Molecular and Physiological Responses of Rice and Weedy Rice to Heat and Drought Stress †
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Material
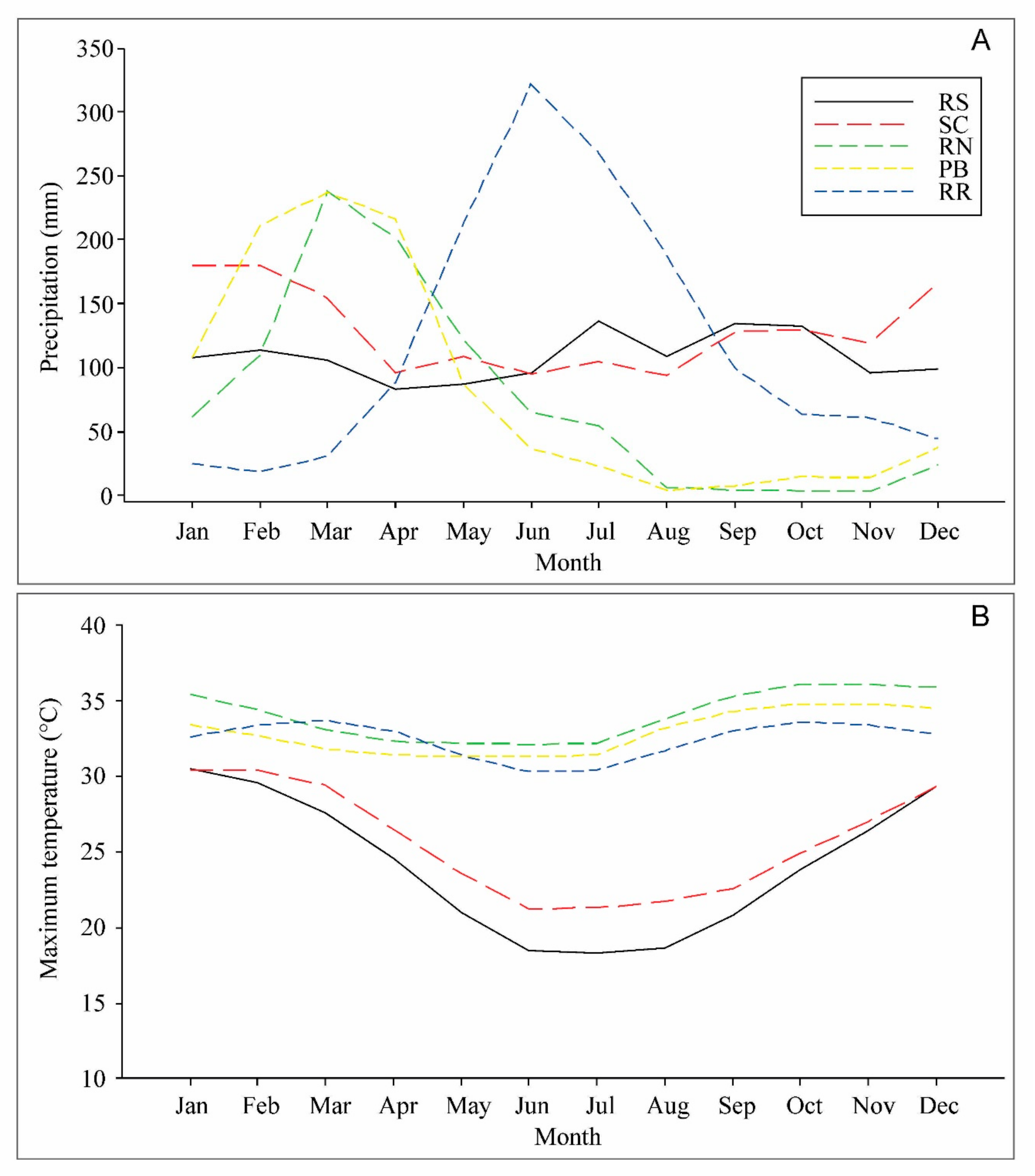
2.2. Drought and Heat Stress Induction
2.3. Determination of Photosynthetic Parameters
2.4. Morphological Data and Yield Determination
2.5. Statistical Analysis
2.6. Gene Expression Analysis by qRT-PCR
3. Results
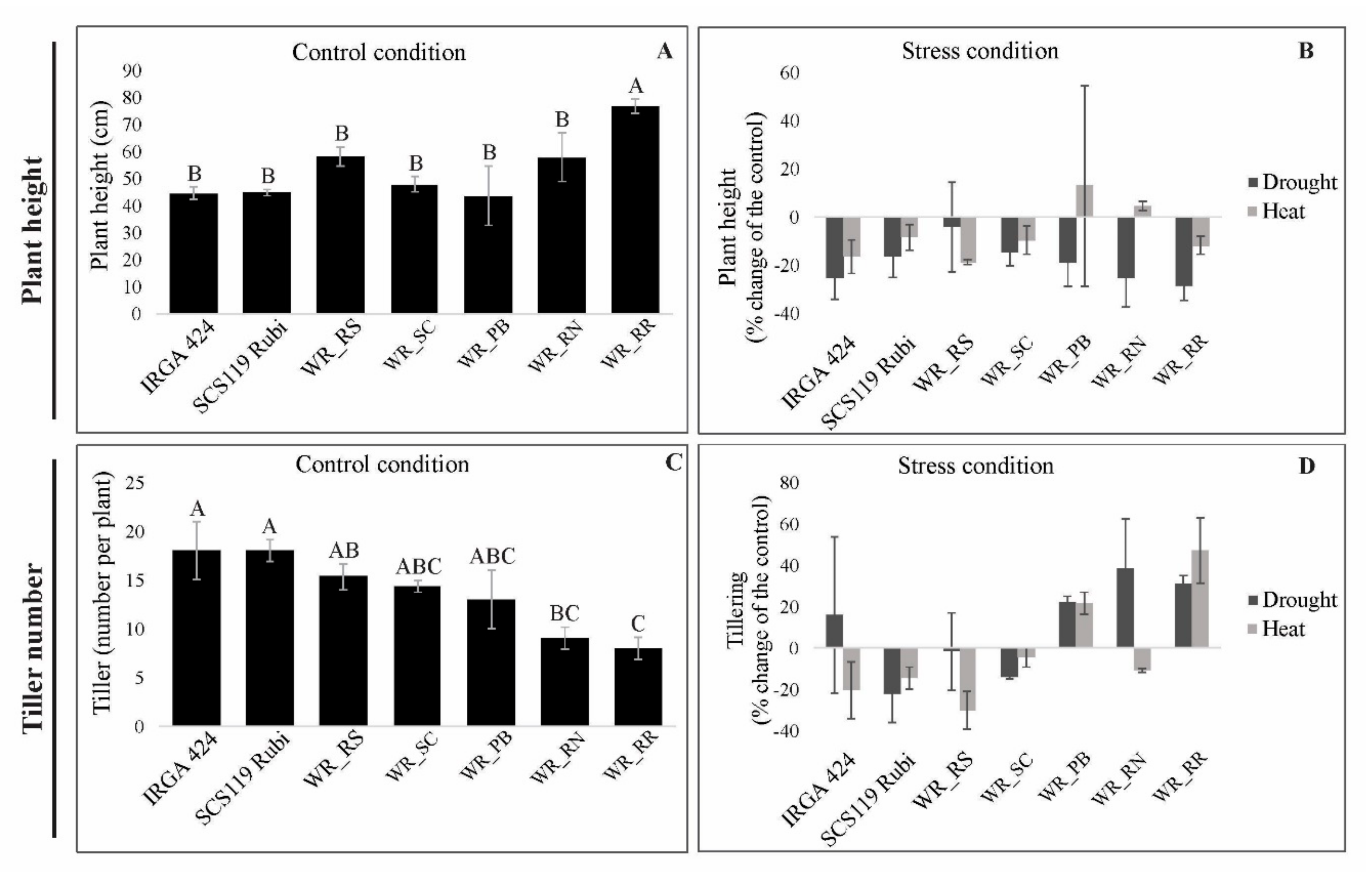
3.1. Morphological Traits under Heat and Drought Stress
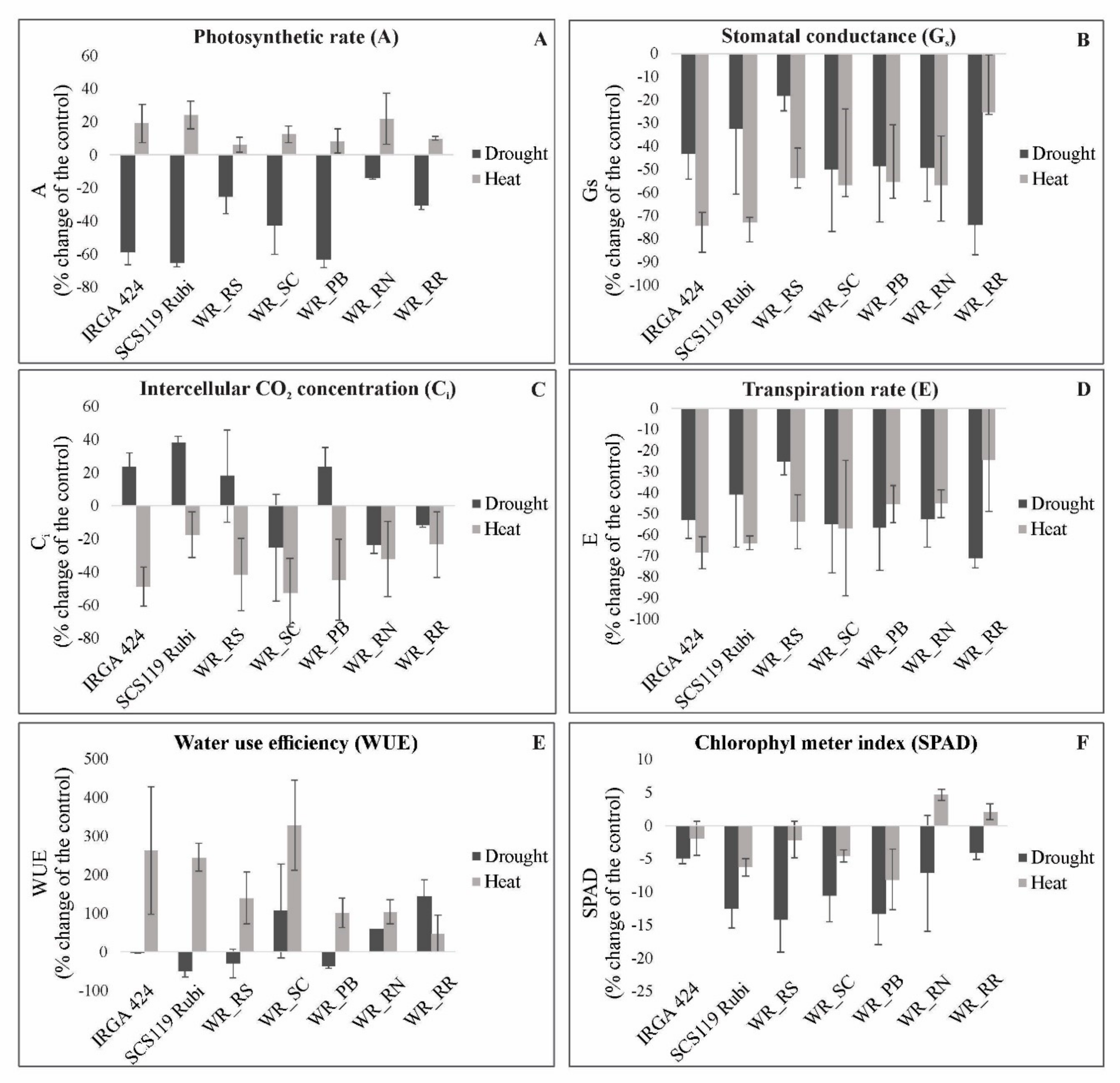
3.2. Photosynthetic Parameters in Heat and Drought Stress
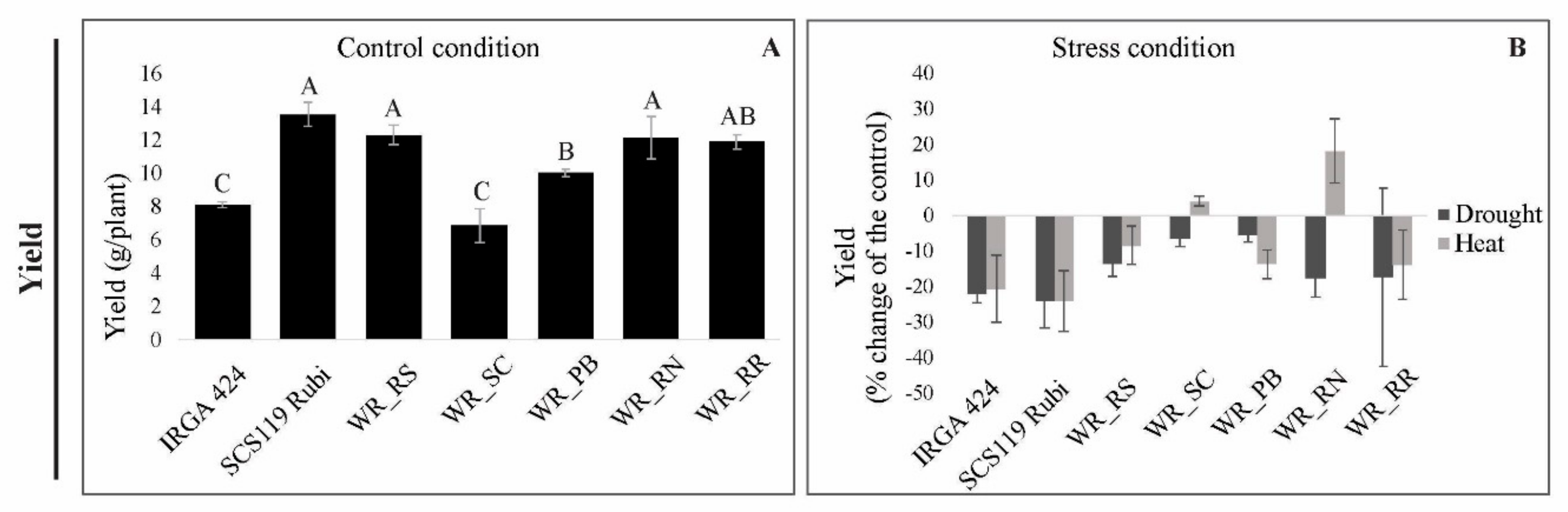
3.3. Grain Yield under Heat and Drought Stress
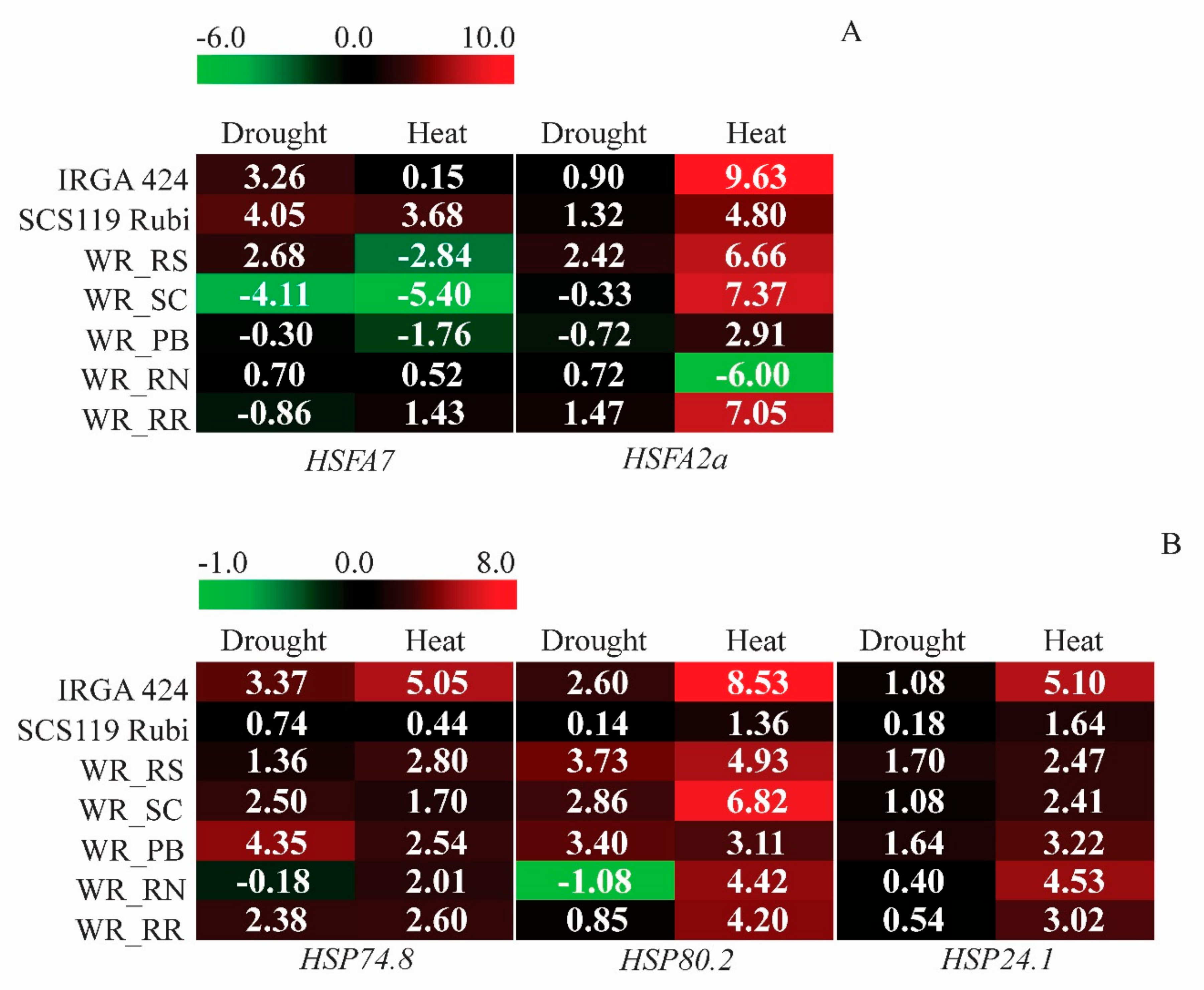
3.4. Gene Expression Analysis under Heat and Drought Conditions
3.4.1. Transcriptional Regulators—HSFs
3.4.2. Stress-Responsive Genes—HSPs
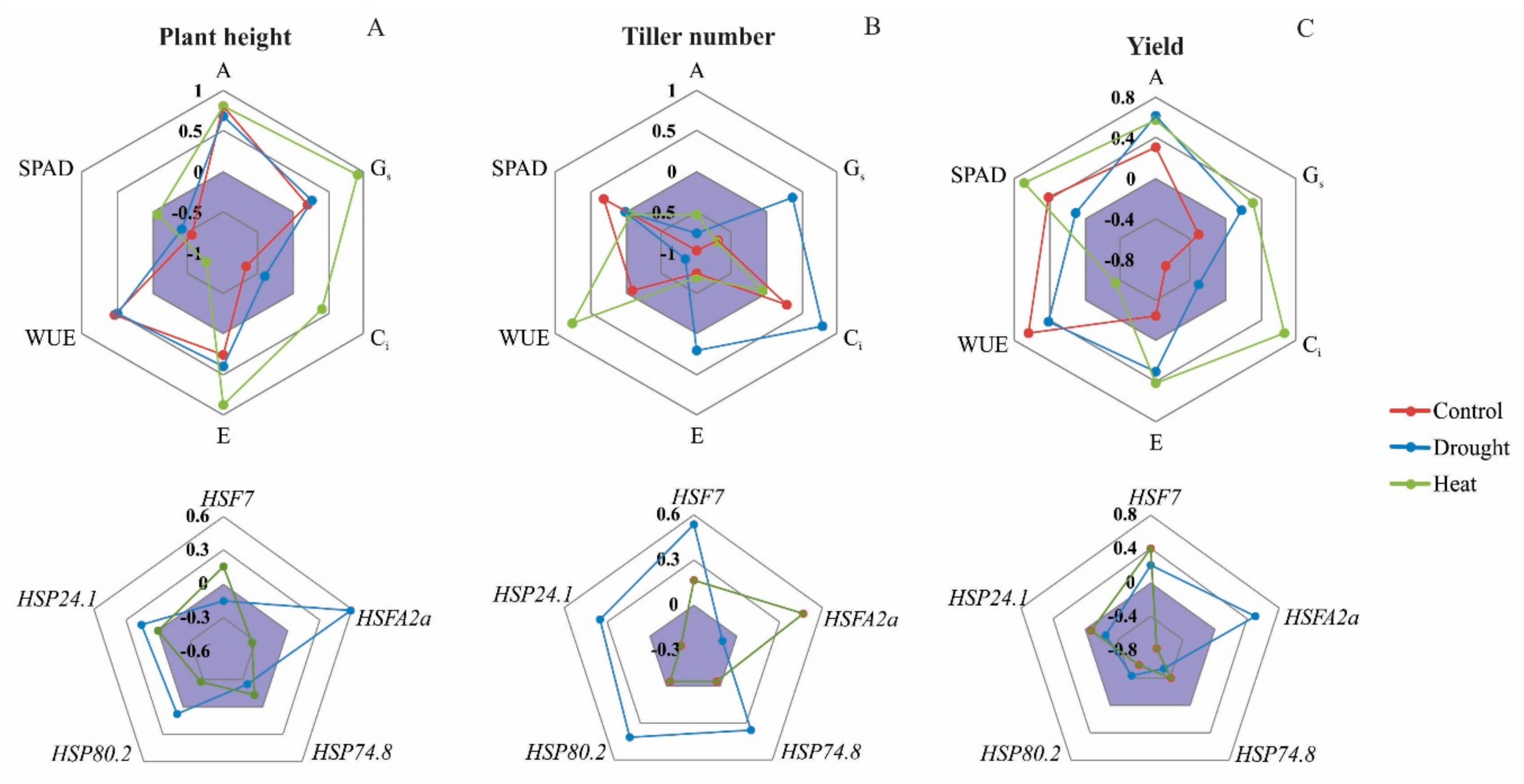
3.5. Relating Physiological and Molecular Responses to Phenotypic Responses
3.5.1. Plant Height
3.5.2. Tiller Number
3.5.3. Yield
4. Discussion
4.1. Physiological Parameters Indicate Genotypes with Suitable Performance under Drought and Heat Stresses
4.2. Relationship between Photosynthetic Parameters and Grain Yield Reveals Genotypes with Suitable Performance under Drought and Heat Stresses
4.3. Modulation of HSFs and HSPs under Drought and Heat Stresses
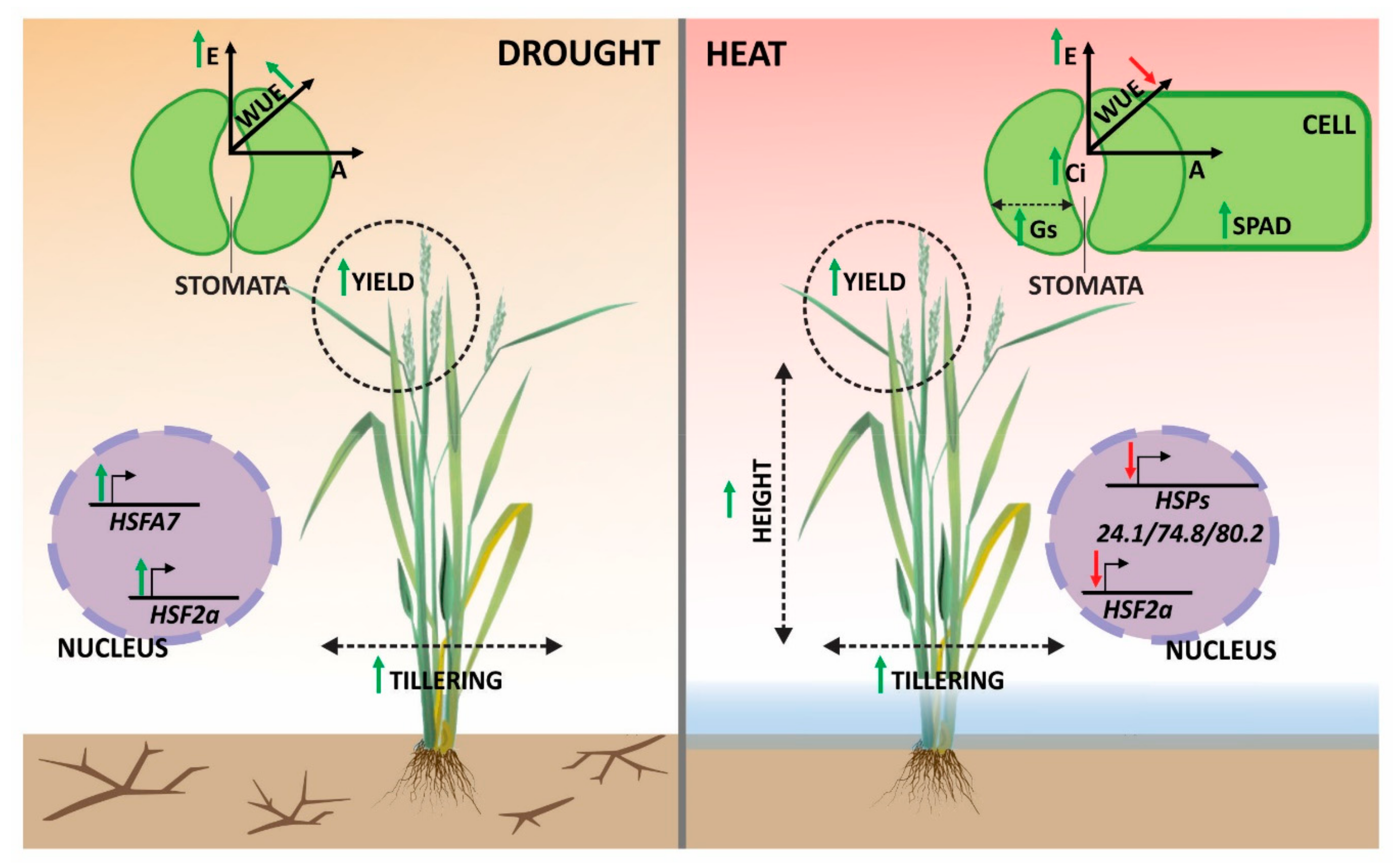
4.4. Modeling Rice and Weedy Rice Responses to Drought and Heat Stresses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The Future of Food and Agriculture: Trends and Challenges; FAO: Rome, Italy, 2017; Volume 4, ISBN 1815-6797. [Google Scholar]
- Menguer, P.K.; Sperotto, R.A.; Ricachenevsky, F.K. A walk on the wild side: Oryza species as source for rice abiotic stress tolerance. Genet. Mol. Biol. 2017, 40, 238–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foley, J.A.; Ramankutty, N.; Brauman, K.A.; Cassidy, E.S.; Gerber, J.S.; Johnston, M.; Mueller, N.D.; O’Connell, C.; Ray, D.K.; West, P.C.; et al. Solutions for a cultivated planet. Nature 2011, 478, 337–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmgren, M.; Edenbrandt, A.K.; Vedel, S.E.; Andersen, M.M.; Landes, X.; Østerberg, J.T.; Falhof, J.; Olsen, L.I.; Christensen, S.B.; Sandøe, P.; et al. Are we ready for back-to-nature crop breeding? Trends Plant Sci. 2015, 20, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Sang, T.; Ge, S. The Puzzle of Rice Domestication. J. Integr. Plant Biol. 2007, 49, 760–768. [Google Scholar] [CrossRef]
- Mueller, N.D.; Gerber, J.S.; Johnston, M.; Ray, D.K.; Ramankutty, N.; Foley, J.A. Closing yield gaps through nutrient and water management. Nat. Cell Biol. 2012, 490, 254–257. [Google Scholar] [CrossRef]
- Xu, J.; Henry, A.; Sreenivasulu, N. Rice yield formation under high day and night temperatures—A prerequisite to ensure future food security. Plant Cell Environ. 2020, 43, 1595–1608. [Google Scholar] [CrossRef]
- Ambavaram, M.M.R.; Basu, S.; Krishnan, A.; Ramegowda, V.; Batlang, U.; Rahman, L.; Baisakh, N.; Pereira, A. Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat. Commun. 2014, 5, 5302. [Google Scholar] [CrossRef]
