessArticle
Development of Plasma Protein Classification Models for Alzheimer’s Disease Using Multiple Machine Learning Approaches
by
Amy Tsurumi, Catherine M. Cahill, Andy J. Liu, Pranam Chatterjee, Sudeshna Das and Ami Kobayashi
Int. J. Mol. Sci. 2025, 26(23), 11673; https://doi.org/10.3390/ijms262311673 - 2 Dec 2025
Abstract
Alzheimer’s Disease (AD) management is challenging due to limitations in detection methods. Currently, cerebrospinal fluid (CSF) biomarkers involve assessing β-amyloid (Aβ) and phosphorylated tau proteins. The lumbar puncture procedure to obtain CSF is invasive and sometimes causes significant anxiety in patients. In contrast,
[...] Read more.
Alzheimer’s Disease (AD) management is challenging due to limitations in detection methods. Currently, cerebrospinal fluid (CSF) biomarkers involve assessing β-amyloid (Aβ) and phosphorylated tau proteins. The lumbar puncture procedure to obtain CSF is invasive and sometimes causes significant anxiety in patients. In contrast, plasma biomarkers would allow rapid, accurate, and cost-effective diagnosis, while minimizing invasiveness and discomfort. Using a dataset involving 120 plasma proteins from clinically diagnosed AD patients versus cognitively normal subjects, we developed classification models by applying various machine learning algorithms (EBlasso, EBEN, XGBoost, LightGBM, TabNet, and TabPFN) to plasma proteomic measurements. Gene ontology and pathway enrichment, and a literature review were used to evaluate the potential relevance of the biomarkers identified in AD-related mechanisms. Biomarkers identified were also evaluated for the enrichment of aging-related biomarkers. The models developed yielded high AUROC and accuracy, mostly >0.9. Proteins selected as predictors by all the models included Angiopoietin-2 (ANG-2), epidermal growth factor (EGF), Interleukin 1α (IL-1α), and platelet growth factor subunit B (PDGF-BB). Ample previous literature supported their relevance in AD. The pool of all the biomarkers identified was significantly enriched with known aging-related biomarkers (p = 0.040). Applying cutting-edge algorithms is expected to be advantageous for developing AD prediction models with plasma proteomic data, and future large studies to externally validate the constructed models in other populations to assess their generalizability is important. The proteins uncovered may represent novel preventative or therapeutic targets.
Full article
(This article belongs to the Special Issue Advances in Molecular Mechanisms of Neurodegenerative Diseases)
►▼
Show Figures
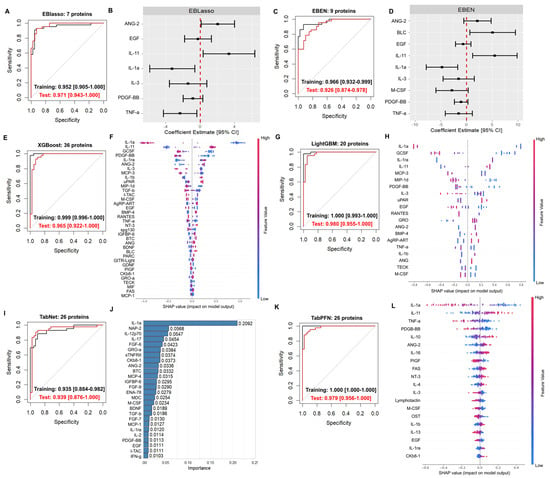
Figure 1
Open AccessArticle
Insulin Deficiency Exacerbates Muscle Atrophy and Osteopenia in Chrebp Knockout Mice
by
Chihiro Ushiroda, Mioko Ito, Risako Yamamoto-Wada, Kanako Deguchi, Shihomi Hidaka, Toshinori Imaizumi, Yusuke Seino, Atsushi Suzuki, Daisuke Yabe and Katsumi Iizuka
Int. J. Mol. Sci. 2025, 26(23), 11672; https://doi.org/10.3390/ijms262311672 - 2 Dec 2025
Abstract
Type 1 diabetes mellitus is a major risk factor for both sarcopenia and osteoporosis, primarily due to the body’s inability to utilize glucose as a result of insulin deficiency. Impairments in insulin and glucose signaling can accelerate the decline in muscle and bone
[...] Read more.
