Mathematical Modeling Using Gaussian Functions and Chaotic Attractors: A Hybrid Approach for Realistic Representation of the Intrinsic Dynamics of Heartbeats
Abstract
1. Introduction
Related Works on Methods for the Generation of ECG Data
2. Materials and Methods
- A—Amplitude, determining the peak height of the wave;
- μ—Central position in time (defines when the wave appears in the cycle);
- σ—Standard deviation (defines the width and smoothness of the wave);
- t—Time variable.
- P-wave—Models the initial depolarization of the atria. It is smooth and rounded and can typically be well-described by a single Gaussian function.
- QRS complex—This is a fast and sharp transition, requiring three separate Gaussian functions to model its shape:
- Q-wave—A short, small negative deflection;
- R-wave—A tall and sharp peak, and the most prominent part of the signal;
- S-wave—A brief negative drop following the R-wave;
- T wave—It can be well-described with two Gaussian functions due to its wider and more rounded shape, which requires greater flexibility for accurate modeling.
- μ—Temporal location, defining the position of the wave along the time axis;
- —Width of the first half-Gaussian function, defining the spread of the initial part of the wave;
- —Width of the second half-Gaussian function, defining the spread of the latter part of the wave;
- —Plateau duration, determining the length of the horizontal segment between the two half-Gaussian functions;
- A—Amplitude, defining the wave height.
- -
- The QRS complex in ECG exhibits a sharp peak but is not perfectly symmetrical; a single Gaussian function would result in an idealized, less realistic shape;
- -
- GMF allows for a more realistic modeling of the QRS complex, where two Gaussian components with different parameters can better represent the steep ascending phase and the less steep descending phase of the signal;
- -
- This method reduces reconstruction errors when generating synthetic ECG signals in real-time.
2.1. Numerical Integration: Runge–Kutta Methods
2.2. Attractor–Timing Coupling: A Dimensionless Derivation
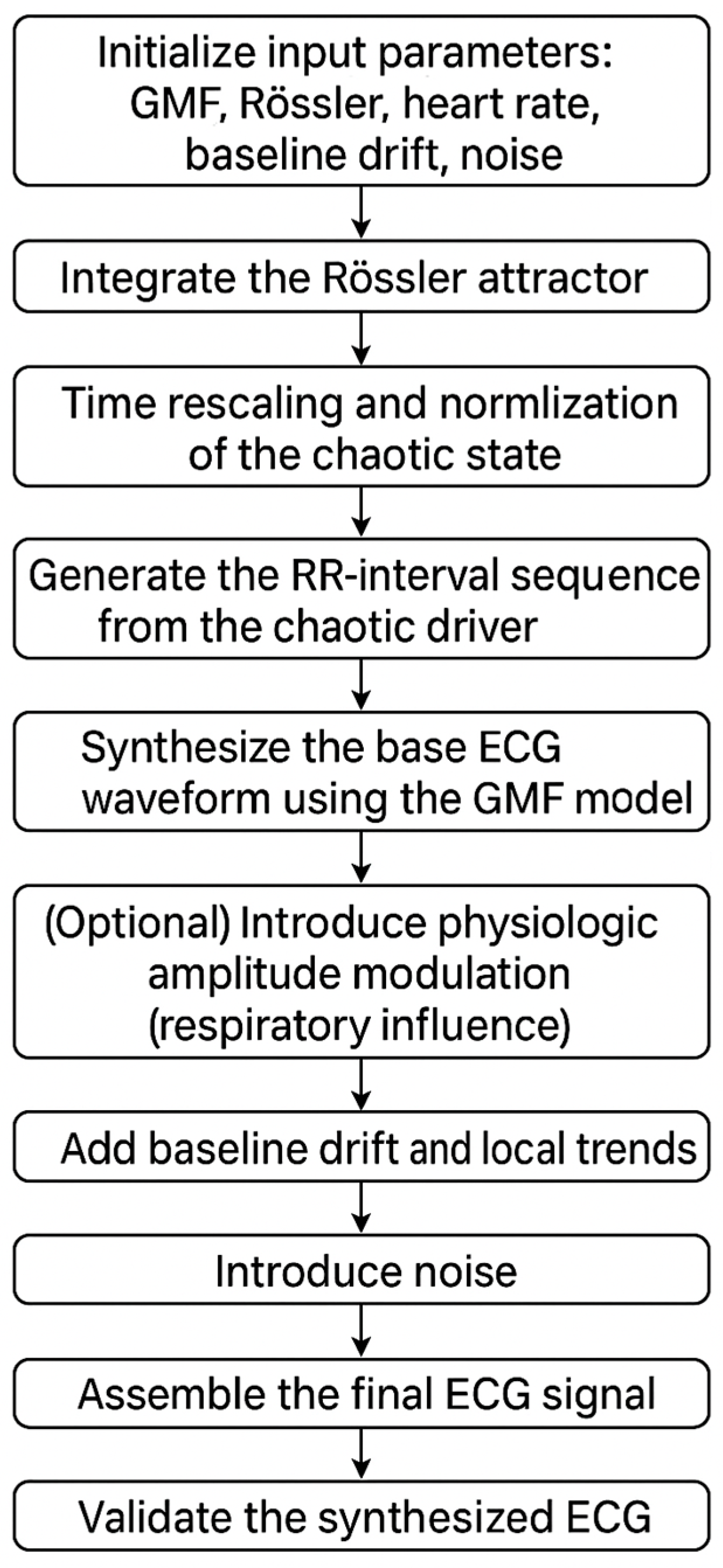
- 1.
- Initialize input parameters:Morphological (GMF) parameters for each wave :where is amplitude, , is temporal offset, control asymmetric rise/decay, and is the plateau duration.Rössler attractor parameters:where a, b, c are system parameters,are initial conditions, is the time-scaling factor, and α controls the strength of HRV modulation.Heart rate and sampling parameters:mean RR interval umber of heartbeats N, and sampling frequency .Baseline drift and noise parameters:, for drift,, for high- and low-frequency components, and the variance of additive Gaussian noise.(Optional) Respiratory amplitude modulation parameters: .
- 2.
- Integrate the Rössler attractor.Solve the Rössler system (4) numerically using a Runge–Kutta scheme with time step Δt generating the trajectory over the desired simulation horizon.
- 3.
- Time rescaling and normalization of the chaotic state—with Equations (8) and (9).
- 4.
- Generate the RR-interval sequence from the chaotic driver—with Equations (10) and (11).
- 5.
- Synthesize the base ECG waveform using the GMF model—with Equation (13).
- 6.
- (Optional) Introduce physiologic amplitude modulation (respiratory influence)To emulate slow, non-chaotic amplitude variability due to respiration, modulate the GMF amplitudes by a low-frequency sinusoid:.The time-dependent amplitudes replace the constant it Step 5 (Equation (12)).
- 7.
- Add Baseline Drift and Local Trends:
- -
- Introduce slow baseline drift using the follows:where is random phase, is a Gaussian step, and controls the strength of the unstable drift.