- Noldin, J.; Chandler, J.M.; McCauley, G.N. Seed longevity of red rice ecotypes buried in soil. Planta Daninha 2006, 24, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Andres, A.; Theisen, G.; Conceno, G.; Galon, G.C.A.L. Weed Resistance to Herbicides in Rice Fields in Southern Brazil. In Herbicides—Current Research and Case Studies in Use; IntechOpen: London, UK, 2013; p. 13. [Google Scholar]
- Goulart, I.C.G.D.R.; Borba, T.C.O.; Menezes, V.G.; Merotto, A. Distribution of Weedy Red Rice (Oryza sativa) Resistant to Imidazolinone Herbicides and its Relationship to Rice Cultivars and Wild Oryza Species. Weed Sci. 2014, 62, 280–293. [Google Scholar] [CrossRef]
- Santos, L.; Pinto, J.J.O.; Piveta, L.B.; Noldin, J.; Galon, L.; Concenço, G. Carryover Effect of Imidazolinone Herbicides for Crops Following Rice. Am. J. Plant Sci. 2014, 5, 1049–1058. [Google Scholar] [CrossRef] [Green Version]
- Merotto, A.; Goulart, I.C.G.D.R.; Nunes, A.L.; Kalsing, A.; Markus, C.; Menezes, V.G.; Wander, A.E. Evolutionary and social consequences of introgression of nontransgenic herbicide resistance from rice to weedy rice in Brazil. Evol. Appl. 2016, 9, 837–846. [Google Scholar] [CrossRef] [PubMed]
- Burgos, N.R.; Singh, V.; Tseng, T.M.; Black, H.; Young, N.D.; Huang, Z.; Hyma, K.E.; Gealy, D.R.; Caicedo, A.L. The Impact of Herbicide-Resistant Rice Technology on Phenotypic Diversity and Population Structure of United States Weedy Rice. Plant Physiol. 2014, 166, 1208–1220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gealy, D.R.; Roma-Burgos, N.; Yeater, K.M.; Jackson, A.K.; Yeater, K.M. Outcrossing Potential between U.S. Blackhull Red Rice and Indica Rice Cultivars. Weed Sci. 2015, 63, 647–657. [Google Scholar] [CrossRef]
- Ziska, L.H.; Gealy, D.R.; Burgos, N.; Caicedo, A.L.; Gressel, J.; Lawton-Rauh, A.L.; Avila, L.A.; Theisen, G.; Norsworthy, J.; Ferrero, A.; et al. Weedy (Red) Rice: An Emerging Constraint to Global Rice Production; Elsevier Ltd.: Amsterdam, The Netherlands, 2015; Volume 129, ISBN 9780128021385. [Google Scholar]
- Kraehmer, H.; Jabran, K.; Mennan, H.; Chauhan, B.S. Global distribution of rice weeds—A review. Crop. Prot. 2016, 80, 73–86. [Google Scholar] [CrossRef]
- Busi, R.; Nghia, K.N.; Chauhan, B.S.; Vidotto, F.; Tabacchi, M.; Powles, S.B. Can herbicide safeners allow selective control of weedy rice infesting rice crops? Pest Manag. Sci. 2017, 73, 71–77. [Google Scholar] [CrossRef]
- Schmidt, F. Crescimento e produção de arroz irrigado de pericarpo colorido em função da aplicação de nitrogênio e potássio. Sci. Agrar. 2017, 18, 34. [Google Scholar] [CrossRef]
- Simontacchi, M.; Egalatro, A.; Artuso, F.E.; Santa-María, G.E. Plant Survival in a Changing Environment: The Role of Nitric Oxide in Plant Responses to Abiotic Stress. Front. Plant Sci. 2015, 6, 977. [Google Scholar] [CrossRef]
- Ehong, Y.; Ezhang, H.; Ehuang, L.; Eli, D.; Song, F. Overexpression of a Stress-Responsive NAC Transcription Factor Gene ONAC022 Improves Drought and Salt Tolerance in Rice. Front. Plant Sci. 2016, 7, 4. [Google Scholar] [CrossRef]
- Divya, K.; Bhatnagar-Mathur, P.; Sharma, K.K.; Reddy, P.S. Heat Shock Proteins (Hsps) Mediated Signalling Pathways during Abiotic Stress Conditions; Elsevier BV: Amsterdam, The Netherlands, 2019; pp. 499–516. [Google Scholar]
- Gilroy, S.; Suzuki, N.; Miller, G.; Choi, W.-G.; Toyota, M.; Devireddy, A.R.; Mittler, R. A tidal wave of signals: Calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 2014, 19, 623–630. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 2018, 162, 2–12. [Google Scholar] [CrossRef] [Green Version]
