Self-Assembly and Gelation Study of Dipeptide Isomers with Norvaline and Phenylalanine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and General Methods
2.2. Dipeptide Synthesis and Purification
2.3. Circular Dichroism
2.4. Self-Assembly Tests
2.5. Transmission Electron Microscopy (TEM)
2.6. Raman Microspectroscopy
2.7. Single-Crystal X-ray Diffraction (XRD)
3. Results and Discussion
3.1. Dipeptide Preparation and Spectroscopic Characterization
3.2. Self-Assembly Tests
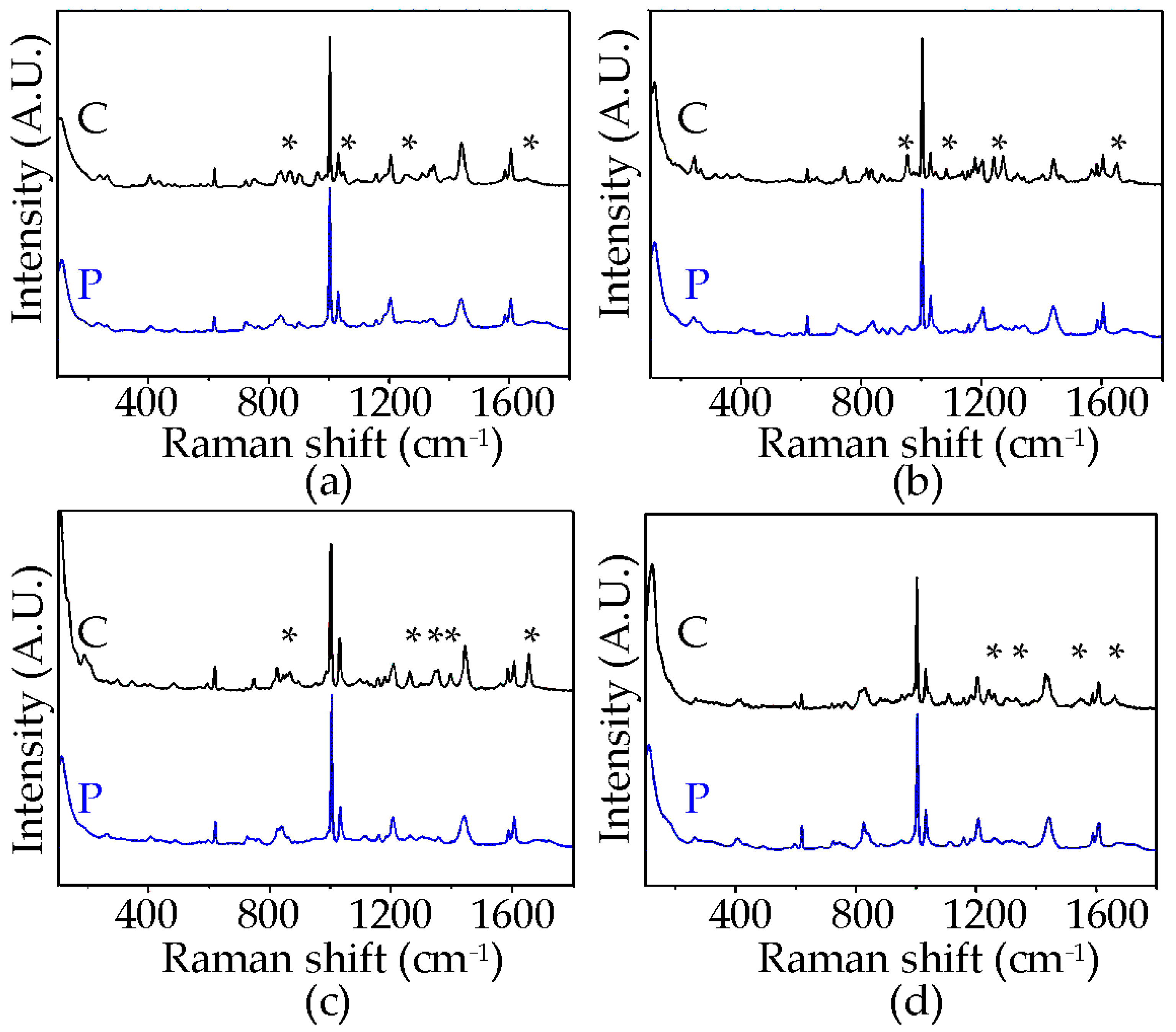
3.3. Raman Spectroscopy
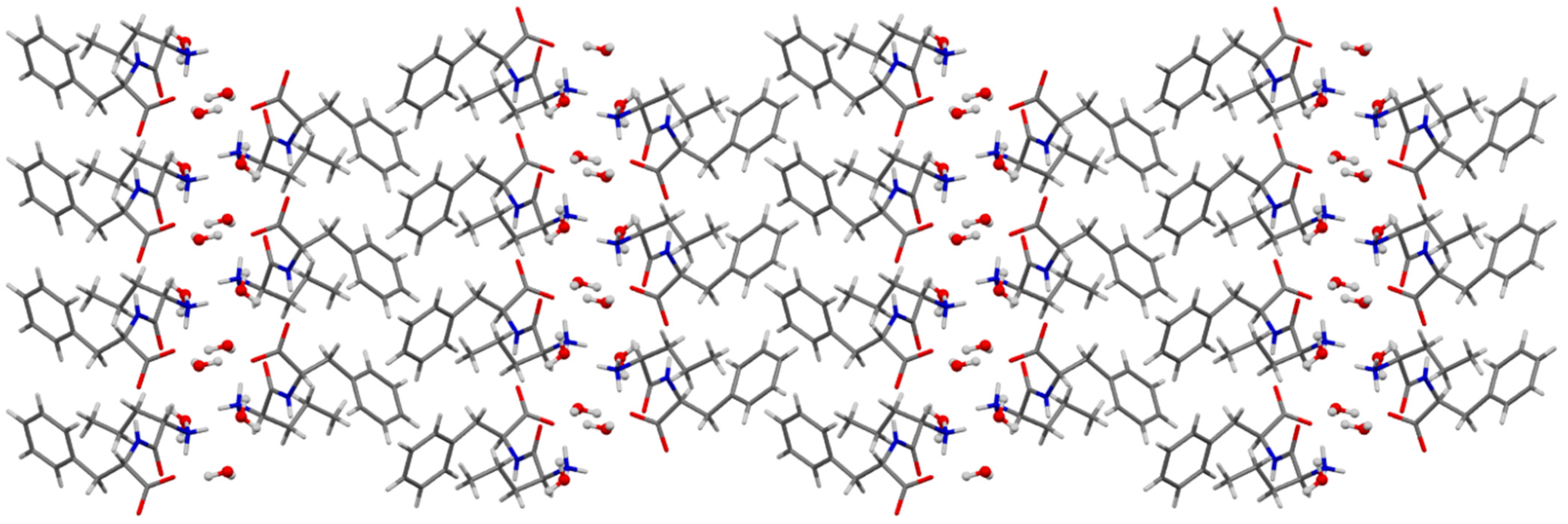
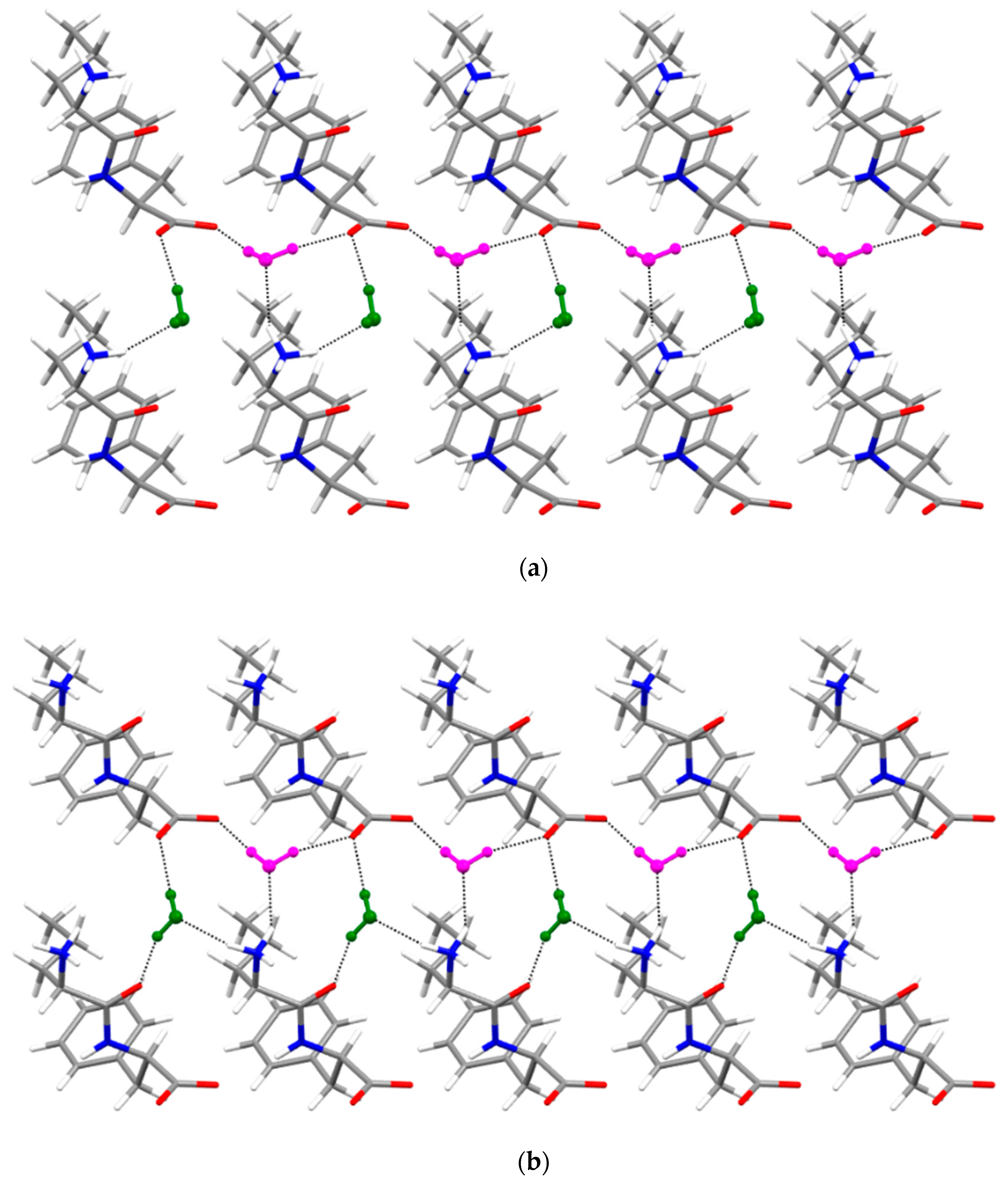
3.4. Single-Crystal X-ray Diffraction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adams, D.J. Dipeptide and tripeptide conjugates as low-molecular-weight hydrogelators. Macromol. Biosci. 2011, 11, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tao, K.; Ji, W.; Makam, P.; Rencus-Lazar, S.; Gazit, E. Self-assembly of cyclic dipeptides: Platforms for functional materials. Prot. Pept. Lett. 2020, 27, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Rissanou, A.N.; Simatos, G.; Siachouli, P.; Harmandaris, V.; Mitraki, A. Self-assembly of alanine-isoleucine and isoleucine-isoleucine dipeptides through atomistic simulations and experiments. J. Phys. Chem. B 2020, 124, 7102–7114. [Google Scholar] [CrossRef] [PubMed]
- Balachandra, C.; Padhi, D.; Govindaraju, T. Cyclic dipeptide: A privileged molecular scaffold to derive structural diversity and functional utility. ChemMedChem 2021, 16, 2558–2587. [Google Scholar] [CrossRef] [PubMed]
- Scarel, M.; Marchesan, S. Diketopiperazine gels: New horizons from the self-assembly of cyclic dipeptides. Molecules 2021, 26, 3376. [Google Scholar] [CrossRef]
- Reches, M.; Gazit, E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef] [Green Version]