- -
- Choose ∈ [0.01, 0.3] mV and ∈ [0.01, 0.1] Hz.
- -
- Add a linear local trend if simulating long-term ECG signals.
- 8.
- Introduce Noise:Generate and add different types of noise:High-frequency noise (electrical interference, EMG artifacts) is as follows:where is white Gaussian noise passed through an HPF (high-pass filter) (cutoff: 20–30 Hz) to mimic broadband EMG fluctuations.Low-frequency noise (respiratory influences, baseline wander) is as follows:where is white Gaussian noise passed through an LPF (low-pass filter) to reflect slow, irregular respiratory oscillations.Additive stochastic noise ε(t), typically zero-mean Gaussian.The total noise term is as follows:
- 9.
- Assemble the Final ECG Signal.Combine all components:Normalize and scale the signal to fit standard ECG ranges (−1.5 to 2.5 mV).
- 10.
- Validate the Synthesized ECGHRV analysis: Compute time-domain and frequency-domain metrics.Spectral analysis: Apply STFT and CWT to confirm realistic frequency components.
- 11.
- Save and Export the Synthetic ECG for Further Analysis.
- Parameter Justification and Sensitivity Considerations
- Boundedness and Stability Analysis
- 1.
- Dimensionless chaotic input (timing only).The Rössler state is normalized (Equation (9)) and used solely to modulate RR intervals (see Equations (8)–(14)), and hence it does not scale the ECG amplitude. This removes any direct route for unbounded growth via chaos.
- 2.
- Positive and bounded RR intervals.RR intervals are generated by the exponential map extended with a stochastic component (Equation (11)), where is zero main physiological noise.This is due to the following: for all is constrained such that ; we have ensuring strictly increasing beat times in Equation (12). Thus, the temporal dynamics of the ECG remain well-defined.Additionally, the model uses ∣α∣ ≪ and clipping (e.g., ), which yields ∈ [, ] with > 0, so beat times are well-defined, and the number of overlapping beats around any t is finite.This guarantees both positivity and boundedness of the inter-beat timing.
- 3.
- Bounded morphology.
- 4.
- Controlled overlap of successive beats.Let W be the effective support of a beat (plateau plus 5 max ()). Choosing } (rest and fatigue) implies no inter-beat overlap ( = 1). For stress, where may approach W, we upper-bound the local overlap by (empirically valid for our parameter sets). Thus, at any time, we have the following:
- 5.
- Bounded drift and noise.
- 6.
- Global amplitude bound.By the triangle inequality, we have the following:Hence, imposing the following:guarantees [−2, 2] mV.
2.3. Spectral Analysis
- Short-Time Fourier Transform (STFT)
- Power Spectral Density (PSD)
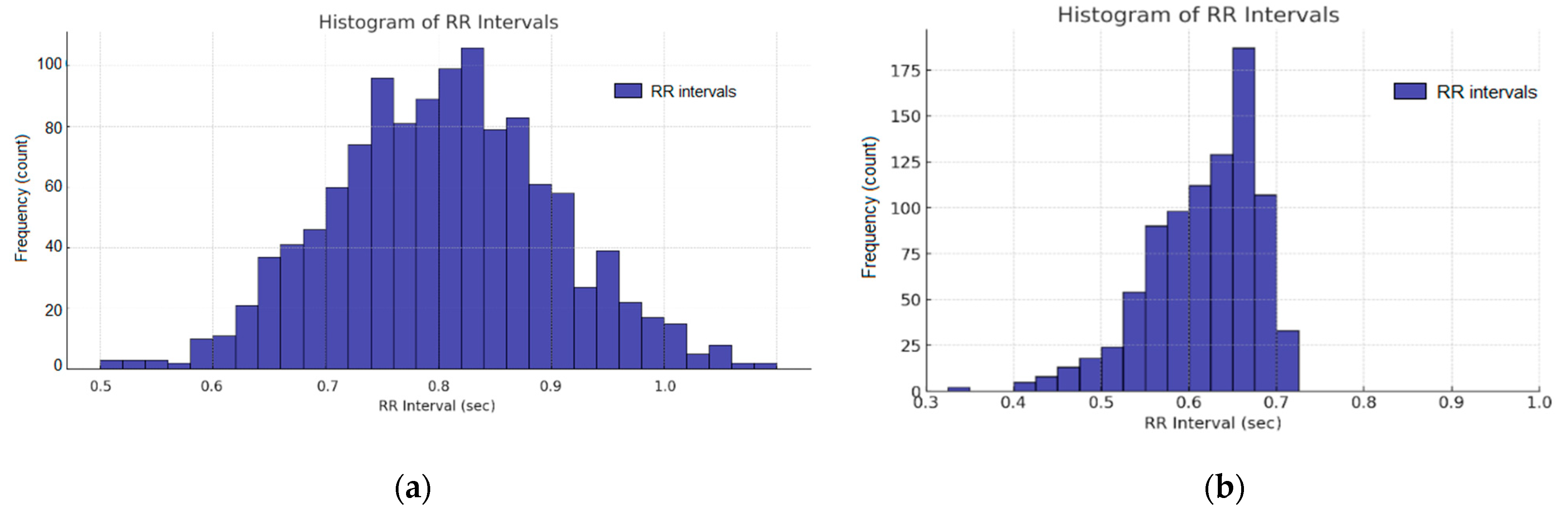
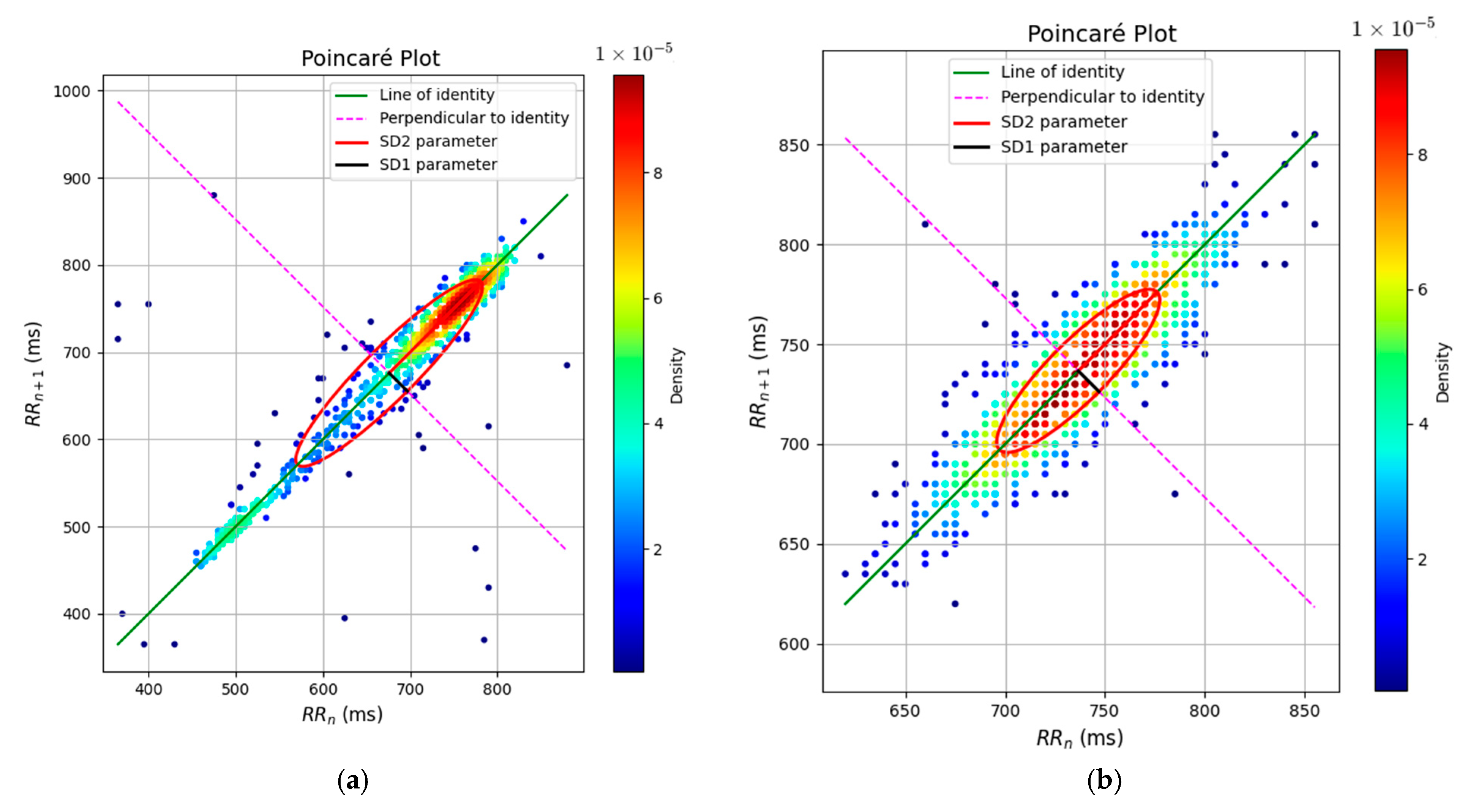
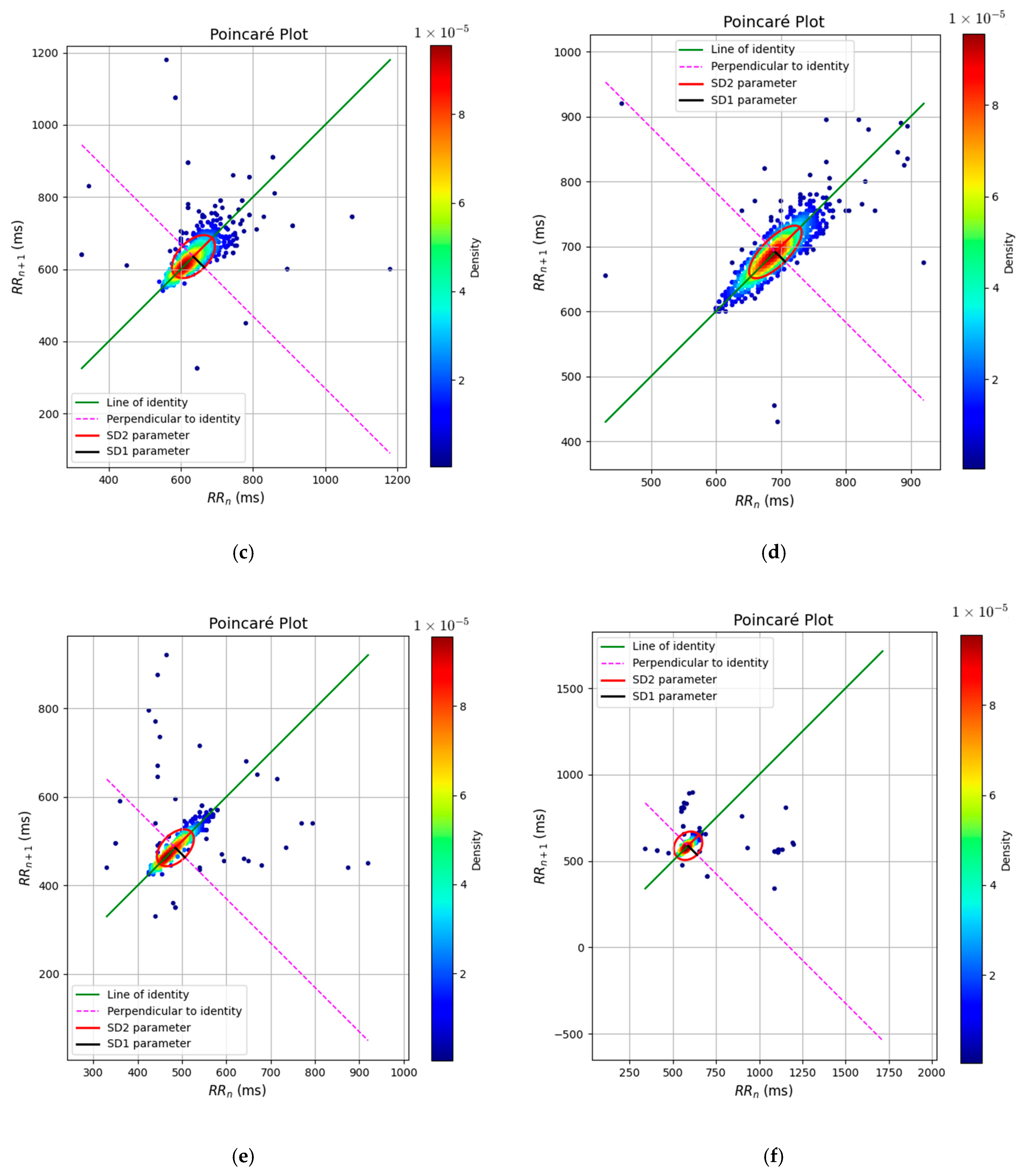
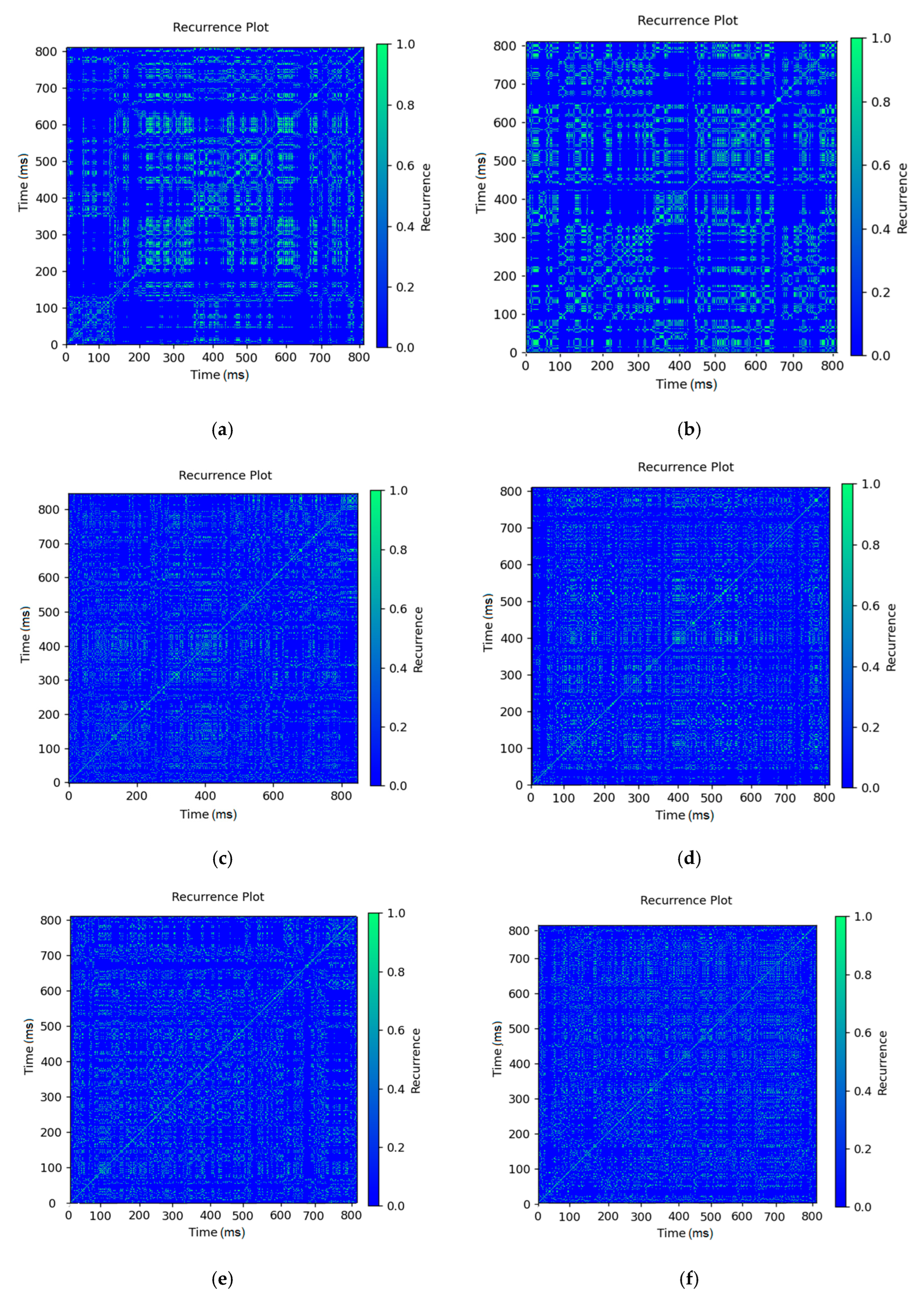
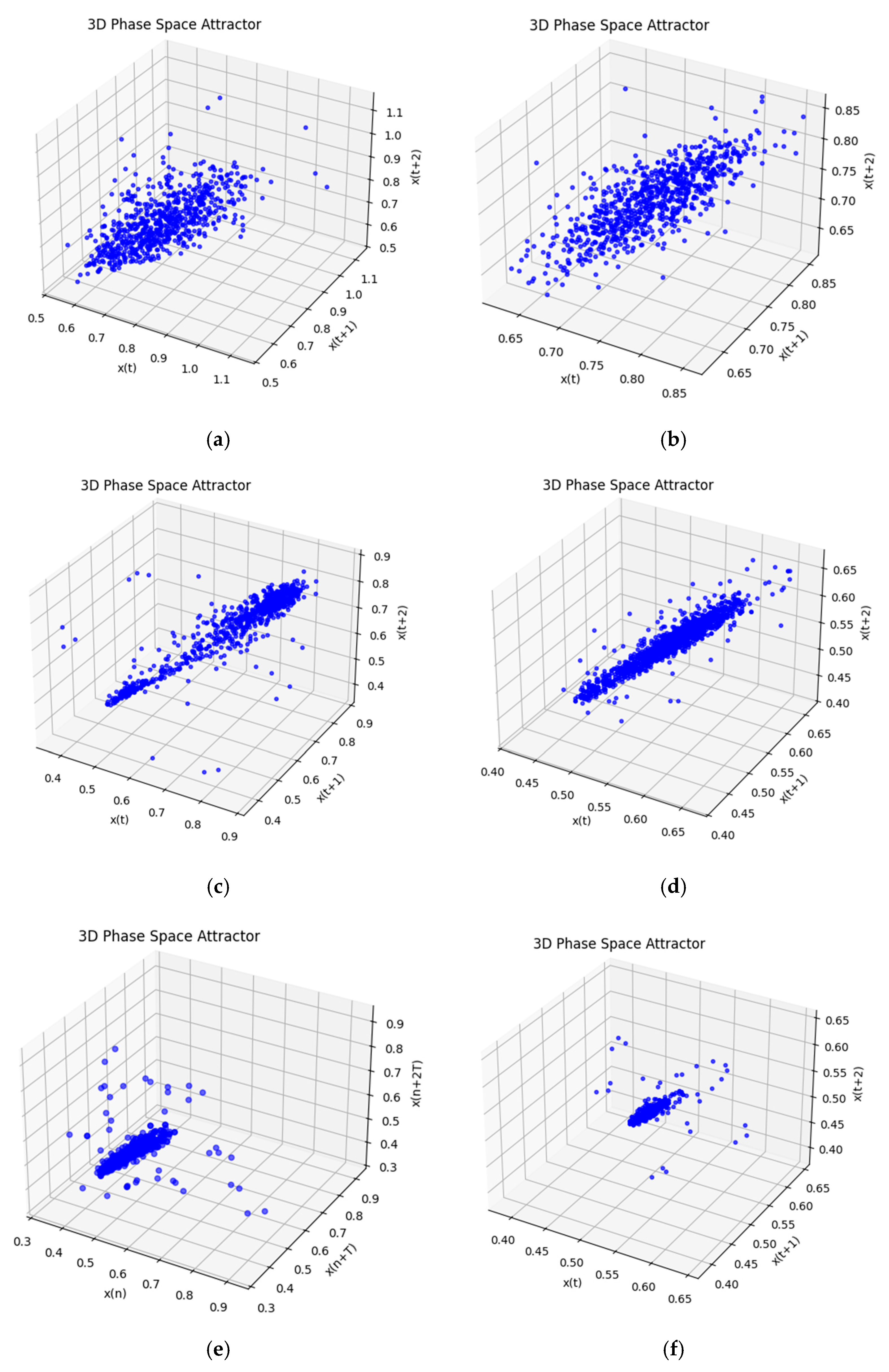
3. Results
- Computational Complexity Analysis
- (1)
- Numerical integration of the Rössler attractor using the fourth-order Runge–Kutta (RK4) method;
- (2)
- Beat-wise synthesis of the ECG morphology using Gaussian mesa functions (GMFs).
4. Discussion
- Sampling frequency (fs): 500 Hz.
- Recording duration: 5 min (300 s).
- Average heart rate (HRmean): 60 bpm (1 Hz).
- Gaussian noise level: 0.03.
- Low-frequency modulation: 0.05 (for resting state).
- Baseline/Drift: amplitude 0.11, and frequency 0.04 Hz (simulates slow variations).
- McSharry and Zeeman: Ordinary differential equation parameters according to the original models, no change, and only amplitude scaling for benchmarking.