- IPCC. Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; IPCC: Geneva, Switzerland, 2014; Volume 9781107025, ISBN 9781139177245. [Google Scholar]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic Stress Responses and Microbe-Mediated Mitigation in Plants: The Omics Strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- Lavania, D.; Kumar, R.; Goyal, I.; Rana, S.; Grover, A. Genetic improvement of rice crop under high temperature stress: Bridging plant physiology with molecular biology. Indian J. Plant Physiol. 2016, 21, 391–408. [Google Scholar] [CrossRef]
- Haslbeck, M.; Vierling, E. A First Line of Stress Defense: Small Heat Shock Proteins and Their Function in Protein Homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef] [Green Version]
- Usman, M.G.; Rafii, M.Y.; Ismail, M.R.; Malek, M.A.; Latif, M.A.; Oladosu, Y. Heat Shock Proteins: Functions And Response Against Heat Stress In Plants. Int. J. Sci. Technol. Res. 2014, 3, 204–218. [Google Scholar]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Li, P.-S.; Yu, T.-F.; He, G.-H.; Chen, M.; Zhou, Y.-B.; Chai, S.-C.; Xu, Z.; Ma, Y. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom. 2014, 15, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Aneja, B.; Yadav, N.R.; Kumar, N.; Yadav, R.C. Hsp transcript induction is correlated with physiological changes under drought stress in Indian mustard. Physiol. Mol. Biol. Plants 2015, 21, 305–316. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Hu, G.; Han, B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant Sci. 2009, 176, 583–590. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, B.; Li, J.; Zhang, L.; Wang, Y.; Zheng, H.; Lu, M.; Chen, J. Hsf and Hsp gene families in Populus: Genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genom. 2015, 16, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Nakai, A. New aspects in the vertebrate heat shock factor system: Hsf3 and Hsf4. Cell Stress Chaperones 1999, 4, 86–93. [Google Scholar] [CrossRef]
- Chandel, G.; Dubey, M.; Meena, R.K. Differential expression of heat shock proteins and heat stress transcription factor genes in rice exposed to different levels of heat stress. J. Plant Biochem. Biotechnol. 2013, 22, 277–285. [Google Scholar] [CrossRef]
- Jin, G.-H.; Gho, H.-J.; Jung, K.-H. A systematic view of rice heat shock transcription factor family using phylogenomic analysis. J. Plant Physiol. 2013, 170, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.-L.; Zou, J.; Liu, C.-F.; Zhou, X.-Y.; Zhang, X.; Luo, G.-Y.; Chen, X. Over-expression of OsHsfA7 enhanced salt and drought tolerance in transgenic rice. BMB Rep. 2013, 46, 31–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharf, K.D.; Rose, S.; Zott, W.; Schöffl, F.; Nover, L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990, 9, 4495–4501. [Google Scholar] [CrossRef] [PubMed]
- Nover, L.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.-D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperon 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Guo, J.; Wu, J.; Jingkang, G.; Wang, C.; Luo, L.; Yuan, Y.; Wang, Y.; Wang, J. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 35, 105–118. [Google Scholar] [CrossRef]
- Lin, Y.-X.; Jiang, H.; Chu, Z.-X.; Tang, X.-L.; Zhu, S.; Cheng, B. Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom. 2011, 12, 76. [Google Scholar] [CrossRef] [Green Version]