- Brahmachari, S.; Arnon, Z.A.; Frydman-Marom, A.; Gazit, E.; Adler-Abramovich, L. Diphenylalanine as a reductionist model for the mechanistic characterization of β-amyloid modulators. ACS Nano 2017, 11, 5960–5969. [Google Scholar] [CrossRef]
- Safaryan, S.; Slabov, V.; Kopyl, S.; Romanyuk, K.; Bdikin, I.; Vasilev, S.; Zelenovskiy, P.; Shur, V.Y.; Uslamin, E.A.; Pidko, E.A.; et al. Diphenylalanine-based microribbons for piezoelectric applications via inkjet printing. ACS Appl. Mater. Interfaces 2018, 10, 10543–10551. [Google Scholar] [CrossRef]
- Zelenovskiy, P.S.; Domingues, E.M.; Slabov, V.; Kopyl, S.; Ugolkov, V.L.; Figueiredo, F.M.L.; Kholkin, A.L. Efficient water self-diffusion in diphenylalanine peptide nanotubes. ACS Appl. Mater. Interfaces 2020, 12, 27485–27492. [Google Scholar] [CrossRef]
- Erdoğan, H. Cation-based approach to morphological diversity of diphenylalanine dipeptide structures. Soft Matter 2021, 17, 5221–5230. [Google Scholar] [CrossRef]
- Diaferia, C.; Roviello, V.; Morelli, G.; Accardo, A. Self-assembly of pegylated diphenylalanines into photoluminescent fibrillary aggregates. Chemphyschem 2019, 20, 2774–2782. [Google Scholar] [CrossRef] [PubMed]
- Rajbhandary, A.; Nilsson, B.L. Investigating the effects of peptoid substitutions in self-assembly of fmoc-diphenylalanine derivatives. Biopolymers 2017, 108, e22994. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.D.; Wojciechowski, J.P.; Robinson, A.B.; Heu, C.; Garvey, C.J.; Ratcliffe, J.; Waddington, L.J.; Gardiner, J.; Thordarson, P. Controlling self-assembly of diphenylalanine peptides at high ph using heterocyclic capping groups. Sci. Rep. 2017, 7, 43947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karikis, K.; Butkiewicz, A.; Folias, F.; Charalambidis, G.; Kokotidou, C.; Charisiadis, A.; Nikolaou, V.; Nikoloudakis, E.; Frelek, J.; Mitraki, A.; et al. Self-assembly of (boron-dipyrromethane)-diphenylalanine conjugates forming chiral supramolecular materials. Nanoscale 2018, 10, 1735–1741. [Google Scholar] [CrossRef] [PubMed]