Type 1 diabetes mellitus is a major risk factor for both sarcopenia and osteoporosis, primarily due to the body’s inability to utilize glucose as a result of insulin deficiency. Impairments in insulin and glucose signaling can accelerate the decline in muscle and bone health. To investigate this interaction, we examined whether insulin deficiency exacerbates muscle and bone deterioration in Chrebp knockout (KO) mice. Male wild-type (WT) and KO mice, aged 18 weeks, were intraperitoneally treated with 200 mg/kg BW streptozotocin (STZ), which selectively destroys pancreatic beta cells, thereby inducing insulin deficiency. Two weeks after STZ administration, compared with STZ-treated WT mice, STZ-treated KO mice presented significantly greater reductions in body weight and gastrocnemius muscle weight (BW: WT-vehicle vs. WT-STZ; 2.58 [−1.23, 6.39] (p = 0.21); KO-vehicle vs. KO-STZ: 8.03 [5.23, 10.82]; GA muscle: WT vehicle vs. WT STZ: 0.084 [0.047, 0.12], p < 0.0001; KO vehicle vs. KO STZ: 0.084, [0.047, 0.12], p < 0.0001). The decrease in grip strength caused by STZ administration was greater in the KO mice than in the WT mice (mean differences [95% CIs]: WT vehicle—WT STZ, 49.6. [0.9, 98.4], p = 0.046; WT STZ—KO STZ: 71.40 [29.1, 113.7], p = 0.0059; KO vehicle—KO STZ: 84.3 [51.9, 116.8], p = 0.0003). Consistent with these findings, STZ administration reduced IGF-1 expression and increased atrogin mRNA levels, with the highest levels in STZ-treated KO mice. In skeletal muscle, the changes in IGF-1 and Atrogen induced by STZ administration were significantly greater in the KO group than in the WT group (IGF-1: WT vehicle—WT STZ: 0.19 [−0.072, 0.46], p = 0.17; KO vehicle—KO STZ: 0.79 [0.53, 1.06], p < 0.0001; Atrogen: WT vehicle—WT STZ: −2.7 [−3.01, −2.29], p < 0.0001; KO vehicle—KO STZ: −3.35 [−3.71, −2.99], p < 0.0001). The BMD in the Chrebp-deficient group was greater than that in the wild-type group (WT vehicle—KO vehicle: −5.2 [−8.4, −1.9], p = 0.0014); however, the administration of STZ significantly decreased the BMD only in the KO group (WT vehicle—WT STZ: p = 0.45, KO vehicle—KO STZ: 7.2 [3.9, 10.4], p < 0.0001). These results suggest that Chrebp deficiency combined with insulin deficiency aggravates sarcopenia and osteoporosis risk. Therefore, insulin and glucose signals are important for maintaining muscle and bone mass and function. However, further studies are needed to elucidate the mechanisms by which ChREBP deletion and insulin deficiency cause osteosarcopenia.
Full article
(This article belongs to the Section Molecular Endocrinology and Metabolism)
►▼
Show Figures
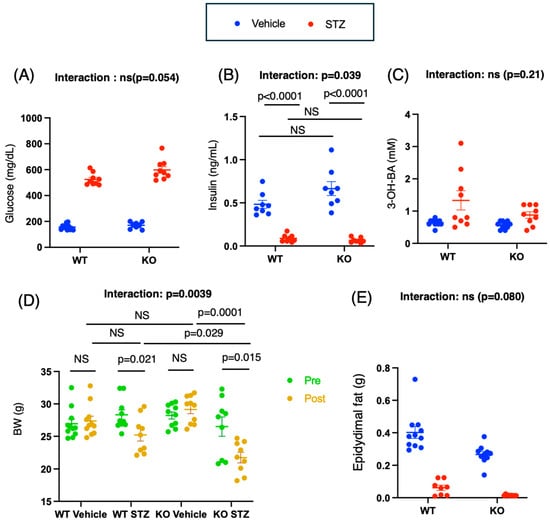
Figure 1
Open AccessEditorial
Special Issue: Protein–Protein Interactions: New Perspectives in Drug Discovery
by
Tibor Szénási
Int. J. Mol. Sci. 2025, 26(23), 11671; https://doi.org/10.3390/ijms262311671 - 2 Dec 2025
Abstract
Protein–protein interactions (PPIs) are the cornerstone of cellular life, forming a vast and dynamic network—the “interactome”—that governs nearly every biological process, from signal transduction and DNA replication to metabolic regulation and immune responses [...]