- Scharf, K.-D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta Bioenerg. 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Liu, A.-L.; Zou, J.; Zhang, X.; Zhou, X.-Y.; Wang, W.-F.; Xiong, X.-Y.; Chen, L.-Y.; Chen, X. Expression Profiles of Class A Rice Heat Shock Transcription Factor Genes Under Abiotic Stresses. J. Plant Biol. 2010, 53, 142–149. [Google Scholar] [CrossRef]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.-D. Crosstalk between Hsp90 and Hsp70 Chaperones and Heat Stress Transcription Factors in Tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [Green Version]
- Barua, D.; Heckathorn, S.A.; Coleman, J.S. Variation in Heat-shock Proteins and Photosynthetic Thermotolerance among Natural Populations of Chenopodium album L. from Contrasting Thermal Environments: Implications for Plant Responses to Global Warming. J. Integr. Plant Biol. 2008, 50, 1440–1451. [Google Scholar] [CrossRef] [PubMed]
- Zatsepina, O.G.; Velikodvorskaia, V.V.; Molodtsov, V.B.; Garbuz, D.; Lerman, D.N.; Bettencourt, B.R.; Feder, M.E.; Evgenev, M.B. A Drosophila melanogaster strain from sub-equatorial Africa has exceptional thermotolerance but decreased Hsp70 expression. J. Exp. Biol. 2001, 204, 1869–1881. [Google Scholar] [PubMed]
- Nakano, K.; Iwama, G.K. The 70-kDa heat shock protein response in two intertidal sculpins, Oligocottus maculosus and O. snyderi: Relationship of hsp70 and thermal tolerance. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2002, 133, 79–94. [Google Scholar] [CrossRef]
- Wickert, E.; Schiocchet, M.A.; Noldin, J.A.; Raimondi, J.V.; De Andrade, A.; Scheuermann, K.K.; Marschalek, R.; Martins, G.N.; Hickel, E.; Eberhardt, D.S.; et al. Exploring Variability: New Brazilian Varieties SCS119 Rubi and SCS120 Onix for the Specialty Rices Market. Open J. Genet. 2014, 4, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Diniz, F.D.A.; Ramos, A.M.; Rebello, E.R.G. Brazilian climate normals for 1981–2010. Pesqui. Agropecu. Bras. 2018, 53, 131–143. [Google Scholar] [CrossRef]
- Bernardo, S.; Soares, A.; Mantovani, E. Manual de Irrigação; Livraria UFV: Viçosa, MG, Brazil, 2009; p. 625. [Google Scholar]
- SOSBAI. Arroz Irrigado: Recomendações Técnicas da Pesquisa para o Sul do Brasil. In Sociedade Sul-Brasileira de Arroz Irrigado; SOSBAI: Cachoeirinha, RS, Brazil, 2018; p. 205. [Google Scholar]
- Ramegowda, V.; Basu, S.; Krishnan, A.; Pereira, A. Rice Growth under Drought Kinase Is Required for Drought Tolerance and Grain Yield under Normal and Drought Stress Conditions. Plant Physiol. 2014, 166, 1634–1645. [Google Scholar] [CrossRef] [Green Version]
- De Freitas, G.P.M.; Basu, S.; Ramegowda, V.; Braga, E.B.; Pereira, A. Comparative analysis of gene expression in response to cold stress in diverse rice genotypes. Biochem. Biophys. Res. Commun. 2016, 471, 253–259. [Google Scholar] [CrossRef]
- Hatfield, J.; Dold, C. Water-Use Efficiency: Advances and Challenges in a Changing Climate. Front. Plant Sci. 2019, 10, 103. [Google Scholar] [CrossRef] [Green Version]
- Pocojeski, E.; Kaefer, S.; Moro, V.J.; Griebeler, G.; Da Silva, L.S. Uso do clorofilômetro no monitoramento nutricional de arroz irrigado com vistas ao manejo da adubação nitrogenada. Rev. Ceres 2015, 62, 310–318. [Google Scholar] [CrossRef] [Green Version]