- Dudukovic, N.A.; Hudson, B.C.; Paravastu, A.K.; Zukoski, C.F. Self-assembly pathways and polymorphism in peptide-based nanostructures. Nanoscale 2018, 10, 1508–1516. [Google Scholar] [CrossRef]
- Diaferia, C.; Avitabile, C.; Leone, M.; Gallo, E.; Saviano, M.; Accardo, A.; Romanelli, A. Diphenylalanine motif drives self-assembling in hybrid pna-peptide conjugates. Chem. Eur. J. 2021, 27, 14307–14316. [Google Scholar] [CrossRef]
- Garcia, A.M.; Melchionna, M.; Bellotto, O.; Kralj, S.; Semeraro, S.; Parisi, E.; Iglesias, D.; D’Andrea, P.; De Zorzi, R.; Vargiu, A.V.; et al. Nanoscale assembly of functional peptides with divergent programming elements. ACS Nano 2021, 15, 3015–3025. [Google Scholar] [CrossRef]
- Li, S.; Liu, Y.; Xing, R.; Yan, X. Covalently assembled dipeptide nanoparticles with adjustable fluorescence emission for multicolor bioimaging. ChemBioChem 2019, 20, 555–560. [Google Scholar] [CrossRef]
- Yuran, S.; Razvag, Y.; Das, P.; Reches, M. Self-assembly of azide containing dipeptides. J. Pept. Sci. 2014, 20, 479–486. [Google Scholar] [CrossRef]
- Kralj, S.; Bellotto, O.; Parisi, E.; Garcia, A.M.; Iglesias, D.; Semeraro, S.; Deganutti, C.; D’Andrea, P.; Vargiu, A.V.; Geremia, S.; et al. Heterochirality and halogenation control phe-phe hierarchical assembly. ACS Nano 2020, 14, 16951–16961. [Google Scholar] [CrossRef]
- Görbitz, C.H. Nanotube formation by hydrophobic dipeptides. Chem. Eur. J. 2001, 7, 5153–5159. [Google Scholar] [CrossRef]
- Görbitz, C.H. The structure of nanotubes formed by diphenylalanine, the core recognition motif of alzheimer’s beta-amyloid polypeptide. Chem. Commun. 2006, 2332–2334. [Google Scholar] [CrossRef] [PubMed]