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Galli, A.; Giorgi, G.; Narduzzi, C. Standardized Gaussian Dictionary for ECG Analysis: A Metrological Approach. IEEE Open J. Instrum. Meas. 2022, 1, 4000209. [Google Scholar] [CrossRef]
- Sameni, R.; Shamsollahi, M.B.; Jutten, C.; Clifford, G.D. A Nonlinear Bayesian Filtering Framework for ECG De-noising. IEEE Trans. Biomed. Eng. 2007, 54, 2172–2185. [Google Scholar] [CrossRef]
- Fadhel, A.A.; Hasan, H.M. Enhancing ECG Signal Classification Accuracy through Gaussian Modeling Method. Trait. Signal 2023, 40, 1425–1434. [Google Scholar] [CrossRef]
- Mishra, A.; Bhusnur, S. Recapitulation of Synthetic ECG Signal Generation Methods and Analysis. Int. J. Signal Process. Syst. 2022, 10, 14–17. [Google Scholar] [CrossRef]
- Maris, D.T.; Goussis, D.A. The “Hidden” Dynamics of the Rössler Attractor. Phys. D Nonlinear Phenom. 2015, 295–296, 66–90. [Google Scholar] [CrossRef]
- Awal, M.A.; Mostafa, S.S.; Ahmad, M.; Alahe, M.A.; Rashid, M.A.; Kouzani, A.Z.; Mahmud, M.A.P. Design and Optimization of ECG Modeling for Generating Different Cardiac Dysrhythmias. Sensors 2021, 21, 1638. [Google Scholar] [CrossRef] [PubMed]
- Parvaneh, S.; Pashna, M. Electrocardiogram Synthesis Using a Gaussian Combination Model (GCM). In Proceedings of the Computers in Cardiology, Durham, NC, USA, 30 September–3 October 2007; pp. 621–624. [Google Scholar]
- Billah, M.S.; Mahmud, T.B.; Snigdha, F.S.; Arafat, M.A. A Novel Method to Model ECG Beats Using Gaussian Functions. In Proceedings of the 4th International Conference on Biomedical Engineering and Informatics (BMEI), Shanghai, China, 15–17 October 2011; pp. 612–616. [Google Scholar]
- Kundu, P.; Gupta, R. Electrocardiogram Synthesis Using Gaussian and Fourier Models. In Proceedings of the IEEE International Conference on Research in Computational Intelligence and Communication Networks (ICRCICN), Kolkata, India, 20–22 November 2015; pp. 312–317. [Google Scholar]
- Dubois, R.; Roussel, P.; Vaglio, M.; Extramiana, F.; Badilini, F.; Maison-Blanche, P.; Dreyfus, G. Efficient Modeling of ECG Waves for Morphology Tracking. In Proceedings of the 36th Annual Computers in Cardiology Conference (CinC), Park City, UT, USA, 13–16 September 2009; pp. 313–316. [Google Scholar]
- Vulaj, Z.; Draganic, A.; Brajovic, M.; Orovic, I. A Tool for ECG Signal Analysis Using Standard and Optimized Hermite Transform. arXiv 2017, arXiv:1703.00446. [Google Scholar] [CrossRef]
- Ray, S.; Chouhan, V. Electrocardiogram Reconstruction Based on Hermite Interpolating Polynomial with Chebyshev Nodes. Int. J. Electr. Comput. Eng. Syst. 2022, 36, 837–845. [Google Scholar] [CrossRef]
- Kubicek, J.; Penhaker, M.; Kahankova, R. Design of a Synthetic ECG Signal Based on the Fourier Series. In Proceedings of the International Conference on Advances in Computing, Communications and Informatics (ICACCI), Delhi, India, 24–27 September 2014; pp. 1881–1885. [Google Scholar]
- Halawani, S.M.; Kari, S.; Bidewi, I.A.; Ahmad, A.R. ECG Simulation Using Fourier Series: From Personal Computers to Mobile Devices. Int. J. Recent Innov. Trends Comput. Commun. 2014, 2, 1803–1811. [Google Scholar]
- Roonizi, E.K.; Sameni, R. Morphological Modeling of Cardiac Signals Based on Signal Decomposition. Comput. Biol. Med. 2013, 43, 1453–1461. [Google Scholar] [CrossRef]
- McSharry, P.E.; Clifford, G.D.; Tarassenko, L.; Smith, L.A. A Dynamical Model for Generating Synthetic Electrocardiogram Signals. IEEE Trans. Biomed. Eng. 2003, 50, 289–294. [Google Scholar] [CrossRef]
- Chang, K.H.; Young, M.S. Design of a Three-Lead Synthetic ECG Generator Using the Simplified McSharry’s Model. Instrum. Sci. Technol. 2009, 37, 397–409. [Google Scholar] [CrossRef]
- Sayadi, O.; Sameni, R.; Shamsollahi, M.B. ECG Denoising Using Parameters of ECG Dynamical Model as the States of an Extended Kalman Filter. In Proceedings of the 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Lyon, France, 22–26 August 2007; pp. 2548–2551. [Google Scholar]
- Soler, A.I.R.; Bonomini, M.P.; Biscay, C.F.; Ingallina, F.; Arini, P.D. Modelling of the Electrocardiographic Signal during an Angioplasty Procedure in the Right Coronary Artery. J. Electrocardiol. 2020, 62, 65–72. [Google Scholar] [CrossRef]
- Takha, A.; Talbi, M.L.; Ravier, P. Fractional Calculus Integration for Improved ECG Modeling: A McSharry Model Expansion. Med. Eng. Phys. 2024, 132, 104237. [Google Scholar] [CrossRef]
- Jafarnia-Dabanloo, N.; McLernon, D.; Zhang, H.; Ayatollahi, A.; Johari-Majd, V. A Modified Zeeman Model for Producing HRV Signals and Its Application to ECG Signal Generation. J. Theor. Biol. 2007, 244, 180–189. [Google Scholar] [CrossRef]
- Abad, S.L.M.; Dabanloo, N.J.; Jameie, S.B.; Sadeghniiat, K. A Developed Zeeman Model for HRV Signal Generation in Different Stages of Sleep. In Proceedings of the 13th International Conference on Biomedical Engineering, Singapore, 3–6 December 2008; IFMBE Proceedings. Springer: Berlin/Heidelberg, Germany, 2009; Volume 23, pp. 1–6. [Google Scholar]