- Cruz, C.D. GENES—A software package for analysis in experimental statistics and quantitative genetics. Acta Sci. Agron. 2013, 35, 271–276. [Google Scholar] [CrossRef]
- Demsar, J.; Curk, T.; Erjavec, A.; Gorup, C.; Hocevar, T.; Milutinovic, M.; Mozina, M.; Polajnar, M.; Toplak, M.; Stari, A.; et al. Orange: Data Mining Toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Zou, J.; Liu, A.; Chen, X.; Zhou, X.; Gao, G.; Wang, W.; Zhang, X. Expression analysis of nine rice heat shock protein genes under abiotic stresses and ABA treatment. J. Plant Physiol. 2009, 166, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Wang, F.; Geng, X.; Zhang, L.; Yao, Y.; Ni, Z.; Peng, H.; Sun, Q. Overexpression of heat stress-responsive TaMBF1c, a wheat (Triticum aestivum L.) Multiprotein Bridging Factor, confers heat tolerance in both yeast and rice. Plant Mol. Biol. 2014, 87, 31–45. [Google Scholar] [CrossRef]
- Ye, S.; Yu, S.; Shu, L.; Wu, J.; Wu, A.; Luo, L. Expression profile analysis of 9 heat shock protein genes throughout the life cycle and under abiotic stress in rice. Chin. Sci. Bull. 2012, 57, 336–343. [Google Scholar] [CrossRef] [Green Version]
- Benneman, D.; Nohato, A.; Vargas, L.; De Avila, L.A.; Agostinetto, D. Identification and Validation of Reference Genes for the Normalization in Real-Time Rt-Qpcr on Rice and Red Rice in Competition, under Different Nitrogen Doses. Planta Daninha 2017, 35. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Saeed, A.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A Free, Open-Source System for Microarray Data Management and Analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [Green Version]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2012, 64, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Qazi, H.A.; Jan, N.; Ramazan, S.; John, R. Protein Modification in Plants in Response to Abiotic Stress; Elsevier BV: Amsterdam, The Netherlands, 2019; pp. 171–201. [Google Scholar]
- Lamaoui, M.; Jemo, M.; Datla, R.; Bekkaoui, F. Heat and Drought Stresses in Crops and Approaches for Their Mitigation. Front. Chem. 2018, 6, 26. [Google Scholar] [CrossRef]
- Islam, R.; Feng, B.; Chen, T.; Tao, L.X.; Fu, G. Role of Abscisic Acid in Thermal Acclimation of Plants. J. Plant Biol. 2018, 61, 255–264. [Google Scholar] [CrossRef]
- Kaur, G.; Asthir, B. Molecular responses to drought stress in plants. Biol. Plant. 2017, 61, 201–209. [Google Scholar] [CrossRef]
- Manickavelu, A.; Nadarajan, N.; Ganesh, S.K.; Gnanamalar, R.P.; Babu, R.C. Drought tolerance in rice: Morphological and molecular genetic consideration. Plant Growth Regul. 2006, 50, 121–138. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.; Roychowdhury, R.; Fujita, M. Physiological, Biochemical, and Molecular Mechanisms of Heat Stress Tolerance in Plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Review article Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.-L.; Chen, J.-H.; He, N.-Y.; Guo, F.-Q. Metabolic Reprogramming in Chloroplasts under Heat Stress in Plants. Int. J. Mol. Sci. 2018, 19, 849. [Google Scholar] [CrossRef] [Green Version]
- Killi, D.; Bussotti, F.; Raschi, A.; Haworth, M. Adaptation to high temperature mitigates the impact of water deficit during combined heat and drought stress in C3 sunflower and C4 maize varieties with contrasting drought tolerance. Physiol. Plant. 2016, 159, 130–147. [Google Scholar] [CrossRef]