- Görbitz, C.H. Microporous organic materials from hydrophobic dipeptides. Chem. Eur. J. 2007, 13, 1022–1031. [Google Scholar] [CrossRef] [PubMed]
- Görbitz, C. L-isoleucyl-L-phenylalanine dihydrate. Acta Cryst. C 2004, 60, o371–o373. [Google Scholar] [CrossRef] [Green Version]
- Görbitz, C. L-phenylalanyl- L-isoleucine 0.88-hydrate. Acta Cryst. C 2004, 60, o810–o812. [Google Scholar] [CrossRef]
- de Groot, N.S.; Parella, T.; Aviles, F.X.; Vendrell, J.; Ventura, S. Ile-phe dipeptide self-assembly: Clues to amyloid formation. Biophys. J. 2007, 92, 1732–1741. [Google Scholar] [CrossRef] [Green Version]
- Bellotto, O.; Kralj, S.; Melchionna, M.; Pengo, P.; Kisovec, M.; Podobnik, M.; De Zorzi, R.; Marchesan, S. Self-assembly of unprotected dipeptides into hydrogels: Water-channels make the difference. ChemBioChem 2022, 23, e202100518. [Google Scholar] [CrossRef]
- Kurbasic, M.; Semeraro, S.; Garcia, A.M.; Kralj, S.; Parisi, E.; Deganutti, C.; De Zorzi, R.; Marchesan, S. Microwave-assisted cyclization of unprotected dipeptides in water to 2,5-piperazinediones and self-assembly study of products and reagents. Synthesis 2019, 51, 2829–2838. [Google Scholar] [CrossRef] [Green Version]
- Görbitz, C. β turns, water cage formation and hydrogen bonding in the structures of L-valyl-L-phenylalanine. Acta Cryst. B 2002, 58, 512–518. [Google Scholar] [CrossRef]
- Görbitz, C. An NH3+…Phenyl interaction in L-phenylalanyl- L-valine. Acta Cryst. C 2000, 56, 1496–1498. [Google Scholar] [CrossRef]
- Bellotto, O.; Pierri, G.; Rozhin, P.; Polentarutti, M.; Kralj, S.; D’Andrea, P.; Tedesco, C.; Marchesan, S. Dipeptide self-assembly into water-channels and gel biomaterial. Org. Biomol. Chem. 2022, 20, 6211–6218. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Carreño, C.; Becerra, A.; Lazcano, A. Norvaline and norleucine may have been more abundant protein components during early stages of cell evolution. Orig. Life Evol. Biosph. 2013, 43, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Pokrovskiy, M.V.; Korokin, M.V.; Tsepeleva, S.A.; Pokrovskaya, T.G.; Gureev, V.V.; Konovalova, E.A.; Gudyrev, O.S.; Kochkarov, V.I.; Korokina, L.V.; Dudina, E.N.; et al. Arginase inhibitor in the pharmacological correction of endothelial dysfunction. Int. J. Hypertens. 2011, 2011, 515047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konovalova, E.A.; Chernomortseva, E.S. The study of the endothelial protective properties of the L-norvaline combination with mexidol in the simulation of L-NAME-induced NO deficiency. Res. Result Pharmacol. 2019, 5, 41–46. [Google Scholar] [CrossRef]
- Bolt, H.L.; Williams, C.E.J.; Brooks, R.V.; Zuckermann, R.N.; Cobb, S.L.; Bromley, E.H.C. Log D versus HPLC derived hydrophobicity: The development of predictive tools to aid in the rational design of bioactive peptoids. Pept. Sci. 2017, 108, e23014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, A.M.; Iglesias, D.; Parisi, E.; Styan, K.E.; Waddington, L.J.; Deganutti, C.; De Zorzi, R.; Grassi, M.; Melchionna, M.; Vargiu, A.V.; et al. Chirality effects on peptide self-assembly unraveled from molecules to materials. Chem 2018, 4, 1862–1876. [Google Scholar] [CrossRef] [Green Version]
- Amdursky, N.; Stevens, M.M. Circular dichroism of amino acids: Following the structural formation of phenylalanine. ChemPhysChem 2015, 16, 2768–2774. [Google Scholar] [CrossRef]
- Mazzier, D.; Mosconi, D.; Marafon, G.; Reheman, A.; Toniolo, C.; Moretto, A. Light-driven topochemical polymerization under organogel conditions of a symmetrical dipeptide-diacetylene system. J. Pept. Sci. 2017, 23, 155–161. [Google Scholar] [CrossRef]