- Zanchi, B.; Monachino, G.; Fiorillo, L.; Conte, G.; Auricchio, A.; Tzovara, A.; Faraci, F.D. Synthetic ECG Signals Generation: A Scoping Review. Comput. Biol. Med. 2025, 184, 109453. [Google Scholar] [CrossRef]
- Ryzhii, E.; Ryzhii, M. Modeling of Heartbeat Dynamics with a System of Coupled Nonlinear Oscillators. In Proceedings of the Biomedical Informatics and Technology (ACBIT 2013), Aizu-Wakamatsu, Japan, 16–17 September 2013; Pham, T.D., Ichikawa, K., Oyama-Higa, M., Coomans, D., Jiang, X., Eds.; Springer: Berlin/Heidelberg, Germany, 2014. CCIS. Volume 404, pp. 1–8. [Google Scholar]
- Li, X.; Xu, S.; Habib, F.; Aminnejad, N.; Gupta, A.; Huang, H. CLEP-GAN: An Innovative Approach to Subject-Independent ECG Reconstruction from PPG Signals. arXiv 2025, arXiv:2502.17536. [Google Scholar] [CrossRef]
- Zama, M.H.; Schwenker, F. ECG Synthesis via Diffusion-Based State Space Augmented Transformer. Sensors 2023, 23, 8328. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Kim, J.; Kwon, J.-M.; Jeon, K.-H.; Lee, M.S.; Choi, E. Text-to-ECG: 12-Lead Electrocardiogram Synthesis conditioned on Clinical Text Reports. arXiv 2023, arXiv:2303.09395. [Google Scholar]
- Pöhl, P.; Schlegel, V.; Li, H.; Bharath, A. Generating Realistic Multi-Beat ECG Signals. arXiv 2025, arXiv:2505.18189. [Google Scholar]
- López Alcaraz, J.M.; Strodthoff, N. Diffusion-based conditional ECG generation with structured state space models. Comput. Biol. Med. 2023, 163, 107115. [Google Scholar] [CrossRef]
- Neifar, N.; Ben-Hamadou, A.; Mdhaffar, A.; Jmaiel, M. DiffECG: A Versatile Probabilistic Diffusion Model for ECG Signals Synthesis. arXiv 2023, arXiv:2306.01875. [Google Scholar]
- Bedin, L.; Cardoso, G.; Duchateau, J.; Dubois, R.; Moulines, E. Leveraging an ECG Beat Diffusion Model for Morphological Reconstruction from Indirect Signals. In Proceedings of the 38th International Conference on Neural Information Processing Systems (NeurIPS 2024), Vancouver, BC, Canada, 9–15 December 2024; Article 2682. pp. 84409–84446. [Google Scholar]
- Qiang, Y.; Dong, X.; Liu, X.; Yang, Y.; Fang, Y.; Dou, J. ECGMamba: Towards Efficient ECG Classification with BiSSM. arXiv 2024, arXiv:2406.10098. [Google Scholar] [CrossRef]
- Berger, L.; Haberbusch, M.; Moscato, F. Generative Adversarial Networks in Electrocardiogram Synthesis: Recent Developments and Challenges. Artif. Intell. Med. 2023, 143, 102632. [Google Scholar] [CrossRef] [PubMed]
- Ikram, S.; Ikram, A.; Singh, H.; Ali Awan, M.D.; Naveed, S.; De la Torre Díez, I.; Gongora, H.F.; Candelaria Chio Montero, T. Transformer-based ECG classification for early detection of cardiac arrhythmias. Front. Med. 2025, 12, 1600855. [Google Scholar] [CrossRef] [PubMed]
- Kopets, E.E.; Rybin, V.G.; Vasilchenko, O.V.; Kurtova, K.A.; Karimov, T.I.; Karimov, A.I.; Butusov, D.N. Memristor-based Chaotic Dynamical Model for Generating Electrocardiogram Signal. Izvestiya VUZ. Appl. Nonlinear Dyn. 2025, 33, 691–708. [Google Scholar] [CrossRef]
- Avanzato, R.; Beritelli, F.; Lombardo, A.; Ricci, C. Heart DT: Monitoring and Preventing Cardiac Pathologies Using AI and IoT Sensors. Future Internet 2023, 15, 223. [Google Scholar] [CrossRef]
- Trayanova, N.A.; Prakosa, A. Up digital and personal: How heart digital twins can transform heart patient care. Heart Rhythm 2024, 21, 89–99. [Google Scholar] [CrossRef]
- Bhagirath, P.; Strocchi, M.; Bishop, M.J.; Boyle, P.M.; Plank, G. From bits to bedside: Entering the age of digital twins in cardiac electrophysiology. Europace 2024, 26, euae295. [Google Scholar] [CrossRef]
- Li, L.; Smith, H.; Lyu, Y.; Camps, J.; Qian, S.; Rodriguez, B.; Banerjee, A.; Grau, V. Personalized Topology-Informed Localization of Standard 12-Lead ECG Electrode Placement from Incomplete Cardiac MRIs for Efficient Cardiac Digital Twins. arXiv 2024, arXiv:2408.13945. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Hu, L.; Li, D.; Yan, J.; Ying, H.; Liang, H.; Wu, J. Personalized Heart Disease Detection via ECG Digital Twin Generation. arXiv 2024, arXiv:2404.11171. [Google Scholar] [CrossRef]
- Tasmurzayev, N.; Amangeldy, B.; Imanbek, B.; Baigarayeva, Z.; Imankulov, T.; Dikhanbayeva, G.; Amangeldi, I.; Sharipova, S. Digital Cardiovascular Twins, AI Agents, and Sensor Data: A Narrative Review from System Architecture to Proactive Heart Health. Sensors 2025, 25, 5272. [Google Scholar] [CrossRef]
- Rudnicka, Z.; Proniewska, K.; Perkins, M.; Pregowska, A. Cardiac Healthcare Digital Twins Supported by Artificial Intelligence-Based Algorithms and Extended Reality—A Systematic Review. Electronics 2024, 13, 866. [Google Scholar] [CrossRef]
- Jæger, K.H.; Tveito, A. Deriving the Bidomain Model of Cardiac Electrophysiology From a Cell-Based Model; Properties and Comparisons. Front. Physiol. 2022, 12, 811029. [Google Scholar] [CrossRef]
- Sundnes, J.; Lines, G.T.; Cai, X.; Nielsen, B.F.; Mardal, K.-A.; Tveito, A. Computing the Electrical Activity in the Heart; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Colli Franzone, P.; Pavarino, L.F.; Scacchi, S. Mathematical Cardiac Electrophysiology; Modeling, Simulation and Applications; MS&A; Springer: Cham, Switzerland, 2014; Volume 13. [Google Scholar] [CrossRef]