- Perdomo, J.A.; Capó-Bauçà, S.; Carmo-Silva, A.E.; Galmés, J. Rubisco and Rubisco Activase Play an Important Role in the Biochemical Limitations of Photosynthesis in Rice, Wheat, and Maize under High Temperature and Water Deficit. Front. Plant Sci. 2017, 8, 490. [Google Scholar] [CrossRef] [Green Version]
- Ashraf, M.; Harris, P.J.C. Photosynthesis under stressful environments: An overview. Photosynthetica 2013, 51, 163–190. [Google Scholar] [CrossRef]
- Kumagai, E.; Araki, A.; Kubota, F. Correlation of Chlorophyll Meter Readings with Gas exchange and Chlorophyll Fluorescence in Flag Leaves of Rice (Oryza sativa L.) Plants. Plant Prod. Sci. 2009, 12, 50–53. [Google Scholar] [CrossRef] [Green Version]
- Nadeem, M.; Li, J.; Wang, M.; Shah, L.; Lu, S.; Wang, X.; Ma, C. Unraveling Field Crops Sensitivity to Heat Stress: Mechanisms, Approaches, and Future Prospects. Agronomy 2018, 8, 128. [Google Scholar] [CrossRef] [Green Version]
- Medrano, H.; Tomás, M.; Martorell, S.; Flexas, J.; Hernández, E.; Rosselló, J.; Pou, A.; Escalona, J.-M.; Bota, J. From leaf to whole-plant water use efficiency (WUE) in complex canopies: Limitations of leaf WUE as a selection target. Crop. J. 2015, 3, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Haq, S.U.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.-X.; Zhang, H.-X.; Wei, A.-M.; Ali, M. Heat Shock Proteins: Dynamic Biomolecules to Counter Plant Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, S.; Huang, J.; Sheehy, J.E.; Laza, R.C.; Visperas, R.M.; Zhong, X.; Centeno, G.S.; Khush, G.S.; Cassman, K.G. Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. USA 2004, 101, 9971–9975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Lawas, L.M.F.; Malo, R.; Glaubitz, U.; Erban, A.; Mauleon, R.; Heuer, S.; Zuther, E.; Kopka, J.; Hincha, D.K.; et al. Metabolic and transcriptomic signatures of rice floral organs reveal sugar starvation as a factor in reproductive failure under heat and drought stress. Plant Cell Environ. 2015, 38, 2171–2192. [Google Scholar] [CrossRef]
- Monson, R.K.; Rawsthorne, S. CO2 Assimilation in C3–C4 Intermediate Plants. In Photosynthesis: Physiology and Metabolism; Leegood, R.C., Sharkey, T.D., von Caemmerer, S., Eds.; Kluwer Academic Publishers: Norwich, UK, 2000; pp. 533–550. [Google Scholar]
- Karki, S.; Rizal, G.; Quick, W.P. Improvement of photosynthesis in rice (Oryza sativa L.) by inserting the C4 pathway. Rice 2013, 6, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, F.J.A.; Portes, T.D.A.; Stacciarini-Seraphin, E. Procurando um arroz c4 mediante exame anatômico foliar. Rev. Bras. Fisiol. Veg. 2000, 12, 246–254. [Google Scholar] [CrossRef] [Green Version]
- Priya, M.; Dhanker, O.P.; Siddique, K.H.M.; Hanumantharao, B.; Nair, R.M.; Pandey, S.; Singh, S.; Varshney, R.K.; Prasad, P.V.V.; Nayyar, H. Drought and heat stress-related proteins: An update about their functional relevance in imparting stress tolerance in agricultural crops. Theor. Appl. Genet. 2019, 132, 1607–1638. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.-H.; Ma, X.; Luo, D.-X.; Gong, Z.-H.; Lu, M.-H. The Plant Heat Stress Transcription Factors (HSFs): Structure, Regulation, and Function in Response to Abiotic Stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, L.; Cao, W.; Wang, J.; Yu, J.; Yang, Z.; Huang, B. Characterization and Functional Analysis of FaHsfC1b from Festuca arundinacea Conferring Heat Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2702. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yu, S.; Zuo, K.; Luo, L.; Tang, K. Identification of Gene Modules Associated with Drought Response in Rice by Network-Based Analysis. PLoS ONE 2012, 7, e33748. [Google Scholar] [CrossRef]