- Profit, A.A.; Vedad, J.; Desamero, R.Z. Peptide conjugates of benzene carboxylic acids as agonists and antagonists of amylin aggregation. Bioconjug. Chem. 2017, 28, 666–677. [Google Scholar] [CrossRef] [Green Version]
- Ronen, M.; Kalanoor, B.S.; Oren, Z.; Ron, I.; Tischler, Y.R.; Gerber, D. Characterization of peptides self-assembly by low frequency Raman spectroscopy. RSC Adv. 2018, 8, 16161–16170. [Google Scholar] [CrossRef]
- Levine, M.S.; Ghosh, M.; Hesser, M.; Hennessy, N.; DiGuiseppi, D.M.; Adler-Abramovich, L.; Schweitzer-Stenner, R. Formation of peptide-based oligomers in dimethylsulfoxide: Identifying the precursor of fibril formation. Soft Matter 2020, 16, 7860–7868. [Google Scholar] [CrossRef] [PubMed]
- Scarel, E.; Bellotto, O.; Rozhin, P.; Kralj, S.; Tortora, M.; Vargiu, A.V.; De Zorzi, R.; Rossi, B.; Marchesan, S. Single-atom substitution enables supramolecular diversity from dipeptide building blocks. Soft Matter 2022, 18, 2129–2136. [Google Scholar] [CrossRef]
- Hernández, B.; Pflüger, F.; Kruglik, S.G.; Ghomi, M. Characteristic Raman lines of phenylalanine analyzed by a multiconformational approach. J. Raman Spectrosc. 2013, 44, 827–833. [Google Scholar] [CrossRef]
- Fischer, W.B.; Eysel, H.H. Polarized raman spectra and intensities of aromatic amino acids phenylalanine, tyrosine and tryptophan. Spectrochim. Acta A Mol. Spectrosc. 1992, 48, 725–732. [Google Scholar] [CrossRef]
- Thakuria, R.; Nath, N.K.; Saha, B.K. The Nature and Applications of π–π Interactions: A Perspective. Cryst. Growth Des. 2019, 19, 523–528. [Google Scholar] [CrossRef]
- Grabowski, S.J.; Lipkowski, P. Characteristics of X-H···π Interactions: Ab Initio and QTAIM Studies. J. Phys. Chem. A 2011, 115, 4765–4773. [Google Scholar] [CrossRef]