- Rössler, O.E. On the Rössler Attractor. Chaos Theory Appl. 2020, 2, 1–2. [Google Scholar]
- Peitgen, H.-O.; Jürgens, H.; Saupe, D. Chaos and Fractals: New Frontiers of Science; Springer: New York, NY, USA, 2004; pp. 636–646. [Google Scholar]
- Alhazmi, M.; Saber, S.; Dridi, B.; Alahmari, A.; Messaoudi, M. Application of a fractal-fractional derivative with a power-law kernel to the glucose–insulin interaction system based on Newton’s interpolation polynomials. Fractals 2025, 2540201. [Google Scholar] [CrossRef]
- Clifford, G.D.; Shoeb, A.; McSharry, P.E.; Janz, B.A. Model-Based Filtering, Compression and Classification of the ECG. Int. J. Bioelectromagn. 2005, 7, 158–161. [Google Scholar]
- Clifford, G.D. A Novel Framework for Signal Representation and Source Separation: Applications to Filtering and Segmentation of Biosignals. J. Biol. Syst. 2006, 14, 169–183. [Google Scholar] [CrossRef]
- Clifford, G.D.; Villarroel, M. Model-Based Determination of QT Intervals. In Proceedings of the Computers in Cardiology, Valencia, Spain, 17–20 September 2006; IEEE: Piscataway, NJ, USA, 2006; Volume 33, pp. 357–360. [Google Scholar]
- Awal, M.A.; Mostafa, S.S.; Ahmad, M. Simplified Mathematical Model for Generating ECG Signal and Fitting the Model Using Non-Linear Least Square Technique. In Proceedings of the International Conference on Mechanical Engineering, Dhaka, Bangladesh, 18–20 December 2011; pp. 1–6. [Google Scholar]
- Shaffer, F.; Ginsberg, J.P. An Overview of Heart Rate Variability Metrics and Norms. Front. Public Health 2017, 5, 258. [Google Scholar] [CrossRef] [PubMed]
- Kachuee, M.; Fazeli, S.; Sarrafzadeh, M. ECG Heartbeat Classification: A Deep Transferable Representation. In Proceedings of the 2018 IEEE International Conference on Healthcare Informatics (ICHI), New York, NY, USA, 4–7 June 2018; pp. 443–444. [Google Scholar] [CrossRef]
- Hannun, A.Y.; Rajpurkar, P.; Haghpanahi, M.; Tison, G.H.; Bourn, C.; Turakhia, M.P.; Ng, A.Y. Cardiologist-Level Arrhythmia Detection and Classification in Ambulatory Electrocardiograms Using a Deep Neural Network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef]
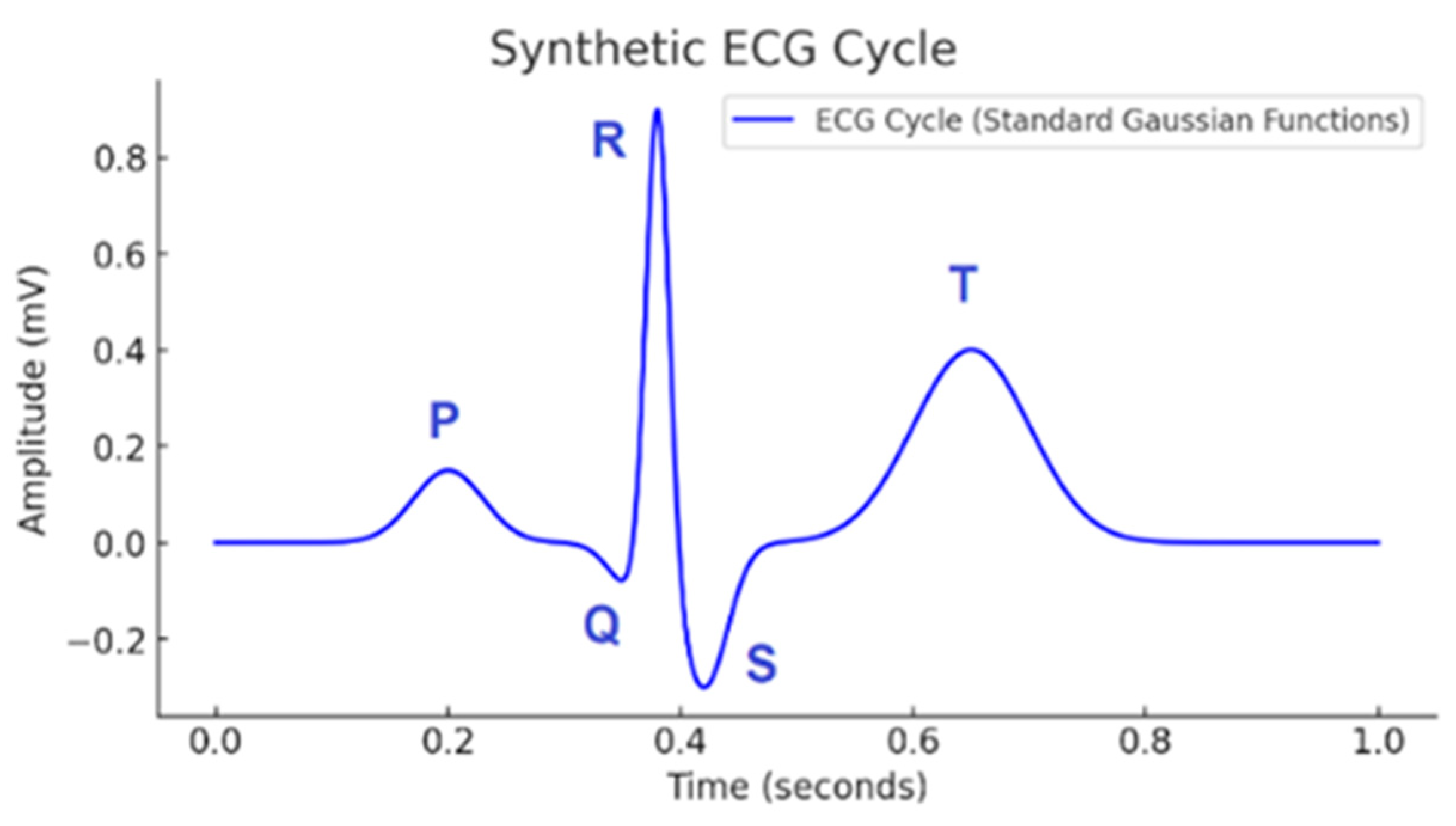
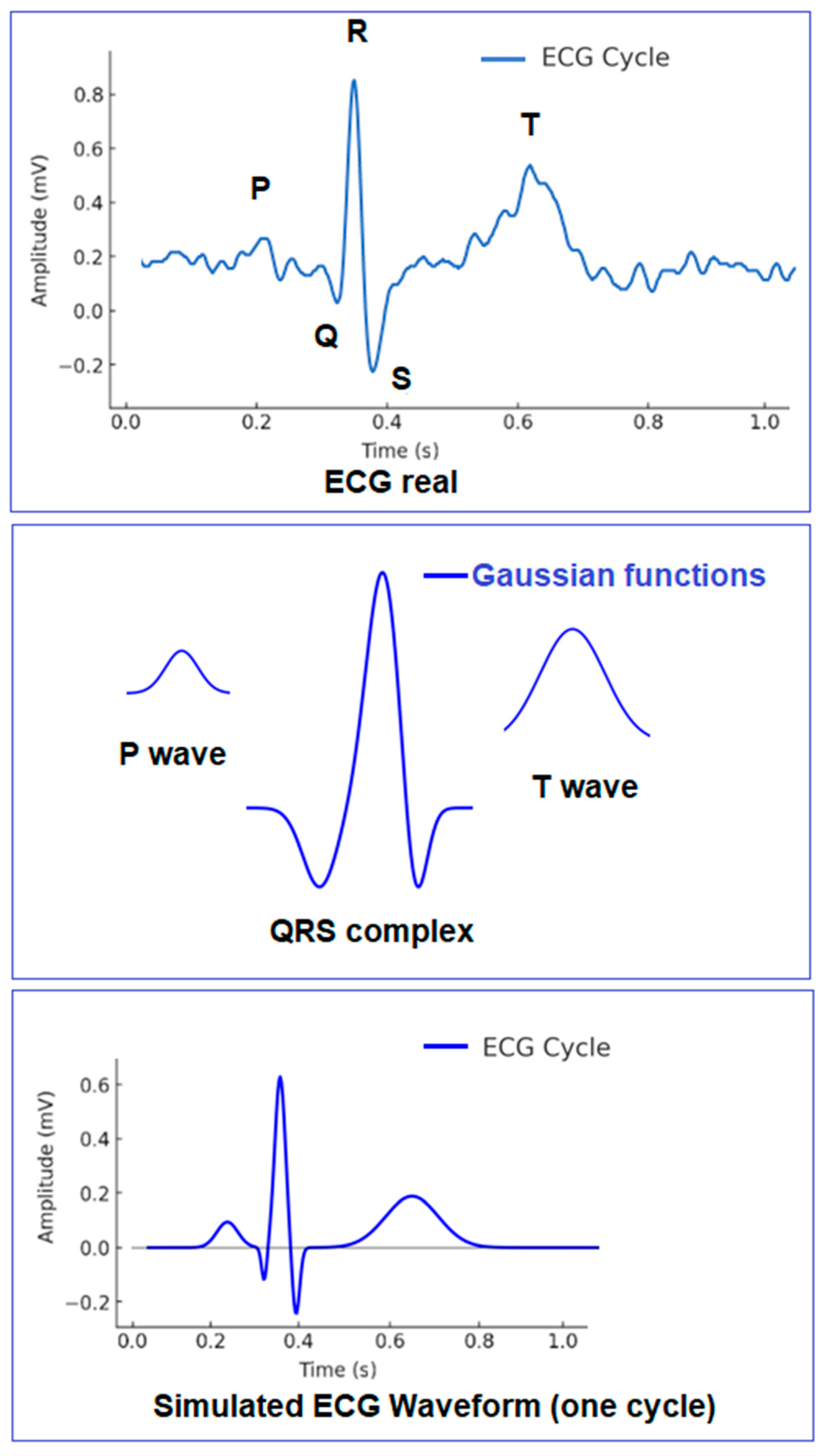
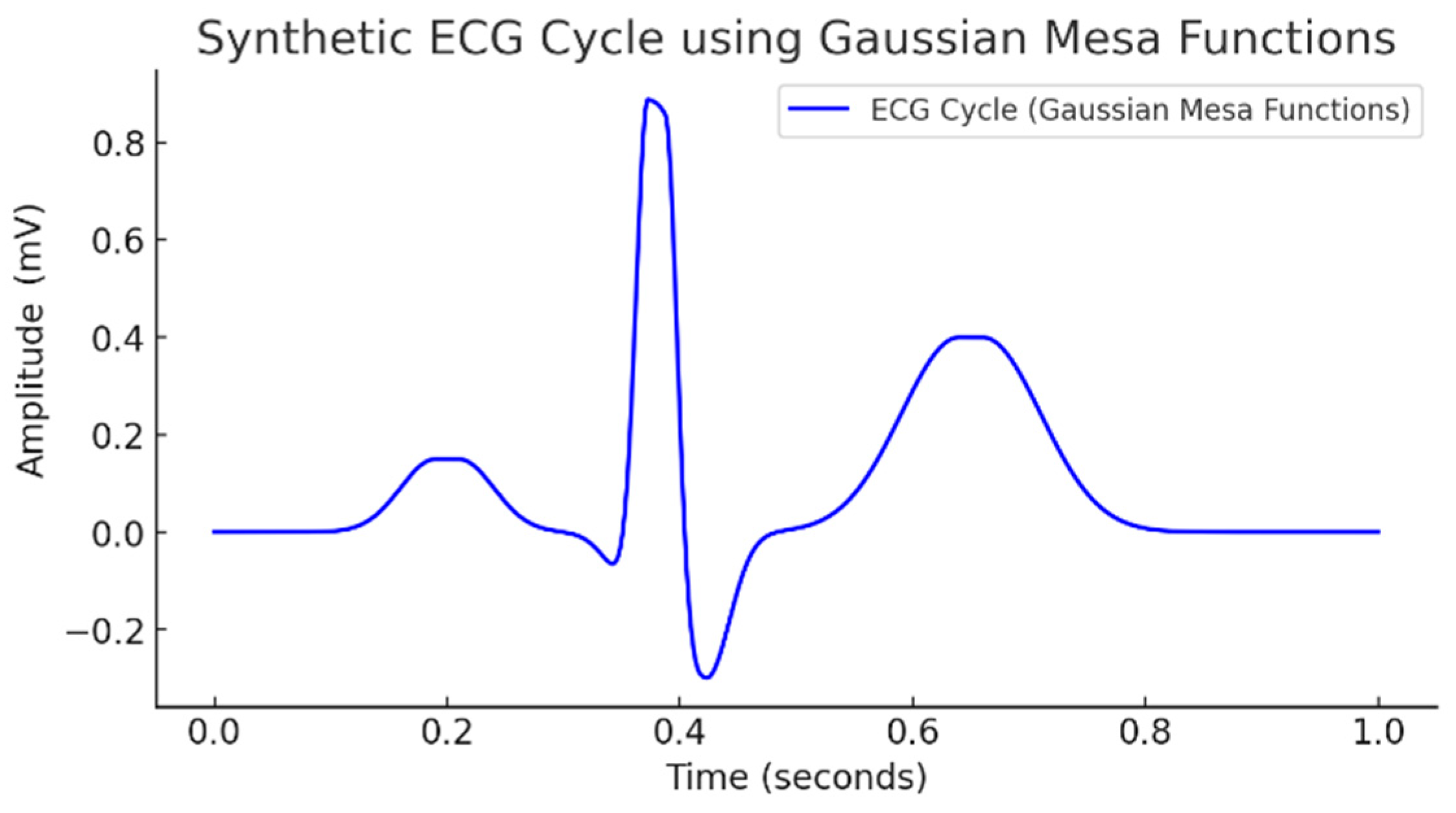
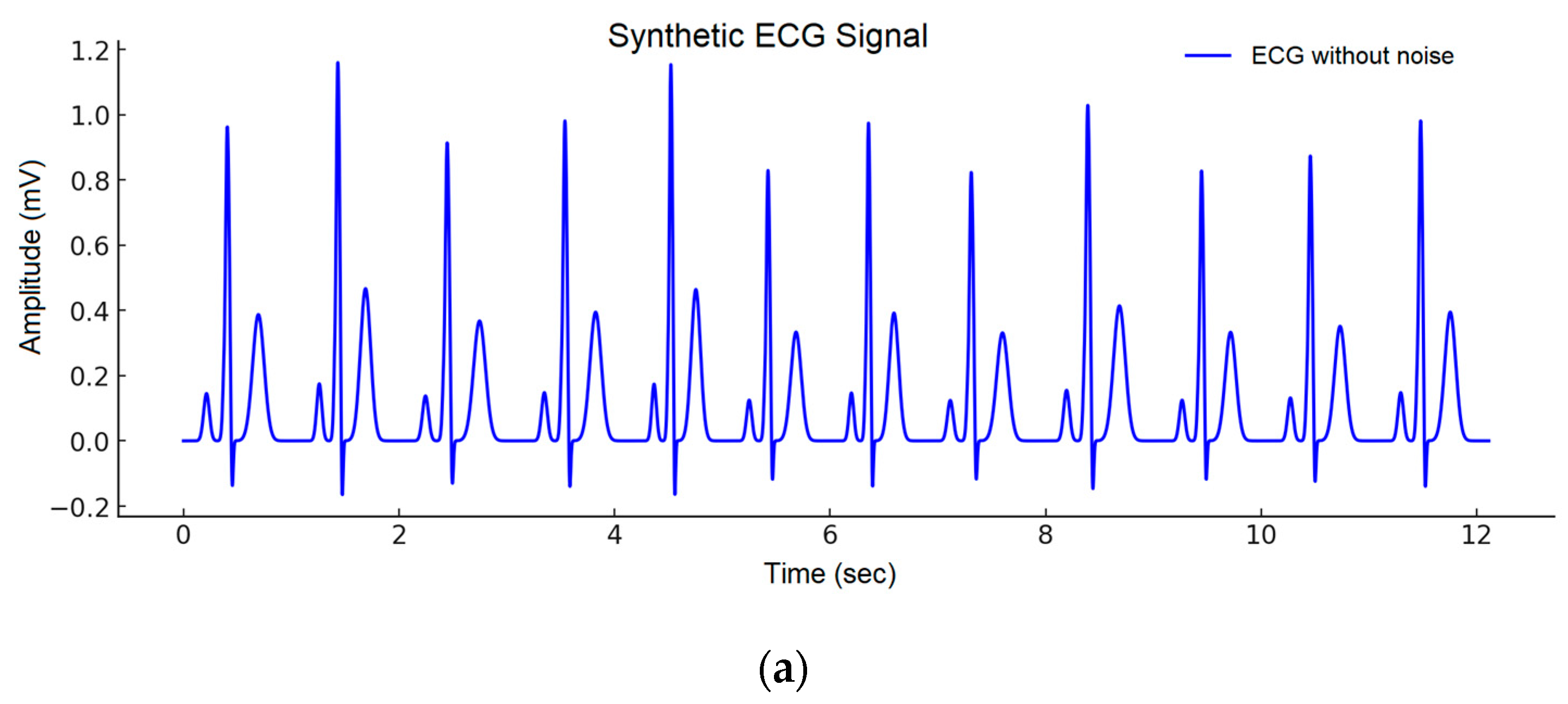
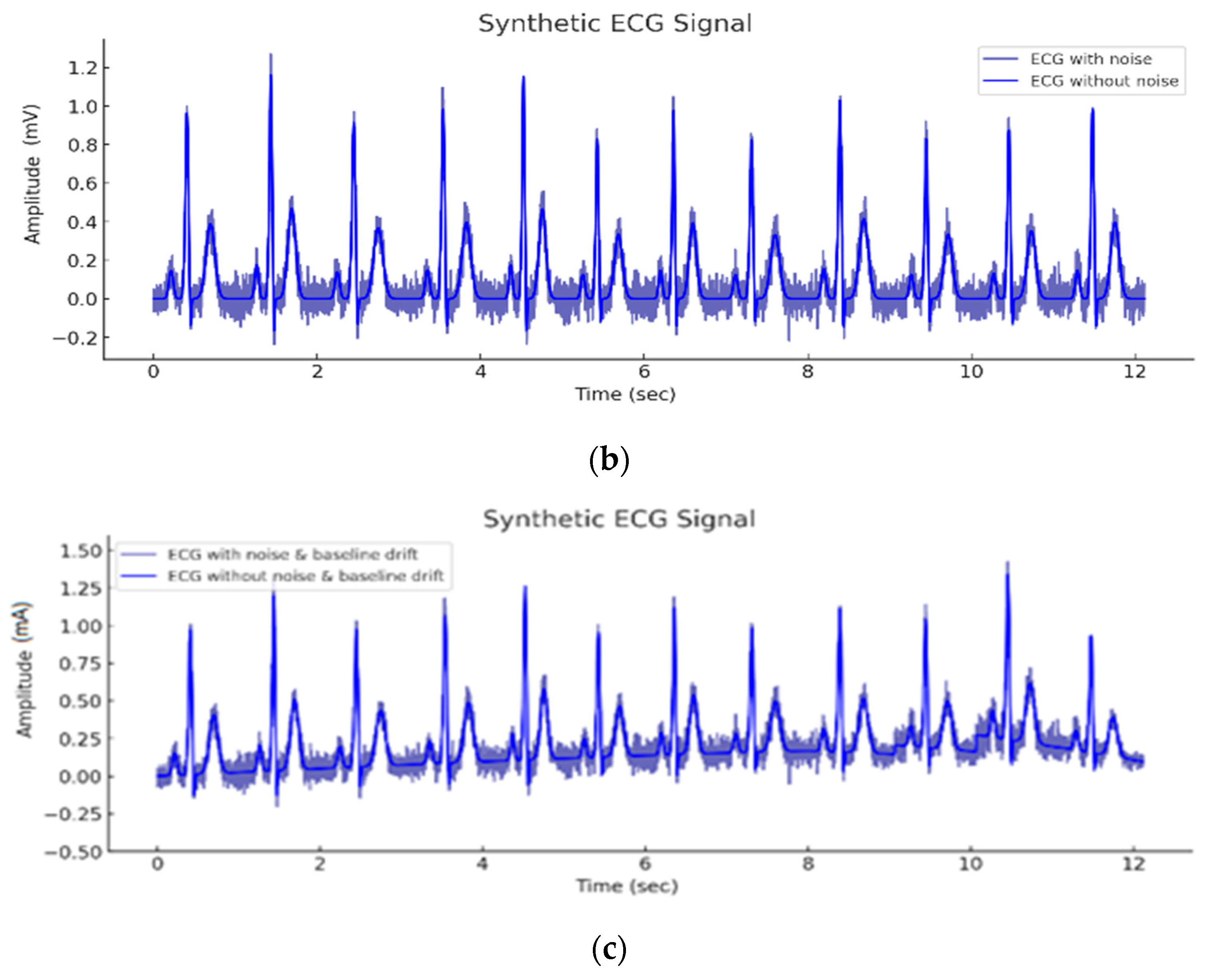
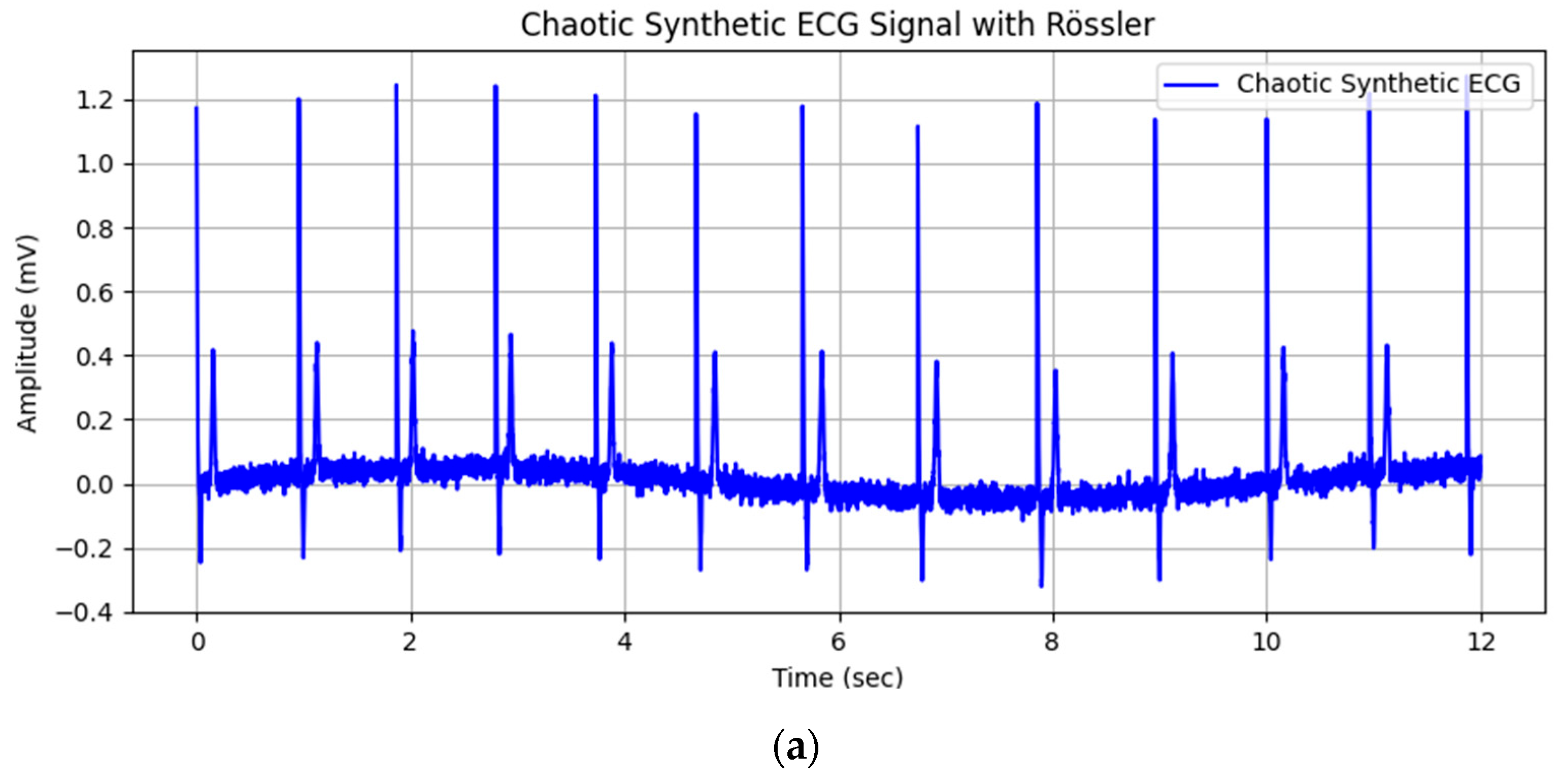
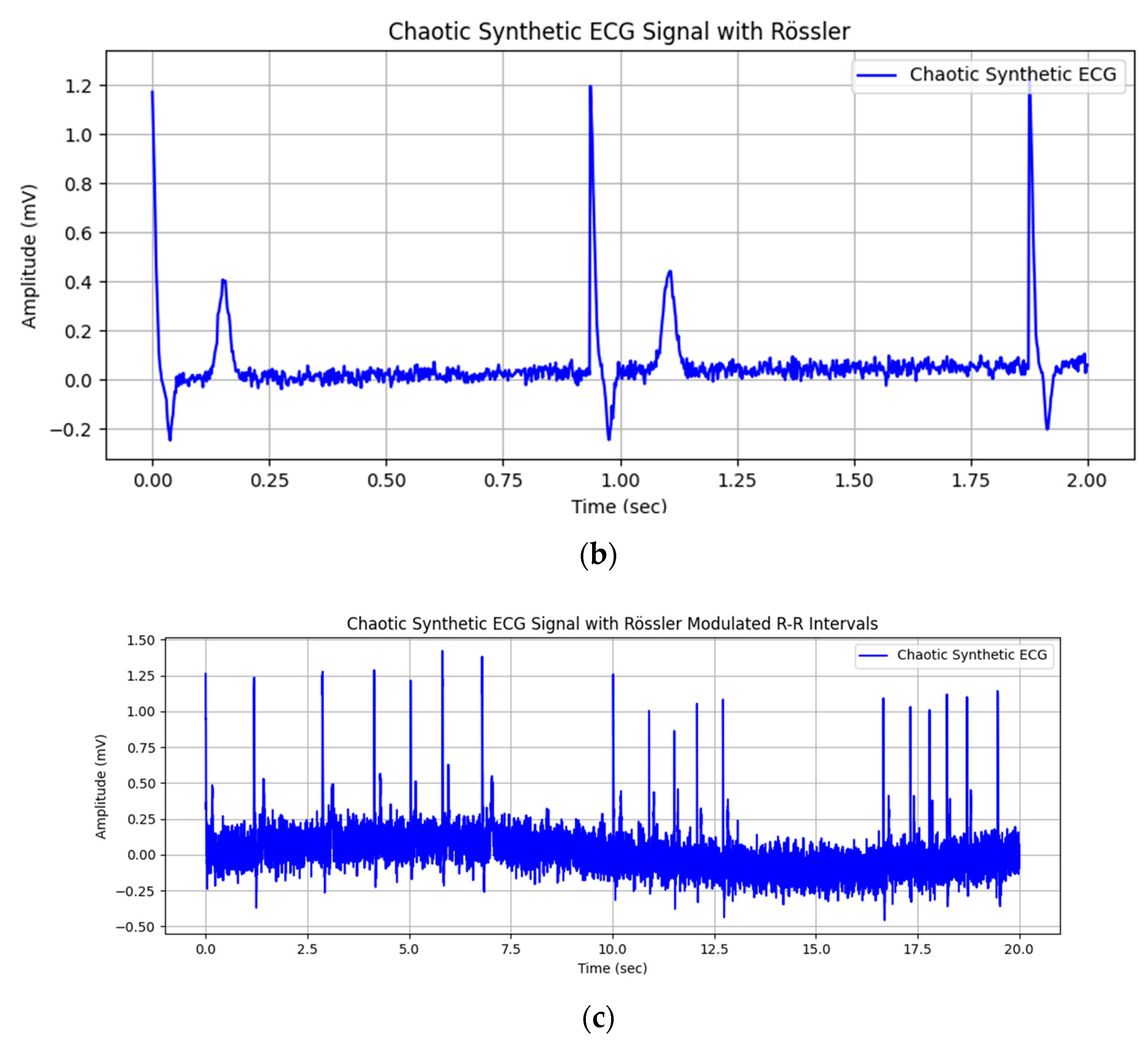
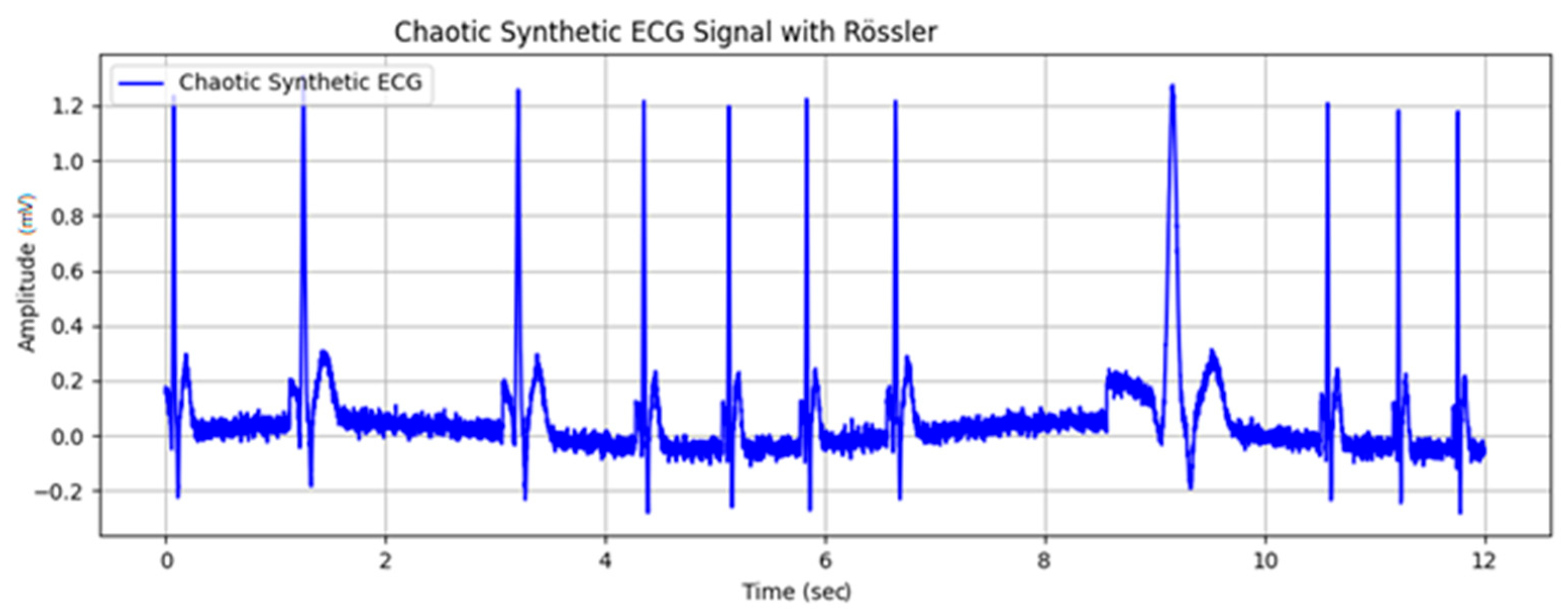
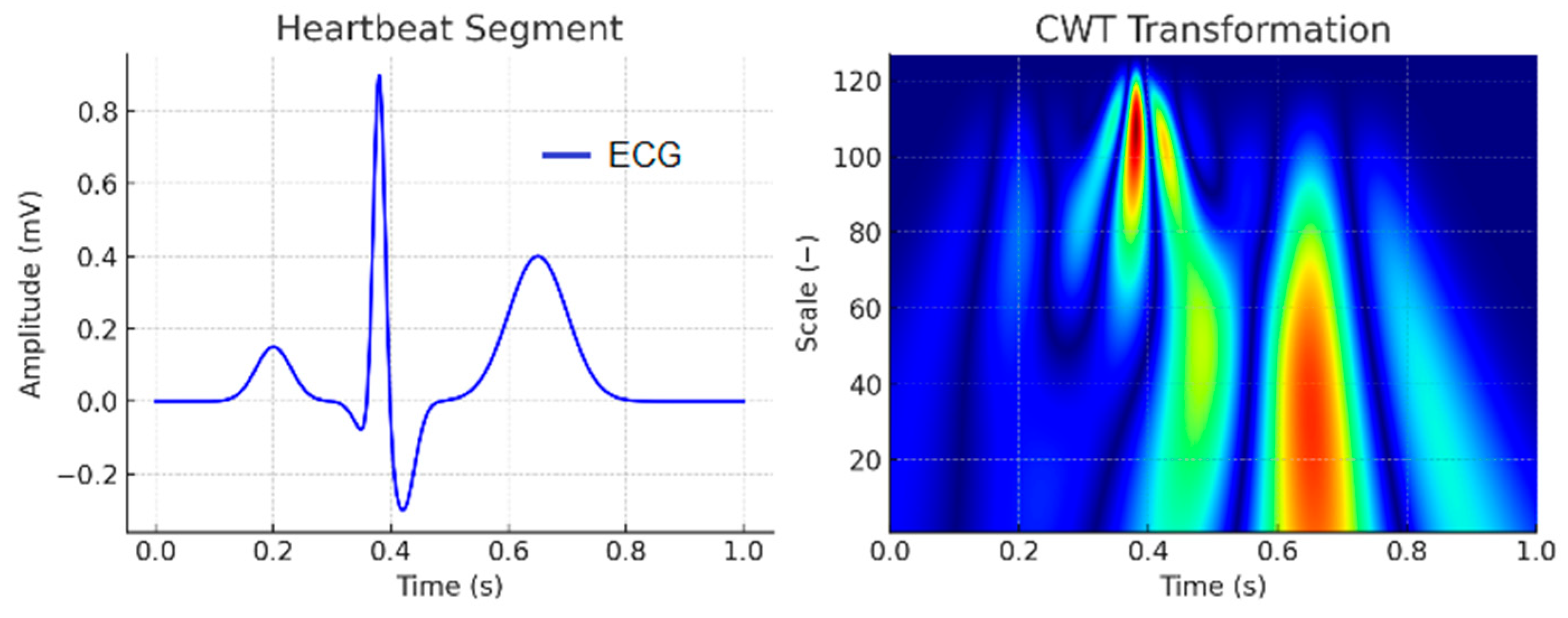
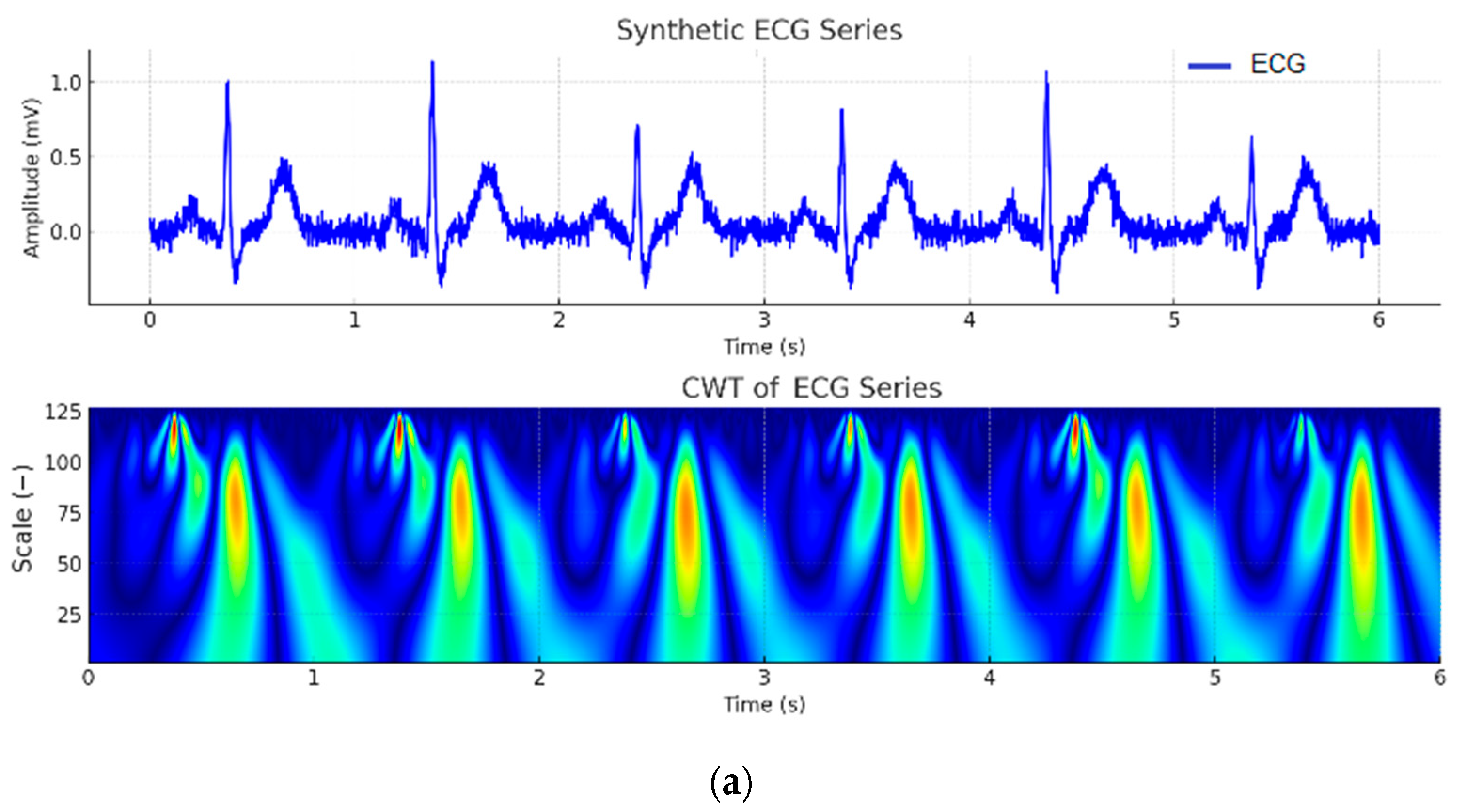
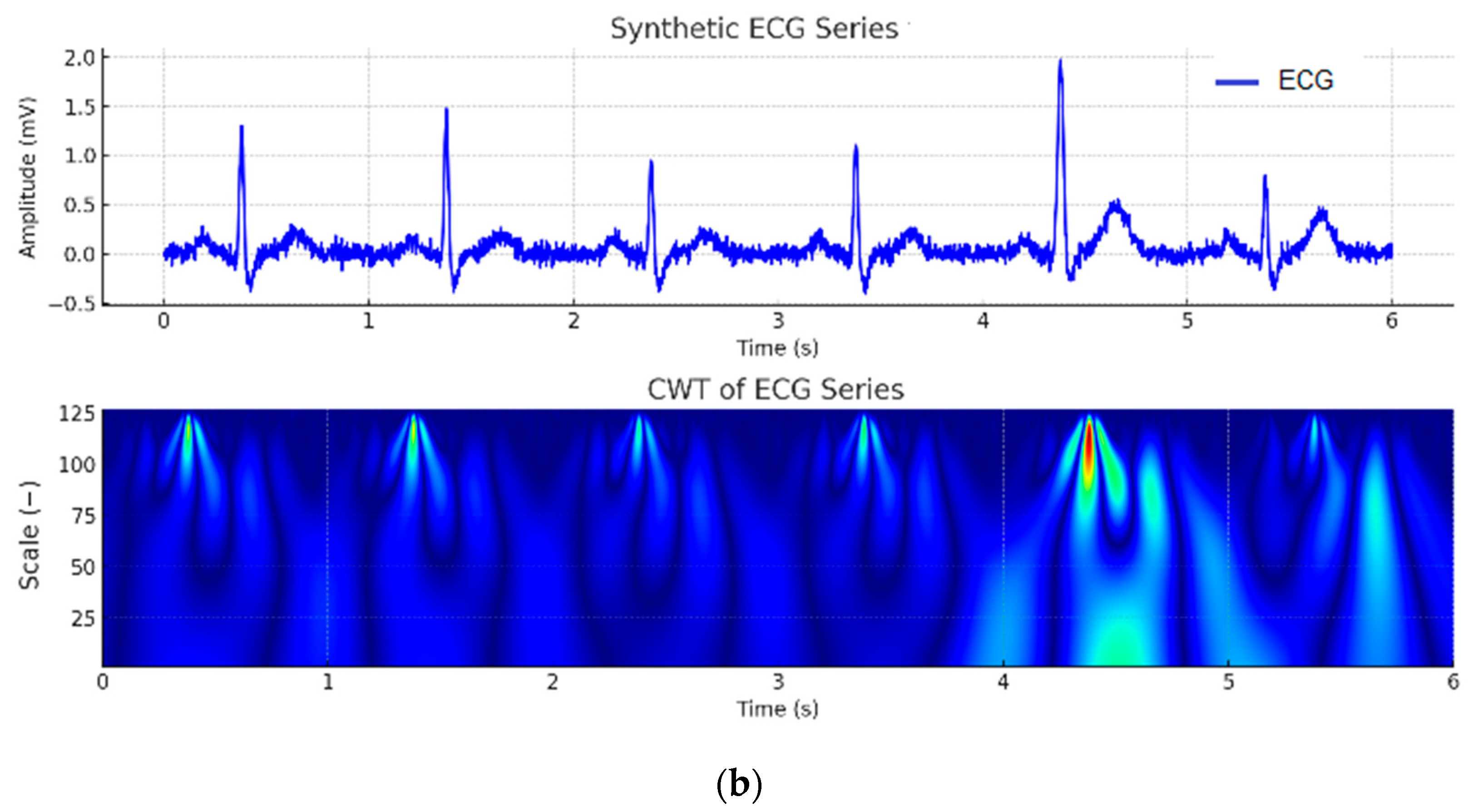
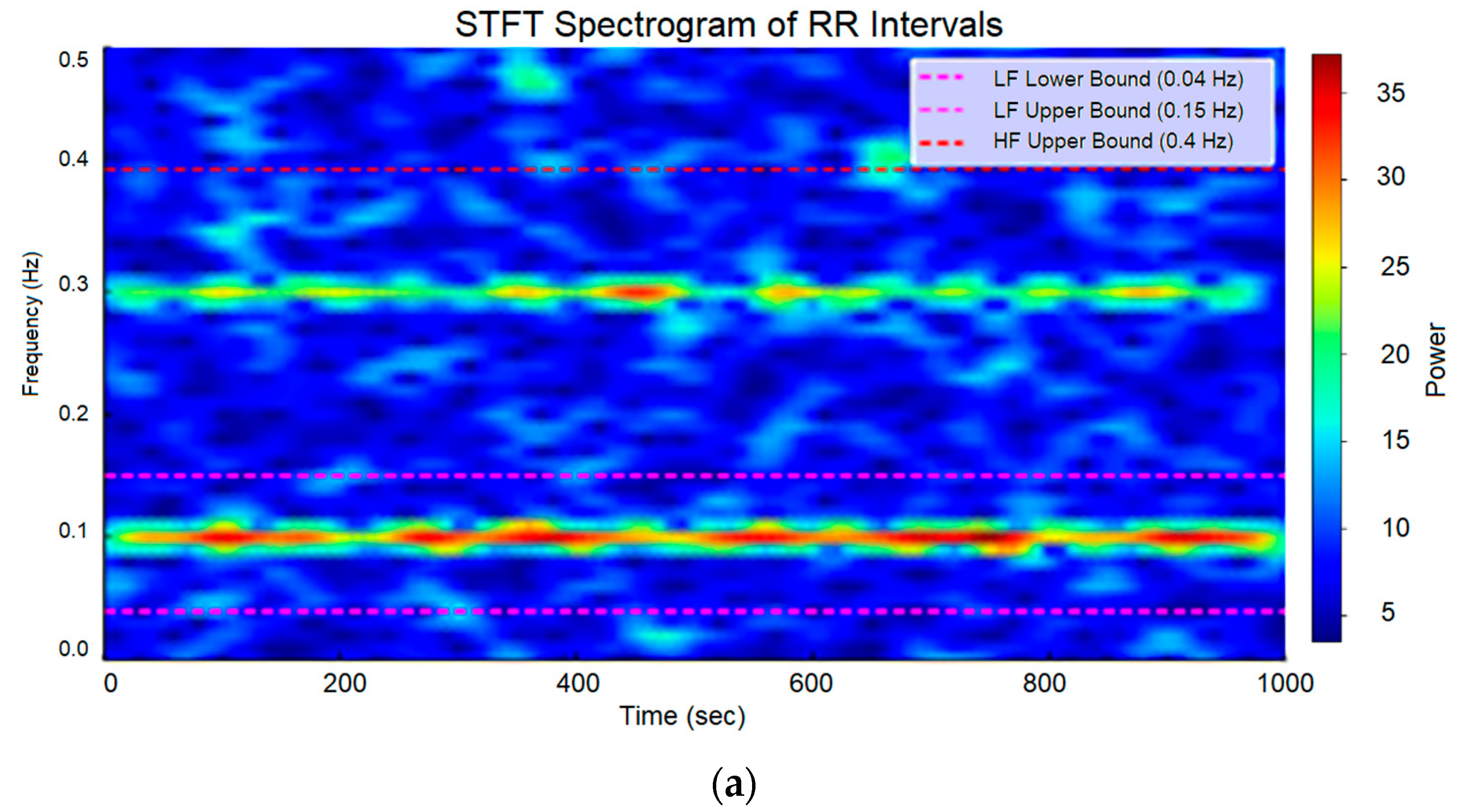
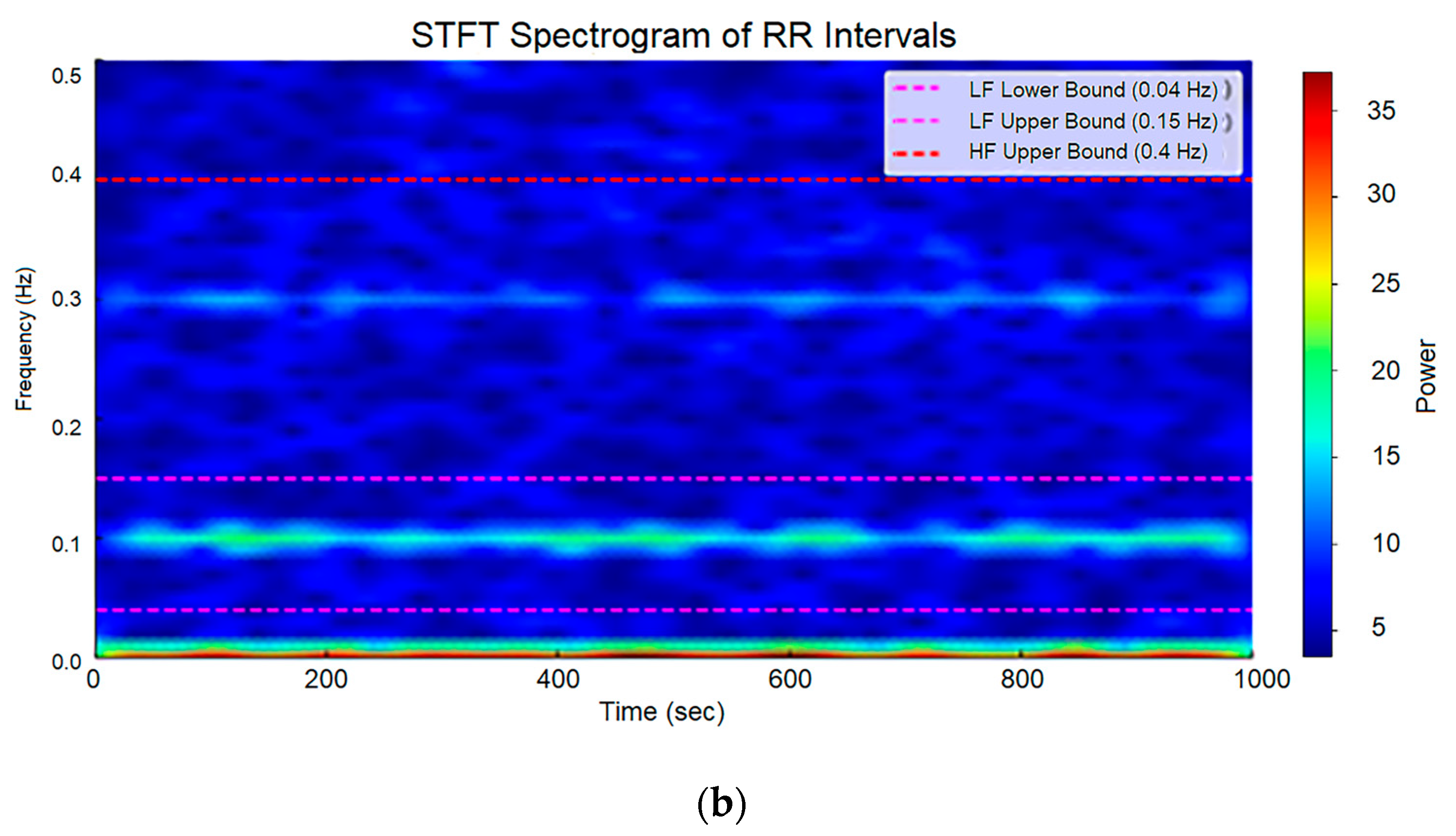
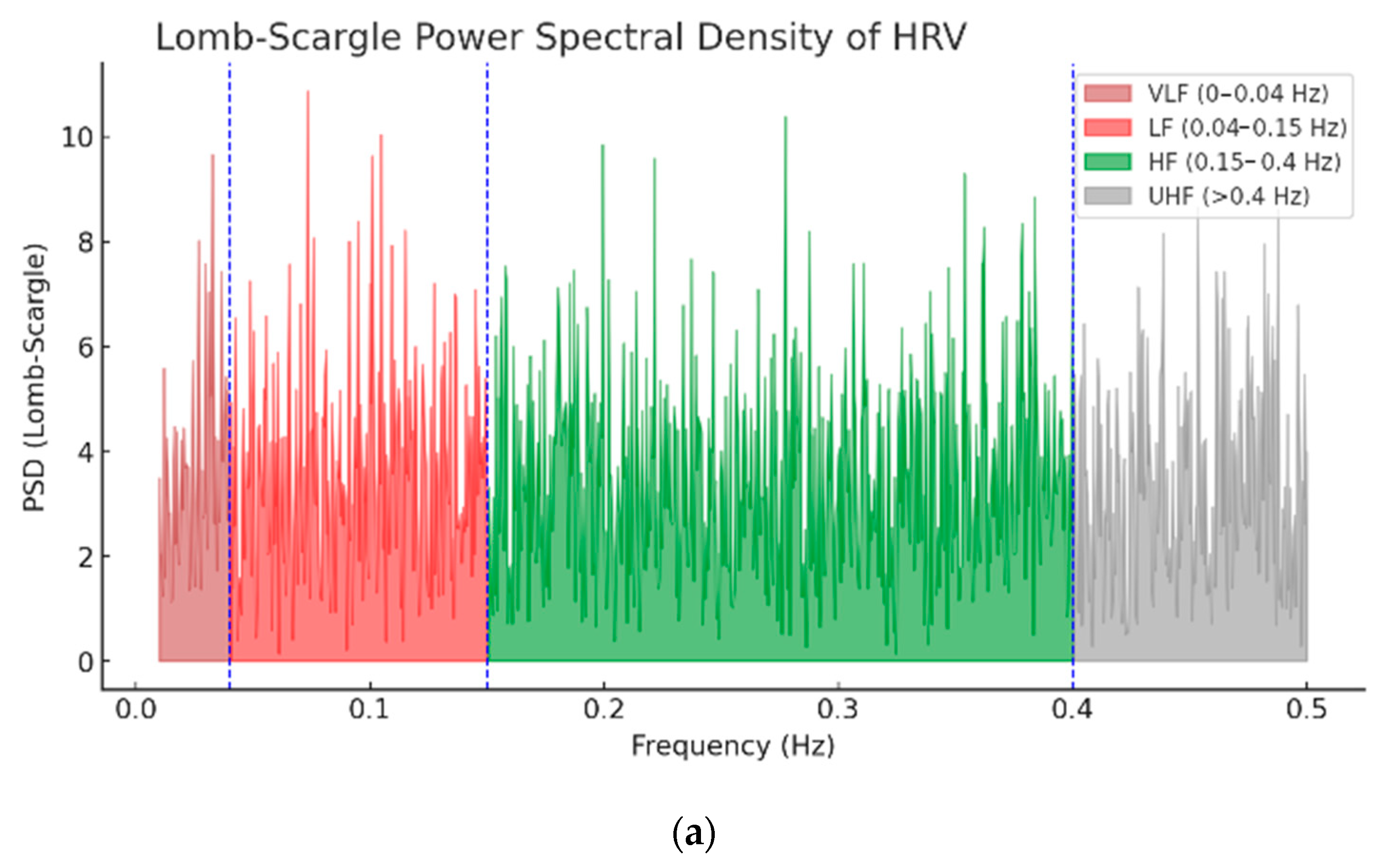
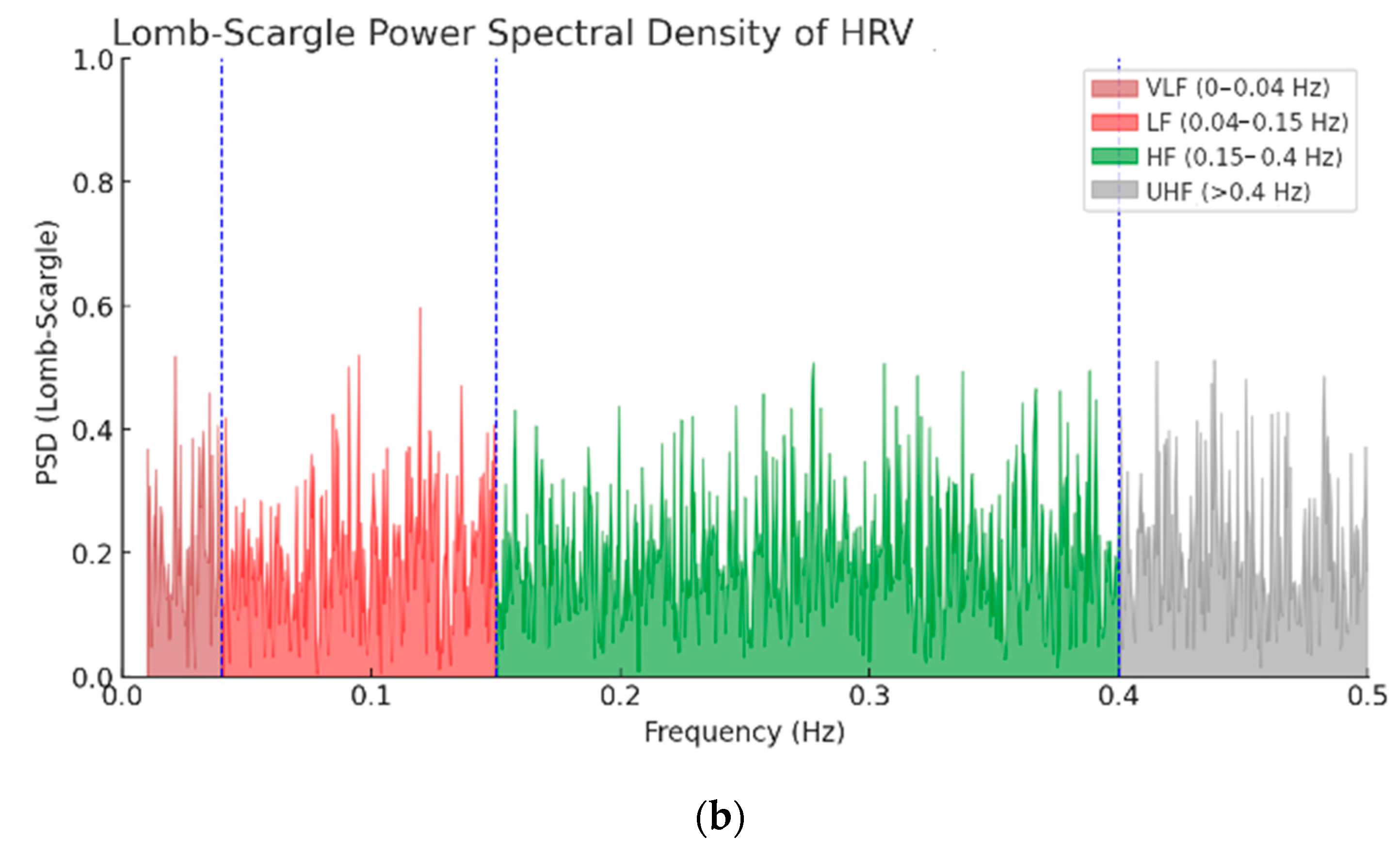
| Wave | Purpose | ||||
|---|---|---|---|---|---|
| P | small | early | wide, symmetric | short | atrial depolarization |
| Q | small negative | before R | narrow | none | initial ventricular activation |
| R | largest positive | center | very steep rise/fall | minimal | main depolarization |
| S | negative | after R | narrow | none | terminal depolarization |
| T | medium positive | late | broader decay | long | ventricular repolarization |
| Parameter | Rest | Fatigue | Stress | Description |
|---|---|---|---|---|
| Mean RR interval [ms] | 800÷1000 | 600÷800 | 300÷600 | Heart rate (HR) increases from ~60 bpm (rest) to ~200 bpm (stress) |
| Standard deviation of RR () [ms] | 60 | 40 | 20 | HRV variations—decrease under stress |
| Attractor parameter a | 0.1 | 0.15 | 0.2 | Influences phase dynamics |
| Attractor parameter b | 0.1 | 0.1 | 0.1 | Standard value for Rössler system |
| Attractor parameter c | 5.7 | 5.9 | 6.2 | Increases chaotic behavior under stress |
| Influence coefficient (α) | 0.05 | 0.10 | 0.15 | Enhances the attractor’s effect on RR |
| amplitude () [mV] | 1.0 | 0.8 | 0.7 | May slightly decrease under fatigue |
| QRS width () [ms] | 90 | 80 | 70 | Narrower complex under stress |
| T wave amplitude () [mV] | 0.4 | 0.25 | 0.3 | T wave decreases under fatigue |
| Baseline drift amplitude () [mV] | 0.1 | 0.15 | 0.2 | More pronounced baseline drift under stress |
| LF/HF (expected value) | 1.5÷2.0 | <1.5 or >2.0 | <0.5 or >2.5 | Autonomic balance ratio |
| Parameters | ECGreal | Real 95% CI | ECGsim | Sim 95% CI | Normal | p-Value | Effect Size |
|---|---|---|---|---|---|---|---|
| MeanRR (ms) | 901.14 ± 102.39 | [835.97, 966.31] | 920.4 ± 100.2 | [856.60, 984.20] | - | NS 1 | 0.19 (negligible) |
| SDNN (ms) | 128.2 ± 51.77 | [95.33, 161.07] | 121.15 ± 20.4 | [108.15, 134.11] | 141 ± 39 | NS | 0.17 (negligible) |
| RMSSD (ms) | 14.09 ± 5.03 | [10.90, 17.28] | 13.98 ± 2.4 | [12.46, 15.50] | 27 ± 12 | NS | 0.03 (negligible) |
| nLF [nu] | 60.31 ± 21.44 | [46.70, 73.92] | 58.75 ± 16.24 | [48.44, 69.06] | - | NS | 0.08 (negligible) |