- Yang, P.S.Y. Transcriptional Network Involved in Drought Response and Adaptation in Cereals. In Abiotic and Biotic Stress in Plants; InTech: London, UK, 2016; pp. 3–30. [Google Scholar] [CrossRef] [Green Version]
- Nagaraju, M.; Reddy, P.S.; Kumar, S.A.; Srivastava, R.K.; Kishor, P.B.K.; Rao, D.M. Genome-wide Scanning and Characterization of Sorghum bicolor L. Heat Shock Transcription Factors. Curr. Genom. 2015, 16, 279–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, R.; Zhu, W.; Song, X.; Lin, X.; Cai, J.; Wang, M.; Yang, Q. Genome-Wide Identification and Function Analyses of Heat Shock Transcription Factors in Potato. Front. Plant Sci. 2016, 7, 490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.-C.; Niu, C.-Y.; Yang, C.-R.; Jinn, T.-L. The heat-stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 2016, 172, 1182–1199. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Xu, L.; Wang, Y.; Cheng, W.; Luo, X.; Xie, Y.; Fan, L.; Liu, L. Genome-wide characterization and evolutionary analysis of heat shock transcription factors (HSFs) to reveal their potential role under abiotic stresses in radish (Raphanus sativus L.). BMC Genom. 2019, 20, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Chai, F.; Wang, Y.; Jiang, J.; Duan, W.; Wang, Y.; Wang, F.; Li, S.; Wang, L. Genome-wide Identification and Classification of HSF Family in Grape, and Their Transcriptional Analysis under Heat Acclimation and Heat Stress. Hortic. Plant J. 2018, 4, 133–143. [Google Scholar] [CrossRef]
- Zha, Q.; Xi, X.; He, Y.; Jiang, A. Transcriptomic analysis of the leaves of two grapevine cultivars under high-temperature stress. Sci. Hortic. 2020, 265, 109265. [Google Scholar] [CrossRef]
- Krishna, P.; Gloor, G. The Hsp90 family of proteins in arabidopsis thaliana. Cell Stress Chaperones 2001, 6, 238–246. [Google Scholar] [CrossRef]
- Aviezer-Hagai, K.; Skovorodnikova, J.; Galigniana, M.; Farchi-Pisanty, O.; Maayan, E.; Bocovza, S.; Efrat, Y.; Von Koskull-Döring, P.; Ohad, N.; Breiman, A. Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol. Biol. 2007, 63, 237–255. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.; Chen, N.; Xiong, Z.; Wolfe, D.; Zou, J. Response of rice production to elevated [CO2] and its interaction with rising temperature or nitrogen supply: A meta-analysis. Clim. Chang. 2015, 130, 529–543. [Google Scholar] [CrossRef]
- Wegele, H.; Müller, L.; Buchner, J. Hsp70 and Hsp90—A Relay Team for Protein Folding Published. Rev. Physiol. Biochem. Pharmacol. 2004, 151, 1–44. [Google Scholar]
- Meiri, D.; Breiman, A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009, 59, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Mani, N.; Ramakrishna, K.; Suguna, K. Characterization of rice small heat shock proteins targeted to different cellular organelles. Cell Stress Chaperon 2015, 20, 451–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Quraan, N.A.; Locy, R.D.; Singh, N.K. Expression of calmodulin genes in wild type and calmodulin mutants of Arabidopsis thaliana under heat stress. Plant Physiol. Biochem. 2010, 48, 697–702. [Google Scholar] [CrossRef] [PubMed]