- Picci, G.; Marchesan, S.; Caltagirone, C. Ion channels and transporters as therapeutic agents: From biomolecules to supramolecular medicinal chemistry. Biomedicines 2022, 10, 885. [Google Scholar] [CrossRef]
- Roy, A.; Talukdar, P. Recent advances in bioactive artificial ionophores. ChemBioChem 2021, 22, 2925–2940. [Google Scholar] [CrossRef]
- Wu, X.; Howe, E.N.W.; Gale, P.A. Supramolecular transmembrane anion transport: New assays and insights. Acc. Chem. Res. 2018, 51, 1870–1879. [Google Scholar] [CrossRef]
- Gao, L.; Zhang, J.H.; Chen, X.X.; Ren, H.L.; Feng, X.L.; Wang, J.L.; Xiao, J.H. Combination of L-arginine and L-norvaline protects against pulmonary fibrosis progression induced by bleomycin in mice. Biomed. Pharmacother. 2019, 113, 108768. [Google Scholar] [CrossRef]
- Javrushyan, H.; Nadiryan, E.; Grigoryan, A.; Avtandilyan, N.; Maloyan, A. Antihyperglycemic activity of L-norvaline and L-arginine in high-fat diet and streptozotocin-treated male rats. Exp. Mol. Pathol. 2022, 126, 104763. [Google Scholar] [CrossRef] [PubMed]
- Polis, B.; Gurevich, V.; Assa, M.; Samson, A.O. Norvaline restores the bbb integrity in a mouse model of Alzheimer’s disease. Int. J. Mol. Sci. 2019, 20, 4616. [Google Scholar] [CrossRef] [Green Version]
- Polis, B.; Srikanth, K.D.; Gurevich, V.; Gil-Henn, H.; Samson, A.O. L-norvaline, a new therapeutic agent against Alzheimer’s disease. Neural Regen. Res. 2019, 14, 1562–1572. [Google Scholar] [CrossRef] [PubMed]
- Gilinsky, M.A.; Polityko, Y.K.; Markel, A.L.; Latysheva, T.V.; Samson, A.O.; Polis, B.; Naumenko, S.E. Norvaline reduces blood pressure and induces diuresis in rats with inherited stress-induced arterial hypertension. BioMed. Res. Int. 2020, 2020, 4935386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battye, T.G.G.; Kontogiannis, L.; Johnson, O.; Powell, H.R.; Leslie, A.G.W. iMOSFLM: A New Graphical Interface for Diffraction-Image