| nHF [nu] | 39.65 ± 9.14 | [33.84, 45.46] | 41.12 ± 8.10 | [35.98, 46.26] | - | NS | 0.17 (negligible) |
| LF/HF (-) | 1.53 ± 0.2 | [1.40, 1.66] | 1.49 ± 0.25 | [1.33, 1.65] | 1.5–2.0 | NS | 0.18 (negligible) |
| SD1 [ms] | 29.09 ± 7.57 | [24.27, 33.91] | 31.74 ± 6.11 | [27.86, 35.62] | - | NS | 0.38 (small) |
| SD2 [ms] | 69.38 ± 29.04 | [50.91, 87.85] | 75.39 ± 13.99 | [66.51, 84.27] | - | NS | 0.28 (small) |
| SD2/SD1 [-] | 2.21 ± 0.39 | [1.96, 2.46] | 2.45 ± 0.39 | [2.20, 2.70] | - | NS | 0.62 (medium) |
| Hurst [-] | 0.78 ± 0.19 | [0.66, 0.90] | 0.85 ± 0.12 | [0.77, 0.93] | 0.5–1.0 | NS | 0.43 (small) |
| SampEn [-] | 1.42 ± 0.39 | [1.17, 1.67] | 1.53 ± 0.37 | [1.29, 1.77] | - | NS | 0.28 (small) |
| Parameters | ECG Arrhythmia | Real 95% CI | ECGsim | Sim 95% CI | p-Value | Effect Size |
|---|---|---|---|---|---|---|
| MeanRR (ms) | 704.09 ± 189.23 | [701.14, 783.48] | 742.31 ± 126.03 | 742.31 ± 126.03 | NS 1 | 0.23 (small) |
| SDNN (ms) | 95.05 ± 34.62 | [82.02, 98.44] | 90.23 ± 25.14 | 90.23 ± 25.14 | NS | 0.16 (small) |
| RMSSD (ms) | 9.02 ± 4.13 | [8.50, 11.72] | 10.11 ± 4.92 | 10.11 ± 4.92 | NS | 0.24 (small) |
| nLF [nu] | 52.81 ± 22.18 | [46.89, 59.35] | 53.12 ± 19.09 | 53.12 ± 19.09 | NS | 0.01 (negligible) |
| nHF [nu] | 48.15 ± 16.38 | [41.34, 53.56] | 47.45 ± 18.73 | 47.45 ± 18.73 | NS | 0.04 (negligible) |
| LF/HF (-) | 1.09 ± 0.3 | [1.04, 1.18] | 1.11±0.2 | 1.11 ± 0.20 | NS | 0.08 (negligible) |
| SD1 [ms] | 12.14 ± 5.84 | [9.60, 13.38] | 11.49 ± 5.78 | 11.49 ± 5.78 | NS | 0.11 (small) |
| SD2 [ms] | 46.51 ± 24.50 | [32.37, 49.33] | 40.85 ± 25.99 | 40.85 ± 25.99 | NS | 0.22 (small) |
| SD2/SD1 [-] | 3.62 ± 0.42 | [3.19, 3.43] | 3.31 ± 0.38 | 3.31 ± 0.38 | NS | 0.77 (moderate) |
| Hurst [-] | 0.56 ± 0.16 | [0.56, 0.64] | 0.60 ± 0.13 | 0.60 ± 0.13 | NS | 0.27 (small) |
| SampEn [-] | 0.69 ± 0.27 | [0.59, 0.73] | 0.66 ± 0.22 | 0.66 ± 0.22 | NS | 0.12 (small) |
| Parameter | Real—Rest (Pre-Training) | Real—Rest (Pre-Training) 95% CI | Simulated—Rest | Simulated—Rest 95% CI | Real—Fatigue (Post-Training) | Real—Fatigue (Post-Training) 95% CI | Simulated—Fatigue | Simulated—Fatigue 95% CI | Real— Stress (After a Competition) | Real— Stress (After a Competition 95% CI | Simulated—Stress | Simulated—Stress 95% CI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MeanRR [ms] | 794.67 ± 223.98 | [721.50, 867.84] | 891.34 ± 240.22 | [812.90, 969.78] | 507.2 ± 127.8 | [465.45, 548.95] | 545.67 ± 128.75 | [502.79, 588.55] | 483.65 ± 120.23 | [443.75, 523.55] | 430.12 ± 125.04 | [387.38, 472.86] |
| SDNN [ms] | 54.7 ± 16.4 | [49.34, 60.06] | 56.30 ± 11.55 | [52.53, 60.07] | 42.6 ± 7.8 | [40.05, 45.15] | 42.87 ± 9.20 | [40.75, 45.00] | 31.24 ± 11.41 | [27.56, 34.92] | 29.12 ± 6.73 | [26.93, 31.31] |
| RMSSD [ms] | 39.89 ± 11.3 | [36.20, 43.58] | 43.45 ± 8.90 | [40.54, 46.36] | 25.8 ± 5.13 | [24.12, 27.48] | 25.94 ± 6.11 | [24.89, 27.00] | 19.22 ± 8.17 | [16.49, 21.95] | 17.44 ± 4.65 | [15.88, 19.00] |
| TP (ms2) | 1921 ± 449 | [1852, 1948] | 1953 ± 426 | [1927, 1973] | 1802 ± 391 | [1773, 1827] | 1852 ± 413 | [1821, 1879] | 1573 ± 483 | [1484, 1516] | 1558 ± 465 | [1535, 1565] |
| nLF [nu] | 62.38 ± 11.8 | [58.64, 66.12] | 58.75 ± 10.23 | [55.53, 61.97] | 68.05 ± 13.72 | [63.57, 72.53] | 66.41 ± 13.54 | [61.89, 70.93] | 72.10 ± 12.41 | [67.95, 76.25] | 76.32 ± 39.88 | [63.22, 89.42] |
| nHF [nu] | 38.04 ± 8.29 | [35.33, 40.75] | 41.12 ± 8.10 | [38.44, 43.80] | 32.14 ± 7.24 | [29.69, 34.59] | 33.87 ± 7.88 | [31.36, 36.38] | 28.42 ± 9.84 | [25.42, 31.42] | 24.23 ± 6.75 | [22.00, 26.46] |
| LF/HF [-] | 1.63 ± 0.28 | [1.54, 1.72] | 1.41 ± 0.25 | [1.33, 1.49] | 2.12 ± 0.79 | [1.86, 2.38] | 2.04 ± 0.41 | [1.91, 2.17] | 2.57 ± 0.79 | [2.31, 2.83] | 3.16 ± 0.50 | [3.00, 3.32] |
| Hurst [-] | 0.76 ± 0.18 | [0.72, 0.84] | 0.82 ± 0.12 | [0.78, 0.86] | 0.62 ± 0.17 | [0.56, 0.68] | 0.63 ± 0.15 | [0.61, 0.65] | 0.53 ± 0.17 | [0.47, 0.59] | 0.51 ± 0.14 | [0.46, 0.56] |
| SampEn [-] | 1.38 ± 0.38 | [1.26, 1.51] | 1.48 ± 0.36 | [1.36, 1.60] | 0.85 ± 0.36 | [0.73, 0.97] | 0.92 ± 0.31 | [0.82, 1.02] | 0.74 ± 0.29 | [0.64, 0.84] | 0.71 ± 0.24 | [0.63, 0.79] |
| Parameter | p-Value Real vs. Sim (Rest) | p-Value Real vs. Sim (Fatigue) | p-Value Real vs. Sim (Stress) | Pearson Correlation Real vs. Sim (Rest) | Pearson Correlation Real vs. Sim (Fatigue) | Pearson Correlation Real vs. Sim (Stress) | Effect Size Real vs. Sim (Rest) | Effect Size Real vs. Sim (Fatigue) | Effect Size Real vs. Sim (Stress) |
|---|---|---|---|---|---|---|---|---|---|
| Mean RR [ms] | 0.894 | 0.598 | 0.086 | 0.82 | 0.89 | 0.76 | 0.416 | 0.300 | −0.432 |
| SDNN [ms] | 0.7603 | 0.4888 | 0.369 | 0.84 | 0.78 | 0.73 | 0.113 | 0.031 | −0.224 |
| RMSSD [ms] | 0.7504 | 0.5237 | 0.289 | 0.78 | 0.75 | 0.81 | 0.350 | 0.027 | −0.266 |
| TP (ms2) | 0.684 | 0.552 | 0.433 | 0.83 | 0.81 | 0.78 | 0.142 | 0.118 | –0.165 |
| nLF [nu] | 0.5769 | 0.8211 | 0.601 | 0.76 | 0.76 | 0.82 | 0.305 | 0.324 | −0.137 |
| nHF [nu] | 0.4985 | 0.3956 | 0.289 | 0.79 | 0.74 | 0.81 | 0.656 | 0.035 | −0.254 |