Full article
(This article belongs to the Special Issue Protein–Protein Interactions: New Perspectives in Drug Discovery)

Journal Menu
► ▼ Journal Menu-
- IJMS Home
- Aims & Scope
- Editorial Board
- Reviewer Board
- Topical Advisory Panel
- Instructions for Authors
- Special Issues
- Topics
- Sections & Collections
- Article Processing Charge
- Indexing & Archiving
- Most Cited & Viewed
- Journal Statistics
- Journal History
- Journal Awards
- Society Collaborations
- Conferences
- Editorial Office
Journal Browser
► ▼ Journal Browser-
arrow_forward_ios
Forthcoming issue
arrow_forward_ios Current issue - Vol. 26 (2025)
- Vol. 25 (2024)
- Vol. 24 (2023)
- Vol. 23 (2022)
- Vol. 22 (2021)
- Vol. 21 (2020)
- Vol. 20 (2019)
- Vol. 19 (2018)
- Vol. 18 (2017)
- Vol. 17 (2016)
- Vol. 16 (2015)
- Vol. 15 (2014)
- Vol. 14 (2013)
- Vol. 13 (2012)
- Vol. 12 (2011)
- Vol. 11 (2010)
- Vol. 10 (2009)
- Vol. 9 (2008)
- Vol. 8 (2007)
- Vol. 7 (2006)
- Vol. 6 (2005)
- Vol. 5 (2004)
- Vol. 4 (2003)
- Vol. 3 (2002)
- Vol. 2 (2001)
- Vol. 1 (2000)
Highly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
Biophysica, CIMB, Diagnostics, IJMS, IJTM
Molecular Radiobiology of Protons Compared to Other Low Linear Energy Transfer (LET) Radiation
Topic Editors: Francis Cucinotta, Jacob RaberDeadline: 20 December 2025
Topic in
Antioxidants, Chemistry, IJMS, Molecules, Pharmaceutics
Natural Bioactive Compounds as a Promising Approach to Mitigating Oxidative Stress
Topic Editors: Andrea Ragusa, Filomena CorboDeadline: 31 December 2025
Topic in
Agriculture, Agronomy, Crops, Plants, IJMS, IJPB
Plant Responses and Tolerance to Salinity Stress, 2nd Edition
Topic Editors: Ricardo Aroca, Pablo CornejoDeadline: 15 January 2026
Topic in
Chemistry, Inorganics, IJMS, Pharmaceuticals
Natural Coumarin and Metal Complexes: Pharmacological Properties and Potential ApplicationsTopic Editors: Dušan Dimić, Edina Avdović, Dejan MilenkovićDeadline: 31 January 2026

Conferences
Special Issues
Special Issue in
IJMS
Biophysical Characterization and Molecular Engineering of Multidomain Proteins (4th Edition)
Guest Editors: Koichi Kato, Takayuki UchihashiDeadline: 10 December 2025
Special Issue in
IJMS
Molecular Mechanisms of Obesity and Metabolic Diseases
Guest Editor: Liliya MihaylovaDeadline: 15 December 2025
Special Issue in
IJMS
Pathogenesis and Molecular Therapy of Brain Tumor
Guest Editor: Theo KrausDeadline: 15 December 2025
Topical Collections
Topical Collection in
IJMS
Genetics and Molecular Breeding in Plants
Collection Editor: Pedro Martínez-Gómez
Topical Collection in
IJMS
Pharmaceutical Nanoimaging and Nanoengineering
Collection Editor: Yuri Lyubchenko
Topical Collection in
IJMS
Latest Review Papers in Molecular Genetics and Genomics
Collection Editor: Salvatore Saccone
Topical Collection in
IJMS
State-of-the-Art Macromolecules in Japan
Collection Editors: Ryoichi Arai, Shunsuke Tomita, Tamotsu Zako, Masafumi Odaka