| Cultivar/Biotype | City/State | Brazil Region | GPS Coordinates | |
|---|---|---|---|---|
| Latitude | Longitude | |||
| IRGA 424 | Pelotas, RS | South | 31°46′19″ S | 52°20′33″ W |
| SCS119 Rubi | Itajaí, SC | South | 26°54′28″ S | 48°39′43″ W |
| WR_SC | Gaspar, SC | South | 26°54′41″ S | 48°50′60″ W |
| WR_RS | Dom Pedrito, RS | South | 31°02′07″ S | 54°52′02″ W |
| WR_PB | São José do Rio do Peixe, PB | Northeastern | 6°44′11″ S | 38°26′39″ W |
| WR_RN | Apodi, RN | Northeastern | 5°44′55″ S | 37°46′27″ W |
| WR_RR | Bonfim, RR | North | 3°19′56″ N | 59°54′18″ W |
| Gene | Family/Name | Oligonucleotide Sequences (5′→3′) | References |
|---|---|---|---|
| OsHSP80.2 | HSP90 | F: CGACGACGAGCAGTATGT | [59] |
| R: CCAGATGTTCCTCCCAGT | |||
| OsHSP74.8 | HSP90 | F: CGAGCAGTTCGAGTACCAGG | [60] |
| R: TCAGCCATAGCTTCCCATAC | |||
| OsHSP24.15 | sHSP | F: GATCAAGGCGGAGATGAAGAAC | [61] |
| R: ACTCGACGTTGACCTGGAAGA | |||
| OsHsfA7 | HSF | F: TTCGCCAGCTCAACACCTA | [38] |
| R: TCCATCAGCCGTTGTCTT | |||
| OsHsfA2a | HSF | F: TTCGTAGGGTGACGTAATCG | [44] |
| R: TCGAAGCCACCGTCCTAG | |||
| UBC-E2 | Ubiquitin-conjugating enzyme E2 | F: CCGTTTGTAGAGCCATAATTGCA | [62] |
| R: AGGTTGCCTGAGTCACAGTTAAGTG | |||
| UBQ5 | Ubiquitin 5 | F: ACCACTTCGACCGCCACTACT | [62] |
| R: ACGCCTAAGCCTGCTGGTT | |||
| ACT11 | Actin 11 | F: CAGCCACACTGTCCCCATCTA | [62] |
| R: AGCAAGGTCGAGACGAAGGA |
| Genotype | A (µmol CO2 m−2 s−1) | Gs (mol H2O m−2 s−1) | Ci (µmol mol−1) | E (mmol H2O m−2 s−1) | WUE (A/E Ratio) | SPAD |
|---|---|---|---|---|---|---|
| IRGA 424 | 18.87 (2.06) B1 | 0.19 (0.03) A | 272.10 (28.16) A | 3.06 (0.62) A | 6.35 (1.70) A | 40.68 (0.64) A |
| SCS119 Rubi | 19.75 (0.35) AB | 0.16 (0.01) A | 248.57 (12.68) A | 2.58 (0.27) A | 7.69 (0.67) A | 43.52 (1.72) A |
| WR_RS | 20.68 (1.45) AB | 0.19 (0.04) A | 259.44 (39.39) A | 3.15 (0.79) A | 6.85 (2.26) A | 41.49 (2.52) A |
| WR_SC | 20.88 (0.08) AB | 0.20 (0.06) A | 266.60 (38.43) A | 3.39 (1.02) A | 6.51 (2.27) A | 40.91 (0.58) A |
| WR_PB | 22.75 (1.02) AB | 0.24 (0.02) A | 276.77 (11.62) A | 3.94 (0.35) A | 5.80 (0.68) A | 41.80 (2.95) A |
| WR_RN | 24.16 (2.13) AB | 0.22 (0.02) A | 260.60 (19.00) A | 3.65 (0.43) A | 6.68 (1.09) A | 42.96 (1.62) A |
| WR_RR | 26.65 (3.38) A | 0.21 (0.03) A | 244.30 (28.71) A | 3.57 (0.62) A | 7.58 (1.64) A | 39.09 (0.66) A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piveta, L.B.; Roma-Burgos, N.; Noldin, J.A.; Viana, V.E.; Oliveira, C.d.; Lamego, F.P.; Avila, L.A.d. Molecular and Physiological Responses of Rice and Weedy Rice to Heat and Drought Stress. Agriculture 2021, 11, 9. https://doi.org/10.3390/agriculture11010009
Piveta LB, Roma-Burgos N, Noldin JA, Viana VE, Oliveira Cd, Lamego FP, Avila LAd. Molecular and Physiological Responses of Rice and Weedy Rice to Heat and Drought Stress. Agriculture. 2021; 11(1):9. https://doi.org/10.3390/agriculture11010009
Chicago/Turabian StylePiveta, Leonard Bonilha, Nilda Roma-Burgos, José Alberto Noldin, Vívian Ebeling Viana, Claudia de Oliveira, Fabiane Pinto Lamego, and Luis Antonio de Avila. 2021. "Molecular and Physiological Responses of Rice and Weedy Rice to Heat and Drought Stress" Agriculture 11, no. 1: 9. https://doi.org/10.3390/agriculture11010009