| LF/HF [-] | 0.5721 | 0.0476 | 0.588 | 0.81 | 0.89 | 0.87 | 0.414 | 0.613 | −0.150 |
| Hurst [-] | 0.6297 | 0.6310 | 0.646 | 0.79 | 0.81 | 0.75 | 0.392 | 0.062 | −0.133 |
| SampEn [-] | 0.6110 | 0.7305 | 0.654 | 0.76 | 0.80 | 0.72 | 0.270 | 0.197 | −0.117 |
| Parameter | Real—Rest (Pre-Training) | Real—Rest (Pre-Training) 95% CI | Simulated—Rest | Simulated—Rest 95% CI | Real—Fatigue (Post-Training) | Real—Fatigue (Post-Training) 95% CI | Simulated—Fatigue | Simulated—Fatigue 95% CI | Real– Stress (After a Competition) | Real–Stress (After a Competition 95% CI | Simulated—Stress | Simulated—Stress 95% CI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SD1 [ms] | 28.24 ± 7.35 | [25.80, 30.68] | 30.73 ± 5.91 | [28.80, 32.66] | 17.81 ± 1.97 | [17.17, 18.45] | 17.91 ± 4.01 | [16.58, 19.24] | 13.06 ± 6.28 | [11.03, 15.09] | 12.34 ± 6.21 | [10.35, 14.33] |
| SD2 [ms] | 67.36 ± 28.19 | [58.15, 76.57] | 72.98 ± 13.55 | [68.55, 77.41] | 56.83 ± 9.64 | [53.68, 59.98] | 58.13 ± 10.33 | [54.75, 61.51] | 50.01 ± 26.34 | [41.37, 58.65] | 43.87 ± 27.92 | [34.55, 53.19] |
| SD2/SD1 [-] | 2.14 ± 0.38 | [2.12, 2.28] | 2.37 ± 0.38 | [2.25, 2.49] | 2.99 ± 0.25 | [2.91, 3.07] | 3.24 ± 0.35 | [3.18, 3.30] | 3.89 ± 0.45 | [3.74, 4.04] | 3.55 ± 0.41 | [3.41, 3.69] |
| Parameter | p-Value Real vs. Sim (Rest) | p-Value Real vs. Sim (Fatigue) | p-Value Real vs. Sim (Stress) | Pearson Correlation Real vs. Sim (Rest) | Pearson Correlation Real vs. Sim (Fatigue) | Pearson Correlation Real vs. Sim (Stress) | Effect Size Real vs. Sim (Rest) | Effect Size Real vs. Sim (Fatigue) | Effect Size Real vs. Sim (Stress) |
|---|---|---|---|---|---|---|---|---|---|
| SD1 [ms] | 0.7573 | 0.4035 | 0.37 | 0.84 | 0.72 | 0.84 | 0.373 | 0.033 | −0.116 |
| SD2 [ms] | 0.6874 | 0.6815 | 0.646 | 0.86 | 0.79 | 0.82 | 0.254 | 0.134 | −0.236 |
| SD2/SD1 [-] | 0.9208 | 0.0971 | 0.37 | 0.84 | 0.85 | 0.78 | 0.605 | 0.788 | −0.79 |
| Parameter | Real—Rest (Pre-Training) | Real—Rest (Pre-Training) 95% CI | Simulated—Rest | Simulated—Rest 95% CI | Real—Fatigue (Post-Training) | Real—Fatigue (Post-Training) 95% CI | Simulated—Fatigue | Simulated—Fatigue 95% CI | Real—Stress (After a Competition) | Real—Stress (After a Competition 95% CI | Simulated—Stress | Simulated—Stress 95% CI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| REC [%] | 3.82 ± 0.54 | [3.64, 4.00] | 3.88 ± 0.49 | [3.71, 4.05] | 4.76 ± 0.60 | [4.56, 4.96] | 4.69 ± 0.57 | [4.50, 4.88] | 5.93 ± 0.68 | [5.70, 6.16] | 5.88 ± 0.64 | [5.67, 6.09] |
| DET [%] | 91.6 ± 2.3 | [90.9, 92.3] | 90.9 ± 2.5 | [90.1, 91.7] | 86.2 ± 2.8 | [85.3, 87.1] | 85.4 ± 2.4 | [84.6, 86.2] | 79.1 ± 3.1 | [78.1, 80.1] | 78.3 ± 2.9 | [77.4, 79.2] |
| LAM [%] | 71.9 ± 3.5 | [70.8, 73.0] | 70.8 ± 3.2 | [69.8, 71.8] | 65.4 ± 3.1 | [64.4, 66.4] | 64.7 ± 2.9 | [63.8, 65.6] | 59.8 ± 2.8 | [58.9, 60.7] | 58.7 ± 2.6 | [57.9, 59.5] |
| Lmax | 186 ± 24 | [179, 193] | 181 ± 22 | [175, 187] | 159 ± 21 | [153, 165] | 154 ± 19 | [149, 159] | 131 ± 18 | [126, 136] | 128 ± 17 | [123, 133] |
| TT (Trapping Time) | 21.6 ± 2.7 | [20.8, 22.4] | 20.9 ± 2.5 | [20.2, 21.6] | 17.4 ± 2.2 | [16.8, 18.0] | 16.9 ± 2.0 | [16.4, 17.4] | 14.1 ± 1.9 | [13.6, 14.6] | 13.7 ± 1.8 | [13.2, 14.2] |
| ENTR | 2.89 ± 0.40 | [2.76, 3.02] | 2.82 ± 0.38 | [2.70, 2.94] | 2.43 ± 0.34 | [2.32, 2.54] | 2.37 ± 0.32 | [2.27, 2.47] | 2.05 ± 0.29 | [1.96, 2.14] | 2.02 ± 0.27 | [1.94, 2.10] |
| Parameter | p-Value Real vs. Sim (Rest) | p-Value Real vs. Sim (Fatigue) | p-Value Real vs. Sim (Stress) | Pearson Correlation Real vs. Sim (Rest) | Pearson Correlation Real vs. Sim (Fatigue) | Pearson Correlation Real vs. Sim (Stress) | Effect Size Real vs. Sim (Rest) | Effect Size Real vs. Sim (Fatigue) | Effect Size Real vs. Sim (Stress) |
|---|---|---|---|---|---|---|---|---|---|
| RQA: REC [%] | 0.521 | 0.487 | 0.441 | 0.77 | 0.80 | 0.79 | 0.115 | 0.122 | 0.081 |
| RQA: DET [%] | 0.338 | 0.410 | 0.367 | 0.81 | 0.79 | 0.78 | 0.144 | 0.098 | 0.121 |
| RQA: LAM [%] | 0.276 | 0.352 | 0.295 | 0.80 | 0.78 | 0.77 | 0.182 | 0.097 | 0.105 |
| RQA: Lmax | 0.446 | 0.391 | 0.418 | 0.78 | 0.76 | 0.75 | 0.210 | 0.243 | 0.173 |
| RQA: TT (Trapping Time) | 0.389 | 0.421 | 0.338 | 0.81 | 0.80 | 0.79 | 0.177 | 0.159 | 0.120 |
| RQA: ENTR | 0.277 | 0.312 | 0.268 | 0.82 | 0.83 | 0.81 | 0.173 | 0.185 | 0.143 |
| Parameter | Real—Rest (Pre-Training) | Real—Rest (Pre-Training) 95% CI | Simulated—Rest | Simulated—Rest 95% CI | Real—Fatigue (Post-Training) | Real—Fatigue (Post-Training) 95% CI | Simulated—Fatigue | Simulated—Fatigue 95% CI | Real— Stress (After a Competition) | Real—Stress (After a Competition 95% CI | Simulated—Stress | Simulated—Stress 95% CI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lyapunov λ1 [bits/s] | 0.102 ± 0.018 | [0.095, 0.108] | 0.099 ± 0.020 | [0.092, 0.106] | 0.138 ± 0.024 | [0.129, 0.147] | 0.132 ± 0.022 | [0.124, 0.140] | 0.168 ± 0.028 | [0.157, 0.179] | 0.162 ± 0.025 | [0.152, 0.172] |
| Correlation Dimension D2 [-] | 2.41 ± 0.32 | [2.30, 2.52] | 2.38 ± 0.29 | [2.28, 2.48] | 2.12 ± 0.27 | [2.03, 2.21] | 2.09 ± 0.25 | [2.00, 2.18] | 1.86 ± 0.22 | [1.78, 1.94] | 1.82 ± 0.20 | [1.75, 1.89] |