Processing with MOSFLM. Acta Crystallogr. Sect. D 2011, 67, 271–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, P.R. Scaling and assessment of data quality. Acta Crystallogr. Sect. D 2006, 62, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1204–1214. [Google Scholar] [CrossRef]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
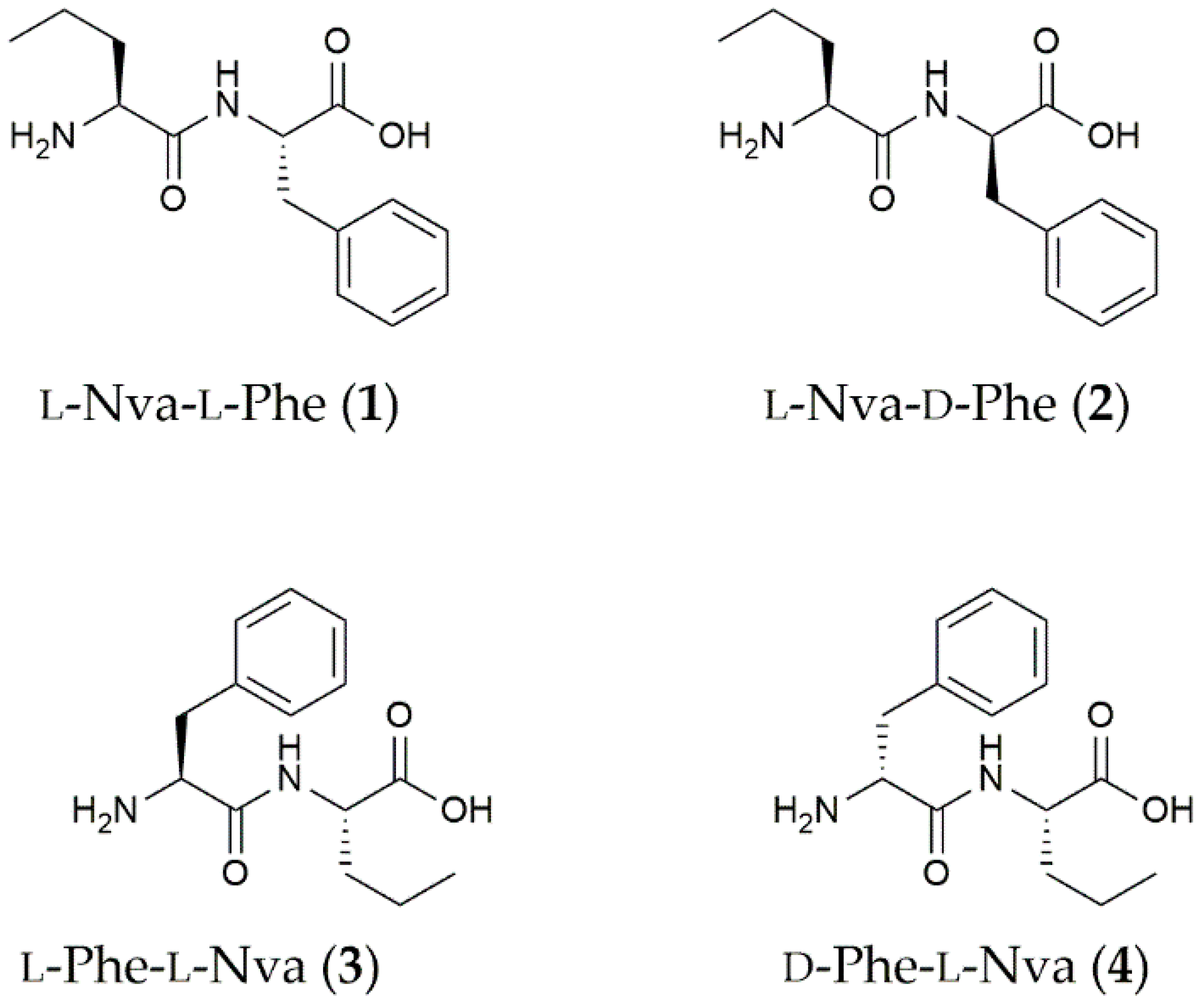


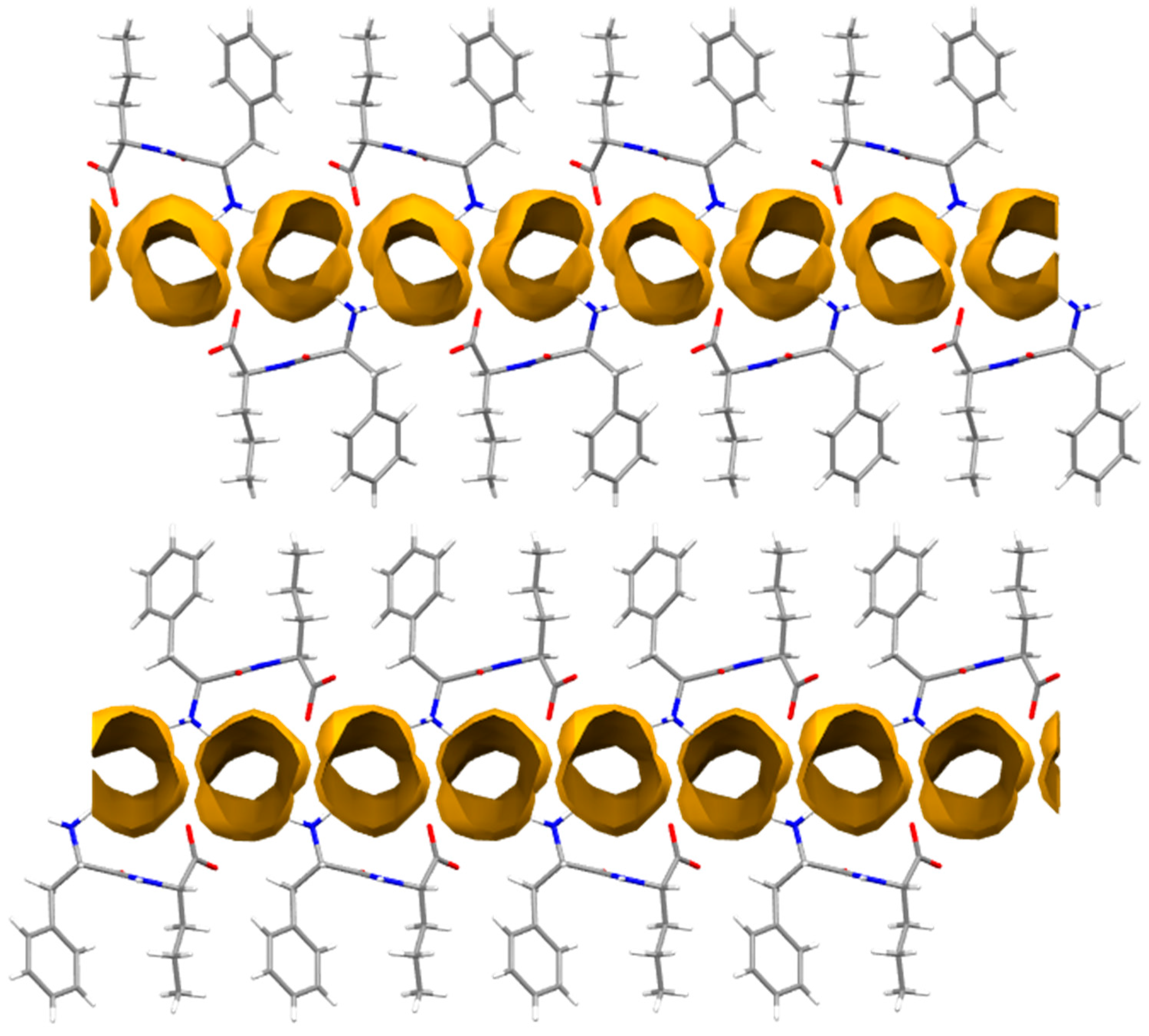

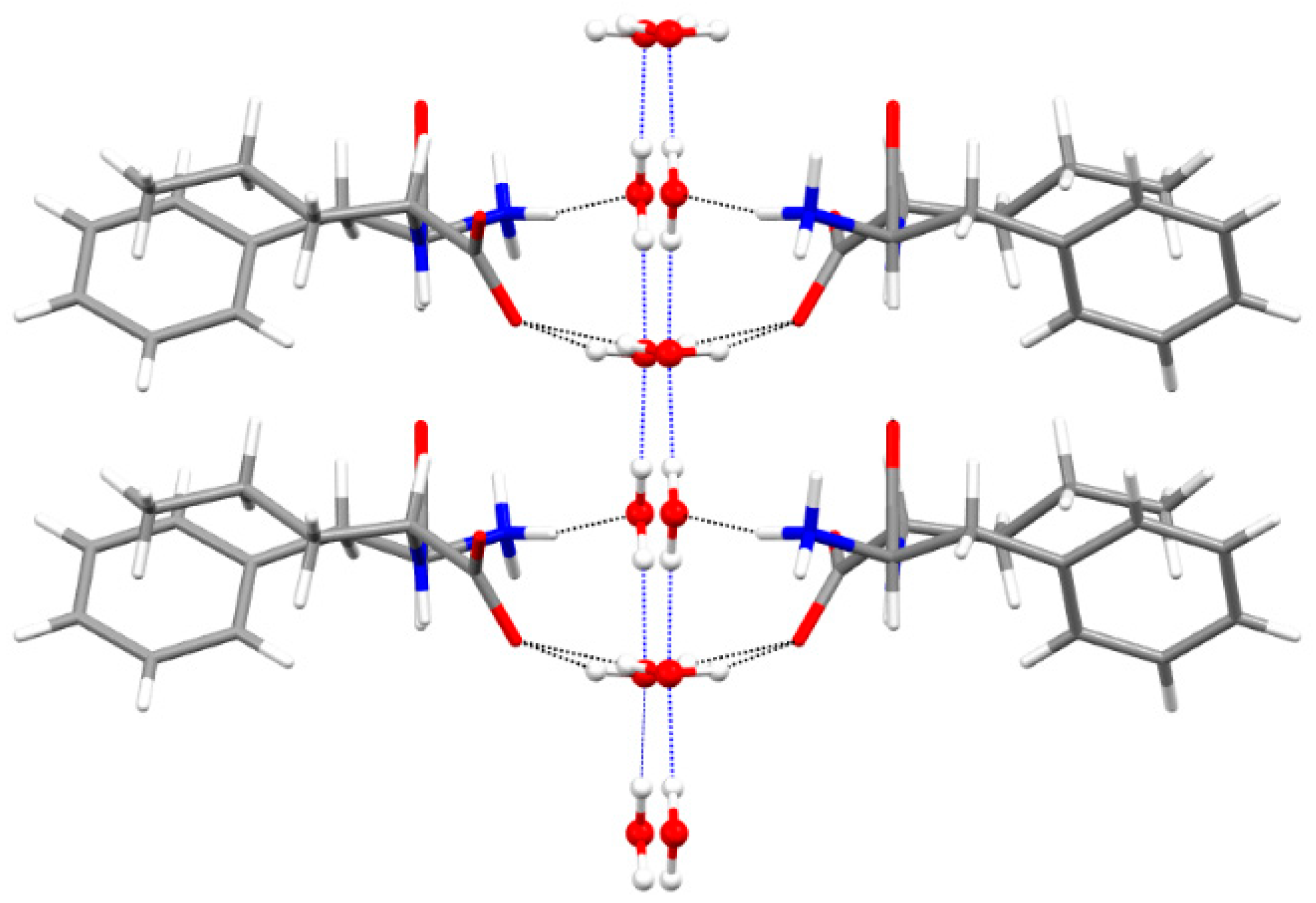
| Solvent | L-Nva-L-Phe (1) | L-Nva-D-Phe (2) | L-Phe-L-Nva (3) | D-Phe-L-Nva (4) |
|---|---|---|---|---|
| PBS buffer | Crystal | Crystal | Crystal | Crystal |
| MeOH | Crystal | Crystal | Crystal | Crystal |
| EtOH | Crystal | Crystal | Crystal | Crystal |
| iPrOH | Crystal | Crystal | Crystal | Crystal |
| MeCN | Crystal | Crystal | Crystal | Gel 1 |
| Acetone | Precipitate | Precipitate | Precipitate | Precipitate |
| Argan oil | Precipitate | Precipitate | Precipitate | Precipitate |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scarel, E.; Pierri, G.; Rozhin, P.; Adorinni, S.; Polentarutti, M.; Tedesco, C.; Marchesan, S. Self-Assembly and Gelation Study of Dipeptide Isomers with Norvaline and Phenylalanine. Chemistry 2022, 4, 1417-1428. https://doi.org/10.3390/chemistry4040093
Scarel E, Pierri G, Rozhin P, Adorinni S, Polentarutti M, Tedesco C, Marchesan S. Self-Assembly and Gelation Study of Dipeptide Isomers with Norvaline and Phenylalanine. Chemistry. 2022; 4(4):1417-1428. https://doi.org/10.3390/chemistry4040093
Chicago/Turabian StyleScarel, Erica, Giovanni Pierri, Petr Rozhin, Simone Adorinni, Maurizio Polentarutti, Consiglia Tedesco, and Silvia Marchesan. 2022. "Self-Assembly and Gelation Study of Dipeptide Isomers with Norvaline and Phenylalanine" Chemistry 4, no. 4: 1417-1428. https://doi.org/10.3390/chemistry4040093