| Permutation Entropy | 0.84 ± 0.06 | [0.82, 0.86] | 0.81 ± 0.07 | [0.79, 0.83] | 0.72 ± 0.05 | [0.70, 0.74] | 0.69 ± 0.06 | [0.67, 0.71] | 0.61 ± 0.05 | [0.59, 0.63] | 0.58 ± 0.05 | [0.56, 0.60] |
| Parameter | p-Value Real vs. Sim (Rest) | p-Value Real vs. Sim (Fatigue) | p-Value Real vs. Sim (Stress) | Pearson Correlation Real vs. Sim (Rest) | Pearson Correlation Real vs. Sim (Fatigue) | Pearson Correlation Real vs. Sim (Stress) | Effect Size Real vs. Sim (Rest) | Effect Size Real vs. Sim (Fatigue) | Effect Size Real vs. Sim (Stress) |
|---|---|---|---|---|---|---|---|---|---|
| Lyapunov λ1 [bits/s] | 0.284 | 0.311 | 0.267 | 0.82 | 0.84 | 0.81 | 0.156 | 0.250 | 0.223 |
| Correlation Dimension D2 [-] | 0.402 | 0.365 | 0.298 | 0.79 | 0.82 | 0.83 | 0.095 | 0.113 | 0.188 |
| Permutation Entropy | 0.315 | 0.337 | 0.289 | 0.80 | 0.83 | 0.82 | 0.120 | 0.145 | 0.210 |
| Parameter | RMSE Real vs. Sim (Rest) | RMSE Real vs. Sim (Fatigue) | RMSE Real vs. Sim (Stress) | DTW Dist. Real vs. Sim (Rest) | DTW Dist. Real vs. Sim (Fatigue) | DTW Dist. Real vs. Sim (Stress) |
|---|---|---|---|---|---|---|
| Mean RR [ms] | 0.894 | 0.598 | 0.086 | 0.82 | 0.89 | 0.76 |
| SDNN [ms] | 0.7603 | 0.4888 | 0.369 | 0.84 | 0.78 | 0.73 |
| RMSSD [ms] | 0.7504 | 0.5237 | 0.289 | 0.78 | 0.75 | 0.81 |
| nLF [nu] | 0.5769 | 0.8211 | 0.601 | 0.76 | 0.76 | 0.82 |
| nHF [nu] | 0.4985 | 0.3956 | 0.289 | 0.79 | 0.74 | 0.81 |
| LF/HF [-] | 0.5721 | 0.0476 | 0.588 | 0.81 | 0.89 | 0.87 |
| SD1 [ms] | 0.7573 | 0.4035 | 0.37 | 0.84 | 0.72 | 0.84 |
| SD2 [ms] | 0.6874 | 0.6815 | 0.646 | 0.86 | 0.79 | 0.82 |
| SD2/SD1 [-] | 0.9208 | 0.0971 | 0.37 | 0.84 | 0.85 | 0.78 |
| Hurst [-] | 0.6297 | 0.6310 | 0.646 | 0.79 | 0.81 | 0.75 |
| SampEn [-] | 0.6110 | 0.7305 | 0.654 | 0.76 | 0.80 | 0.72 |
| RR Number | CPU (sec) | RR Number | CPU (sec) | RR Number | CPU (sec) |
|---|---|---|---|---|---|
| 1000 | 0.00069 | 10,000 | 0.021 | 100,000 | 2.11 |
| 2000 | 0.0012 | 20,000 | 0.42 | 200,000 | 2.34 |
| 3000 | 0.0019 | 30,000 | 0.63 | 300,000 | 2.52 |
| 4000 | 0.0027 | 40,000 | 0.84 | 400,000 | 2.71 |
| 5000 | 0.0034 | 50,000 | 1.05 | 500,000 | 2.89 |
| 6000 | 0.0041 | 60,000 | 1.25 | 600,000 | 3.11 |
| 7000 | 0.0049 | 70,000 | 1.46 | 700,000 | 3.29 |
| 8000 | 0.0057 | 80,000 | 1.67 | 800,000 | 3.47 |
| 9000 | 0.007 | 90,000 | 1.89 | 900,000 | 3.64 |
| Reference | Num. of Gaussians | Parameters for Wave | CPU | HRV | Additional Approach |
|---|---|---|---|---|---|
| [7] | 6 GDF | 3 | Yes | No | No |
| [49] | 6 GDF | 3 | No | No | No |
| [50] | 6 GDF | 3 | No | No | No |
| [51] | 7 GDF | 3 | No | No | No |
| [10] | 6 GMF | 5 | No | No | or BGF |
| [8] | 8 GDF | 3 | No | No | No |
| [6] | 2 GDF | 3 | No | No | No |
| [52] | 5 GDF | 3 | No | No | No |
| [9] | 2 GDF | 3 | No | No | or Fourier Transform |
| This work | 6 GDF (basic + adjustable number) | 5 for GMF+ 7 for attractor | Yes | Yes | Attractor-based variability |
| Parameters | Normal | McSharry | Zeeman | Gauss-Rössler |
|---|---|---|---|---|
| SDNN [ms] | 141 ± 39 | 138.47 ± 7.2 | 212.47 ± 10.6 | 131.82 ± 6.6 |
| SDANN [ms] | 127 ± 35 | 126.22 ± 6.3 | 180.61 ± 9.0 | 103.98 ± 5.2 |
| RMSSD [ms] | 27 ± 12 | 45.94 ± 4.6 | 66.56 ± 6.7 | 36.99 ± 3.7 |
| pNN50 [%] | 14.09 ± 5.03 | 29.93 ± 3.0 | 28.98 ± 2.9 | 17.73 ± 1.8 |
| Total power [ms2] | 3466 ± 1018 | 2614.59 ± 390 | 7256.46 ± 1231 | 2679.19 ± 402 |
| VLF [ms2] | — | 104.20 ± 10.4 | 10.11 ± 1.1 | 242.62 ± 24.2 |
| LF [ms2] | 1170 ± 416 | 1846.35 ± 185 | 5892.44 ± 589 | 1429.35 ±143 |
| HF [ms2] | 975 ± 203 | 664.04 ± 66 | 1353.91 ± 135 | 1007.22 ± 101 |
| LF/HF [-] | 1.5–2.0 | 2.78 ± 0.14 | 4.29 ± 0.21 | 1.42 ± 0.07 |
| SD1 (ms) | — | 59.87 ± 6.0 | 44.03 ± 4.4 | 30.18 ± 3.1 |
| SD2 (ms) | — | 186.67 ± 18.7 | 35.61 ± 3.6 | 43.63 ± 4.3 |
| SD1/SD2 | — | 0.321 ± 0.032 | 1.24 ± 0.12 | 0.692 ± 0.07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georgieva-Tsaneva, G. Mathematical Modeling Using Gaussian Functions and Chaotic Attractors: A Hybrid Approach for Realistic Representation of the Intrinsic Dynamics of Heartbeats. AppliedMath 2025, 5, 172. https://doi.org/10.3390/appliedmath5040172
Georgieva-Tsaneva G. Mathematical Modeling Using Gaussian Functions and Chaotic Attractors: A Hybrid Approach for Realistic Representation of the Intrinsic Dynamics of Heartbeats. AppliedMath. 2025; 5(4):172. https://doi.org/10.3390/appliedmath5040172
Chicago/Turabian StyleGeorgieva-Tsaneva, Galya. 2025. "Mathematical Modeling Using Gaussian Functions and Chaotic Attractors: A Hybrid Approach for Realistic Representation of the Intrinsic Dynamics of Heartbeats" AppliedMath 5, no. 4: 172. https://doi.org/10.3390/appliedmath5040172
APA StyleGeorgieva-Tsaneva, G. (2025). Mathematical Modeling Using Gaussian Functions and Chaotic Attractors: A Hybrid Approach for Realistic Representation of the Intrinsic Dynamics of Heartbeats. AppliedMath, 5(4), 172. https://doi.org/10.3390/appliedmath5040172





